Abstract
A series of potent, broadly neutralizing HIV antibodies have been isolated from B cells of HIV-infected individuals. VRC01 represents a subset of these antibodies that mediate neutralization with a restricted set of IGHV genes. The memory B cells expressing these antibodies were isolated years after infection; thus, the B-cell subpopulation from which they originated and the extent of participation in the initial HIV antibody response, if any, are unclear. Here we evaluated the frequency of anti-gp120 B cells in follicular (FO) and marginal zone (MZ) B-cell compartments of naïve WT mice and comparable human populations in uninfected individuals. We found that in non–HIV-exposed humans and mice, the majority of gp120-reactive B cells are of naïve and FO phenotype, respectively. Murine FO B cells express a diverse antibody repertoire to recognize gp120. In contrast, mouse MZ B cells recognize gp120 less frequently but preferentially use IGHV1-53 to encode gp120-specific antibodies. Notably, IGHV1-53 shows high identity to human IGHV1-2*02, which has been repeatedly found to encode broadly neutralizing mutated HIV antibodies, such as VRC01. Finally, we show that human MZ-like B cells express IGHV1-2*02, and that IGHV1-53 expression is enriched in mouse MZ B cells. These data suggest that efforts toward developing an HIV vaccine might consider eliciting protective HIV antibody responses selectively from alternative B-cell populations harboring IGHV gene segments capable of producing protective antibodies.
Keywords: B-cell subsets, virus, germ line, polyreactive
Traditional vaccine strategies mediate protection by generating memory B cells and long-lived plasma cells (1). These strategies have thus far failed to elicit broadly neutralizing or protective antibodies to HIV (2, 3). A major impediment to vaccine development is lack of knowledge of the parameters that lead to a successful HIV antibody response. HIV-infected individuals develop high titers of antibody to the envelope glycoprotein gp120 in the primary antibody response, but most often these antibodies are nonneutralizing (4). Some protective epitopes, such as the CD4 binding site, are not easily accessible on the free virus, owing in part to masking of protein epitopes by carbohydrates. This glycosylation is host-derived, potentially inducing tolerance in B cells that cross-react with self-antigens (5). In addition, naïve B cells with low-affinity antigen receptors specific for gp120 might not be efficiently triggered by HIV, owing to the low surface density of gp120 on HIV virions (6, 7).
Antibodies able to broadly neutralize diverse strains of HIV have been isolated, but are rare. These broadly neutralizing antibodies are typically highly mutated and often polyreactive (1). Although numerous epitopes on gp120 have been identified as targets of broadly neutralizing antibodies, recent studies of HIV-infected serum demonstrating HIV broadly neutralizing activity have shown that a major neutralizing epitope is directed against the CD4 binding site of gp120 (8–10). The CD4 binding site is a crucial component of viral attachment and entry into the target cell and is one of the most conserved regions of gp120 (11).
The recently isolated broadly neutralizing antibody VRC01 and related broadly neutralizing antibodies are able to neutralize up to 90% of different HIV strains in vitro (12, 13). Despite the fact that these broadly neutralizing antibodies were isolated from different individuals, this set of antibodies selectively uses the IGHV1-2*02 gene segment to encode the Ig heavy chain. Structural studies have shown that the complementary determining region 2 (CDRH2) of IGHV1-2*02 that encodes VRC01 confers broad neutralization by binding the most vulnerable and conserved portion of the CD4 binding site on gp120 (14). The CDRH2 of VRC01 and related antibodies is considerably mutated from the germ line. These findings suggest that promoting HIV neutralization by targeting B cells bearing this IGHV segment may provide a promising vaccine strategy.
A successful HIV vaccine must be able to promote neutralizing antibody responses over the dominant nonprotective responses. Accomplishing this may require the specific participation of different B-cell subsets. The antibody response to physiological pathogens is a cooperative effort between different B-cell subpopulations (15). The major B-cell populations, CD21+CD23+ follicular (FO) cells in mice and IgD+CD27– naïve B cells in humans, require the help of cognate T cells to respond to protein antigens to produce class-switched, affinity-matured antibodies and memory B cells, a process that takes time to develop. In addition, early after infection, marginal zone (MZ) B cells mount rapid antibody responses to repetitive epitopes displayed by pathogens and are not necessarily dependent on T-cell help. It is widely believed that MZ B cells do not participate in germinal center reactions and thus do not somatically mutate Ig genes, although independent studies have shown the direct ability of mouse MZ B cells to induce germinal centers and undergo somatic hypermutation (16, 17). Furthermore, the human antibody response to the capsular polysaccharide of both Streptococcus pneumoniae and Haemophilus influenzae are dominated by IgM+IgD+CD27+ MZ-like human B cells (18–20) and are often mutated (21–24), indicating that MZ B cells are able to undergo somatic hypermutation when responding to bona fide pathogens.
Qualitatively, antibodies from MZ B cells are more often polyreactive than antibodies from FO B cells (25). This polyreactivity may be particularly beneficial in protecting against HIV, which has an extremely low surface envelope spike density, making heteroligation to a viral spike and another antigen potentially important for virus neutralization (7, 26). This second antigen may be a self-antigen on the surface of an infected cell or an HIV virion. Thus, the rarity of B cells that produce broadly neutralizing antibodies may be in part due to peripheral tolerance mechanisms that would impede the activation and differentiation of polyreactive B cells during the immune response to HIV (27).
In this study, we examined B cells in naïve mice to assess the preimmune repertoire available for a primary antibody response to the HIV envelope protein gp120. We found that the majority of B cells capable of responding to gp120 are FO cells in WT mice and naïve B cells in uninfected human adults, suggesting that these B cells likely dominate the primary response to gp120. We also found that murine MZ B cells are able to recognize gp120, but do so using a restricted antibody repertoire dominated by antibodies harboring a specific IGVH gene segment. Of note, this murine IGHV1-53 gene is most closely related to the IGHV1-2*02 gene used by the broadly neutralizing human VRC01 and related antibodies. Our findings suggest that a protective HIV vaccine might involve eliciting broadly neutralizing antibodies from B-cell populations that use IGHV gene segments known to generate protective antibodies.
Results
Most gp120-Reactive Naïve B Cells Are of Follicular Origin.
The antibody response to HIV gp120 has been studied for decades, largely through analysis of serological data from chronically infected subjects. The antibody response to physiological pathogens normally involves a concerted effort among different types of B cells (15); thus, we set out to investigate the extent to which B-cell subsets are capable of participating in the primary HIV antibody response not only to better understand why this response is typically nonprotective, but also to inform directions for vaccine design.
Given the close parallel in the development and function of human and mouse peripheral B-cell subpopulations, we used WT mice to evaluate the preexposure B-cell repertoire available to respond to an HIV vaccine or infecting virions. Specifically, we characterized the naïve gp120-reactive repertoire of the major B-cell subsets in unchallenged mice. To assess whether naïve B cells in these populations are able to produce anti-gp120 antibodies, we sorted FO and MZ B cells from naïve mice and stimulated them polyclonally with LPS, and 5 d later tested supernatants for total and gp120-reactive IgM. MZ B cells responded more robustly than FO B cells to LPS stimulation, as expected (28, 29), producing more than 10-fold more total IgM (Fig. 1); however, despite the significantly increased total IgM production by MZ B cells, FO and MZ B cells produced relatively similar amounts of gp120-reactive IgM (Fig. 1A). Because FO B cells are the major subpopulation, gp120-specific antibody is more likely to be derived from FO B cells.
Fig. 1.
Most gp120-reactive naïve B cells are of follicular origin. (A) Naïve splenic FO (CD21intCD1dint) and MZ B cells (CD21hiCD1dhi) were stimulated with 50 μg/mL LPS for 5 d. Supernatants were analyzed for total (Left) and gp120-reactive (Right) IgM production by ELISA. Results are from three independent experiments with between five and eight mice each. (B) Splenocytes from naïve mice were stained with gp120-biotin-Alexa Fluor 647 and enriched by magnetic cell sorting using antibiotin beads. Enriched B220+ gp120-binding B cells (Left) were then assessed for subset phenotype (Right). Representative plots of column-bound cells pregated on singlet live B220+ cells are shown. The gate for gp120 binding was set based on the fluorescence level of negative cells that did not bind the column. (C) Percentage of gp120-binding FO (open circles) and MZ (filled circles) B cells from individual mice. Results are from three independent experiments, each with two mice. (D) gp120-binding enrichment as described in B performed with three human spleen samples. Representative dot plot of gp120-binding by column-enriched CD20+ B cells (Left) and the distribution of these gp120-reactive B cells as IgD+CD27− naïve and IgD+CD27+ MZ-like B cells (Right) of each spleen are shown. (E) The percent (Left) and normalized frequency (Right) ) of CD20+ gp120-binding B cells in the naïve (open circles) and MZ-like (filled circles) B-cell populations depicted for each individual. The percentage of gp120-reactive naïve and MZ-like B cells was normalized to the frequency of each population in unenriched spleen. P values are derived from the Student t test.
To analyze gp120 recognition by naïve B cells in the context of surface B-cell receptor binding, we labeled gp120 with biotin and Alexa Fluor 647 and performed flow cytometric analyses. Alexa Fluor 647 was chosen because of its brightness and small molecular weight to maximize detection and minimize B-cell recognition of the fluorophore. Only plasma cells from mice immunized with gp120 bound this reagent (Fig. S1A). The observed gp120 binding by naïve B cells is B-cell receptor specific, with no gp120 staining on B cells expressing an Ig transgene of irrelevant B-cell receptor specificity (Fig. S1B). We used magnetic column enrichment to capture all gp120-binding cells from the spleens of naïve mice (Fig. 1B and Fig. S1B). We further evaluated the phenotype of these B220+gp120+ B cells to identify FO (CD21intCD1dint) and MZ (CD21hiCD1dhi) B cells (Fig. 1B). We found that in an unchallenged WT mouse, 50–60% of splenic B cells that bind to gp120 are FO B cells, whereas only 10–20% are MZ B cells (Fig. 1C).
We also used magnetic column enrichment to examine gp120-reactive human B cells from healthy non–HIV-infected spleens. Representative staining is shown in Fig. 1D. The frequency of gp120 reactivity varied among three individuals (Fig. S2). Similarly, the distribution of these gp120-reactive B cells into the IgD+CD27– naïve and IgD+CD27+ MZ-like B-cell populations was also variable but on average favored the naïve B-cell population (Fig. 1E). Because individuals had varying frequencies of splenic B-cell populations (16–44% naïve and 25–35% MZ phenotype), gp120-binding cells were normalized to the percentage of unenriched populations for each individual. We found that gp120 binding was enriched for naïve B cells almost twofold, whereas gp120 binding did not appear to select for MZ B cells (Fig. 1E). This further indicates that the gp120 precursor frequency is higher in naïve B cells than in the MZ-like B-cell population in humans.
Mouse MZ B Cells Repeatedly Use the Same IGHV Gene to Encode Antibodies That Recognize gp120.
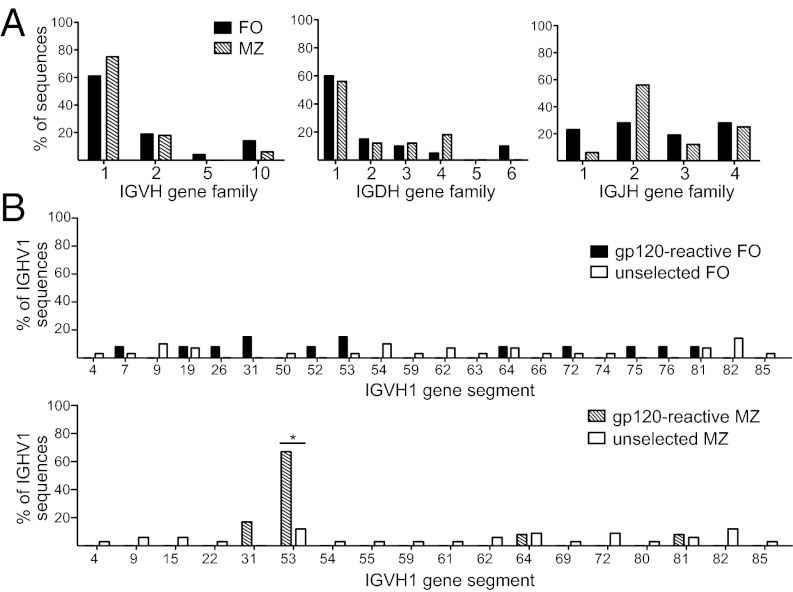
To analyze the naïve anti-gp120 repertoire, we generated hybridomas from naïve FO and MZ B cells isolated from WT mice and screened them for gp120 reactivity by ELISA. Because these hybridomas were generated from mice that had never been exposed to gp120, we anticipated that the antibodies they produced would be germ-line–encoded IgM. As a first line of defense for humoral immunity, germ-line IgM is inherently polyreactive, and accordingly, Ig from naïve B cells is often capable of binding multiple antigens (30, 31). Therefore, it was important in the ELISA screening to ensure that the IgM was bound to the gp120 coated on the plate, not to the protein used for blocking. To accomplish this, we considered hybridomas screened for gp120 reactivity by ELISA positive only if the IgM bound gp120 regardless of the blocking reagent used. From more than 2,500 FO and 1,700 MZ B-cell hybridomas generated, 24 FO and 16 MZ B-cell hybridomas were found to produce antibodies that could recognize gp120. The Ig heavy chains were sequenced from these gp120-reactive hybridomas using degenerate IGHV primers (32), with gene segments assigned according to the International Immunogenetics Information System database (33). These analyses showed that naive FO and MZ gp120-reactive hybridomas were not mutated from germ line and used similar distributions of IGHV and IGHD families (Fig. 2A). However, compared with FO B cells, IGHJ2 was overrepresented among MZ B-cell gp120-reactive antibodies, owing at least in part to a heavily favored VDJ combination (6 of 16 antibodies analyzed) in the MZ population (Fig. 2A). Of these hybridomas expressing the same VDJ rearrangement, the IgL chain VJ rearrangement was amplified from four hybridomas, which revealed that each harbored a unique light chain: IGLVk1-110/IGLJk4, IGLVk5-48/IGLJk5, IGLVk1-135/IGLJk1, and an undefined Igk chain distinct from the other characterized (and fusion partner) IgL chains based on restriction digestion.
Fig. 2.
Mouse MZ B cells repeatedly use the same IGHV gene to encode antibodies that recognize gp120. Hybridomas were generated from LPS-stimulated naïve FO and MZ B cells (five independent fusions from B-cell populations sorted from four to eight mice each). Hybridomas were screened for gp120-reactivity by anti-gp120 ELISA. Reactivity against the blocking reagent was excluded by ELISA performed with BSA in parallel with fish skin gelatin. Only 24 FO and 16 MZ hybridomas showed reactivity to gp120 under both conditions and were considered positive. The Ig heavy chain of each hybridoma was identified using a PCR strategy that selectively amplifies most mouse VDJ rearrangements. (A) Heavy-chain V (Left), D (Center), and J (Right) gene families used by FO (solid) and MZ (hatched) B cells to recognize gp120. (B) IGHV1 family member use in gp120-reactive FO (solid; Upper) and MZ (hatched; Lower) hybridomas and antigen-unselected controls (open). Antigen-unselected controls were generated by sequencing Ig genes from bulk cDNA of LPS-stimulated FO and MZ B cells. *P = 0.0108, Fisher’s exact test.
Because the IGHV1 family is the largest IGHV family in the mouse, and the most frequently represented in gp120 binding, we analyzed the gene segments from this family individually (Fig. 2B). Whereas gp120-reactive Ig heavy chains from FO B cells used diverse IGHV1 family members, gp120-reactive MZ B cells used predominantly IGHV1-53 (Fig. 2B). Furthermore, IGHV1-53 use was not obviously overrepresented in the unselected and antigen-nonspecific Ig heavy chain repertoire from either B-cell population (Fig. 2B). These data indicate that IGHV1-53 is preferentially used by MZ B cells to encode antibodies that recognize gp120.
Mouse IGHV1-53 Is Orthologous to Human IGHV1-2*02.
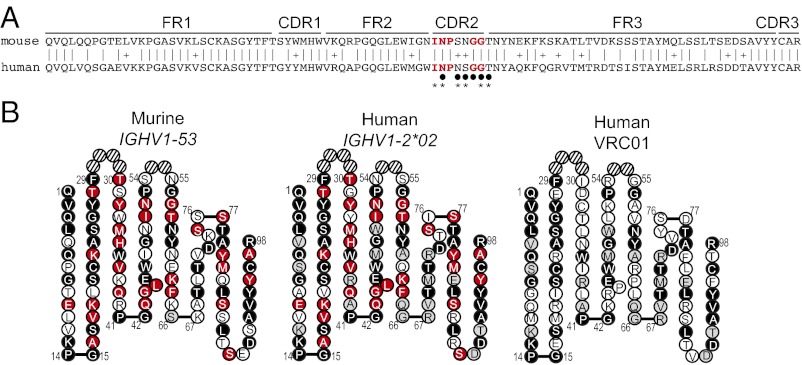
We next asked whether IGHV1-53, used preferentially by murine MZ B cells to generate gp120-reactivity, is similar to any IGHV used in anti-gp120 antibodies found in humans. To address this question, we compared the IGHV1-53 sequence with human IGHV sequences using several databases, including IMGT (33). The IGHV1-53 gene segment shows the highest sequence identity with human IGHV1-2*02 and IGHV1-46*01. Notably, VRC01 and related broadly neutralizing anti-gp120 antibodies are encoded by IGHV1-2*02, whereas other broadly neutralizing antibodies use IGHV1-46*01 (13). At the nucleotide level, mouse IGHV1-53 is 78% identical to human IGHV1-2*02, and the encoded protein sequences are 70% identical and 85% hydrophobically similar (Fig. 3A). In comparison, mouse IGHV1 family members are on average only 63% identical to human IGHV1 family members at the protein level (34), suggesting that this high identity/similarity between murine IGHV1-53 and human IGHV1-2*02 is not strictly a result of family relatedness. Fig. 3B highlights the similarities among the mouse germ-line IGHV1-53, human germ-line IGHV1-2*02, and human mature IGHV of VRC01 based on the Collier de Perles configuration (35). Amino acids common to all three IGHVs are in black, those shared only by IGHV1-2*02 and IGHV1-53 are in red, and those shared only by IGHV1-2*02 and the mature VRC01 are in gray. At the protein level, IGHV1-2*02 and murine IGHV1-53 are 70% identical, whereas IGHV1-2*02 and the mature VRC01 are only 58% identical; more than 40% of amino acids are identical among all three IGHV-encoded sequences.
Fig. 3.
Mouse IGHV1-53 is orthologous to human IGHV1-2*02. (A) Amino acid sequence alignment encoded by germ-line murine IGHV1-53*01 (Upper) and human IGHV1-02*02 (Lower) genes. Identities are indicated by a vertical line; similarities, by “+.” Identical CDRH2 residues are in red. Below the CDRH2 region, an asterisk indicates residues mutated in VRC01, and black circles indicate residues involved in interaction with gp120. (B) Mouse IGHV1-53*01 (Left), human IGHV1-2*02 (Center), and the mature mutated VRC01 VH (Right) regions. Black circles identify amino acids identical in all three, red circles indicate identity between murine IGHV1-53 and human IGHV1-2*02, gray circles indicate identity between IGHV1-2*02 and the mature VRC01, and white circles indicate amino acids not common with IGHV1-2*02.
VRC01 has been crystallized with a gp120 core protein (14). This crystal structure showed that more than one-half of the contact by VRC01 with gp120 is within the IGHV1-2*02-encoded and mutated CDRH2 region, likely accounting for the bias in broadly neutralizing activity for IGHV1-2*02–encoded IgH chains, independent of the IGHD and IGHJ segments (13, 36). In support of the importance of this CDRH2 region, IGHV1-2*02–encoded IgH chains can pair with diverse IgL chains and still provide neutralizing activity (13, 36) and the mouse ortholog IGHV1-53–encoded IgH chains can pair with diverse IgL chains to encode anti-gp120 reactivity. Notably, the CDRH2 protein sequence encoded by the murine germ-line IGHV1-53 is highly similar to IGHV1-2*02, differing only by an inversion of two amino acids to the human germ-line sequence (Fig. 3 A and B).
Germ-Line IGHV1-53 Antibodies Are Polyreactive and Do Not Neutralize HIV.
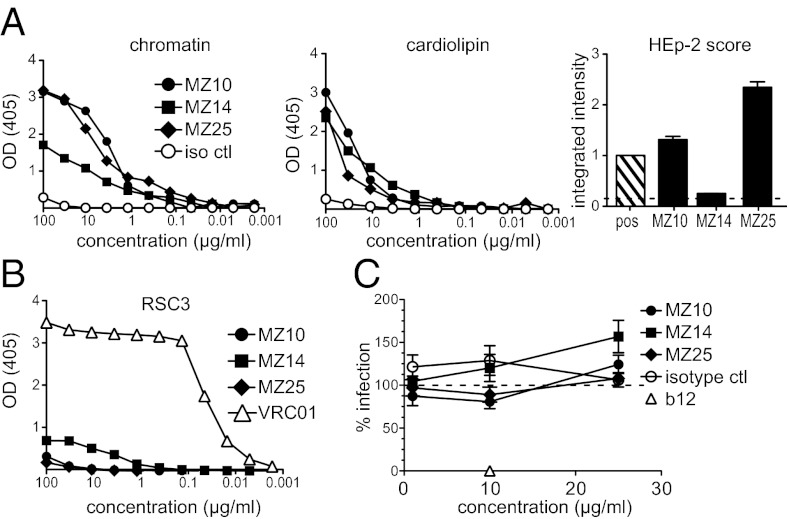
Although VRC01 and its germ-line revertant have not demonstrated polyreactivity, other HIV broadly neutralizing antibodies, including those that use IGHV1-2*02 and IGHV1-46*01 IGHV gene segments, have been shown to exhibit polyreactivity as mature and germ-line antibodies (13). As expected, the mouse MZ gp120-reactive IGHV1-53 antibodies showed polyreactivity by ELISA and HEp-2 reactivity. All three MZ B-cell IGHV1-53–encoded antibodies tested displayed some reactivity with both chromatin and cardiolipin, and two showed strong binding to HEp-2 cells (Fig. 4A). Thus, like other MZ B-cell antibodies and many HIV broadly neutralizing antibodies, the IGHV1-53–encoded antibodies are polyreactive and autoreactive.
Fig. 4.
Germ-line IGHV1-53 antibodies are polyreactive and do not neutralize HIV. (A) The IGHV1-53 MZ antibodies were tested for polyreactivity by binding to chromatin (Left) and cardiolipin (Center) by ELISA or to HEp-2 cells (Right) by indirect immunofluorescence staining. Mouse IgM hybridomas specific for NP (B1-8) or H-2b (3-83) were used as isotype controls. (B) The IGHV1-53 MZ antibodies were tested for CD4 binding site recognition by ELISA against RSC3. VRC01 was used as a positive control. (C) Neutralization ability of IGHV1-53 MZ antibodies was determined using a TZM-bl assay. Mouse IGHV1-53 antibodies were incubated with JR-FL pseudovirus for 30 min before culturing with TZM-bl cells for 48 h. Infection was determined with the Beta-Glo assay and quantitated on a luminometer. Percent infection was calculated in relation to values from wells that received no antibody. Human b12 was used as a positive control for neutralization, and a mouse IgM specific for LPS served as a negative control.
VRC01 and related broadly neutralizing antibodies target the CD4 binding site. To determine whether the gp120-reactive MZ B-cell antibodies also recognize the same epitope on gp120, we measured the ability of these IGHV1-53 antibodies to recognize RSC3, the CD4 binding site epitope used to identify VRC01 (12). Although VRC01 bound very well to RSC3, the MZ IGHV1-53 antibodies did not (Fig. 4B). The modest binding by antibody MZ14 likely is not related to CD4 binding site recognition, given that this antibody also bound the control RSC3Δ, which has a single mutation abolishing the CD4 binding site (Fig. S3).
Finally, we also tested these IGHV1-53 antibodies for their ability to neutralize a JR-FL pseudovirus in a TZM-bl neutralization assay. None of the mouse antibodies tested demonstrated in vitro neutralization (Fig. 4C). Because these MZ B-cell–derived antibodies are unable to neutralize virus, the germ-line IGHV1-53 antibodies do not behave like the heavily mutated VRC01, but are similar to the purported VRC01 germ line (7, 14) and are not protective against HIV in germ-line form.
IGHV1-53 Is Enriched in Mouse MZ, Whereas IGHV1-2*02 Is Comparably Expressed by both Naïve and MZ-Like Human B Cells.
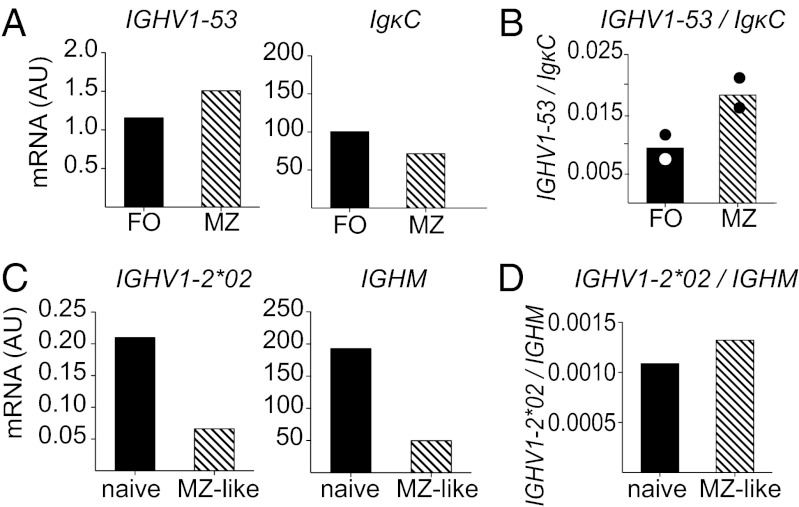
The antigen receptor specificity of newly generated immature B cells has been shown to influence the differentiation of B cells into either MZ or FO B-cell compartments (37). Our data show more frequent IGHV1-53 expression in both MZ B-cell hybridomas recognizing gp120 and unselected antibodies from MZ B cells than in antibodies from the gp120-reactive or unselected FO B-cell population (Fig. 2B). To more accurately determine the expression of this IGHV1-53 gene segment in the different B-cell subsets, we sorted FO and MZ B cells from naïve mice and analyzed IGHV1-53 expression by quantitative RT-PCR. Because FO and MZ B cells express different amounts of IgM and IgD on their surface, IGHV expression was normalized to expression of Ig kappa chain (Fig. 5A). Our results show twofold to threefold greater IGHV1-53 expression by MZ B cells compared with FO B cells (Fig. 5B).
Fig. 5.
IGHV1-53 is enriched in mouse MZ, whereas IGHV1-2*02 is comparably expressed by both naïve and MZ-like human B cells. (A) FO B cells and MZ B cells were sorted from between four and six pooled C57BL/6 mice. Gene expression of IGHV1-53 (Left) and the constant region of the Ig kappa chain (IgκC; Right) were assessed by RT-PCR. The results of one representative experiment of two experiments are shown. (B) IGHV1-53 expression was normalized to IgCκ to control for different amounts of Ig transcript in the different subsets. Circles represent individual experiments, and bars indicate the mean. (C) Naïve and MZ-like B cells were sorted from a human spleen. Gene expression of IGHV1-2*02 (Left) and human IgM (IGHM; Right) was assessed by RT-PCR. (D) IGHV1-2*02 expression by naïve and MZ-like B cells normalized to IgM to control for different amounts of Ig transcript in the different subsets.
We similarly assessed the expression of IGHV1-2*02 in naïve (IgDhiCD27−) and MZ-like B cells (IgDintCD27+) sorted from a healthy human spleen. Because kappa and lambda light chain use varies between individuals (38, 39), IGHV1-2*02 transcript was normalized to IgM expression. IgM transcript is depressed in human MZ-like B cells compared with naive B cells (Fig. 5C), similar to repression reported previously (40). Analysis of the splenic naïve and MZ-like B-cell populations from a single individual revealed that whereas IGHV1-2*02 is indeed expressed by human MZ-like B cells (Fig. 5D), naïve human B cells show comparable expression of this IGHV gene segment.
Discussion
To better understand which B-cell subsets are capable of generating an anti-gp120 antibody response, we analyzed FO and MZ B-cell populations from naïve mice and uninfected humans. Based on frequency and absolute numbers of gp120-reactive B cells, our data indicate that the majority of B cells capable of binding gp120 on their surface are of follicular origin in both humans and mice (Fig. 1). If the primary anti-gp120 response is dominated by FO B cells in humans and is generally nonprotective in acutely infected individuals, then it might be useful to consider promoting the participation of other B-cell subsets as an HIV vaccine strategy. In this regard, a recent study found that repeated treatment of mice with BlyS increased the MZ compartment of mice, followed by a more robust response to Env immunization and increases in the frequency and breadth of neutralizing antibodies (41).
Investigation of the molecular repertoire of gp120-binding antibodies from naïve murine FO and MZ B-cell hybridomas revealed that MZ B cells preferentially used IGHV1-53 to recognize gp120 (Fig. 2B). Because these hybridomas were generated from LPS-stimulated naïve FO and MZ B cells, and because MZ B cells are preferentially stimulated by LPS (28, 29), we independently demonstrated by quantitative RT-PCR that IGHV1-53 is also disproportionately expressed by unstimulated MZ B cells (Fig. 5B). This mouse segment is most closely related to the human IGHV1-2*02, which encodes VRC01 and related broadly neutralizing antibodies. An obvious difference between the two is that the germ-line mouse IGHV1-53 is able to confer binding to gp120, whereas germ-line human IGHV1-2*02 apparently cannot.
To further determine the quality of the mouse IGHV1-53 MZ antibodies, we tested a subset for autoreactivity and neutralization capability (Fig. 4). These IGHV1-53 antibodies bound gp120 and also weakly bound chromatin, cardiolipin, and HEp-2 cells, but were unable to neutralize HIV in vitro. These observations provide further evidence that to be protective, these B cells likely would need to be drawn into a germinal center to undergo somatic hypermutation and affinity maturation.
IGHV1-2*02–encoded broadly neutralizing HIV antibodies have been isolated from circulating B cells from chronically infected subjects (12). It is of interest that the affinity of VRC01 and related antibodies for gp120 is perhaps lower than what might be expected given the degree of somatic mutation displayed (12). This high mutation rate, viewed as a barrier in vaccine design, might be a consequence of chronic antigen stimulation and activation in the HIV disease state of the patient rather than borne out of necessity to enhance function. The original phenotype of the B cells that eventually produced these protective antibodies is unknown, and our findings suggest that the original B cells could have been either FO or MZ B cells; however, we postulate that the B cells generating these broadly neutralizing antibodies originally might have been MZ B cells. Evidence to support this idea includes the much shorter CDRH3 of many IGHV1-2*02–encoded broadly neutralizing antibodies compared with other broadly neutralizing antibodies (12, 13), a characteristic of human and rodent MZ B cells (37, 42). In addition, the IGHV1-2*02 segment has been viewed clinically as a feature of MZ lymphomas (43, 44), indicating that human MZ B cells naturally express this IGHV gene segment. Molecular analyses of a human spleen sample showed no preferential expression for IGHV1-2*02 in MZ B cells, as has been detected in mouse MZ B cells (Fig. 5). The fact that expression appeared relatively even in the two human subsets is interesting, considering that human MZ B cells preferentially use IGHV3 segments in their heavy chains, and that IGHV1 segments are the least-used segments by this subset (45).
Abundant evidence using model antigens in rodent models has indicated that MZ B cells typically mount rapid low-affinity antibody responses devoid of mutation. Although the use of these model antigens clearly has been instructive, the prototypical MZ B-cell antigen, hapten-polysaccharide conjugates, lack a protein epitope, thus precluding the participation of T cells that would be required for germinal center formation (46). Indeed, MZ B cells are capable of seeding germinal centers and undergoing somatic hypermutation (16, 17), and the antibody response in humans to typical MZ B-cell antigens, bacterial capsular polysaccharides, are often mutated (21–24, 42, 47). In reality, the contribution of MZ B cells to pathogen challenge as class-switched, high-affinity antibodies from germinal center reactions is difficult to ascertain and largely undefined, because antibody responses of MZ B cells in an intact mouse are difficult to follow. Nevertheless, there is a rationale for considering the elicitation of MZ B-cell HIV antibody responses in vaccine design. Compared with naïve or FO B cells, MZ B cells exhibit increased expression of costimulatory receptors, including complement receptor 2 and additional Toll-like receptors (TLRs) (48–51). Human MZ B cells respond more robustly to T-cell–like help or TLR stimulation in vitro (52–55). Rodent MZ B cells express CD1d, and human MZ B cells express CD1c (19), allowing these cells to provide cognate help to a wider range of T cells compared with FO B cells. Thus, a carefully selected adjuvant, such as a TLR agonist, feasibly could preferentially target MZ B-cell activation over dominant FO B-cell responses to gp120. If IGHV1-2*02–expressing MZ B cells can be brought into germinal centers to undergo somatic hypermutation, then broadly neutralizing CD4 binding site antibodies may be generated.
Materials and Methods
gp120 Production and ELISAs.
gp120 was produced by transient transfection of COS7 cells with 5 mg of HIV ADA gp120 plasmid (provided by T. M. Ross, University of Pittsburgh) and Lipofectamine (Invitrogen). gp120 was purified on agarose-bound lectin (Vector Laboratories) and stored in PBS and 0.1% azide. For ELISAs, 2 mg/mL gp120 was used as coating material. gp120-reactive IgM was quantitated with a gp120-specific IgM hybridoma developed in-house.
Flow Cytometry and Cell Sorting.
gp120 was labeled with biotin and Alexa Fluor 647 using EZ-Link Biotin (Thermo Scientific) and an Alexa Fluor 647 Microscale Labeling Kit (Thermo Scientific). For analyses of naïve B-cell subsets, splenocytes from unchallenged mice or HIV-uninfected humans were incubated with gp120-biotin-Alexa Fluor 647. After washing, anti-biotin beads (Miltenyi Biotec) were applied to the splenocytes, and the splenocytes were run over an LS Magnetic Column (Miltenyi Biotec), at which point the column-bound and flow-through fractions were collected and stained.
Hybridoma Generation and Screening.
Follicular (FO) and marginal zone (MZ) B cells were purified from spleens of four to eight naïve mice in five independent sorts and stimulated for 3 d with 50 mg/mL LPS (Sigma). Splenocytes were fused to SP/2 fusion partners with polyethylene glycol (ATCC) for 4 h. Cell supernatant was screened for gp120-reactivity by ELISA 7-10 d after selection with hypoxanthine (Sigma) and asazerine (Sigma). Hybridomas were considered gp120-reactive if they had an OD over 2.0 after successive screening and subcloning. Reactivity against the blocking reagent was excluded by blocking with bovine serum albumin (Fisher Scientific) in parallel with fish skin gelatin (Sigma). Hybridomas that bound gp120, regardless of blocking reagent, were considered gp120-reactive. Approximately 2,500 and 1,700 hybridomas were derived from FO and MZ B cells, respectively, and screened for gp120 reactivity. For antigen unselected controls, FO and MZ B cells were sorted from naïve mice and stimulated for 3 d with LPS as they were for the hybridoma fusions. RNA and cDNA were then made from the bulk population, and random heavy chains were amplified by PCR and sequenced.
Polyreactivity Assays.
ELISA plates were coated with 10 mg/mL chromatin (prepared from bovine thymus, gift of L. Wysocki, National Jewish Health) or cardiolipin (Sigma). For the HEp-2 reactivity, 100 mg/mL antibodies were applied to HEp-2 antigen substrate slides (BION Enterprises) and detected with anti-mouse IgM+IgG-Cy5 (Jackson Immunoresearch). Integrated intensity was quantified on an Odyssey Infared Imaging System (LI-COR Biosciences). Samples were normalized to a positive control (a mouse hybridoma that is reactive to chromatin).
CD4 Binding Site Epitope Mapping.
293F cells were transiently transfected with 293Fectin (Invitrogen) and 30 mg of plasmid (gift of J. Mascola, National Institutes of Health) to produce RSC3 and RSC3D as described (12). RSC3 or RSC3D was coated onto ELISA plates at 2 mg/mL. The positive control, VRC01, was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from J. Mascola.
HIV Neutralization Assay.
HIV pseudovirus constructs were a gift of P. Clapham (University of Massachusetts). Pseudovirus was prepared by cotransfecting JRFL subtype B envelope and pNL4.3∆env backbone into 293T cells. Two hundred TCID50/mL pseudovirus was mixed with different concentrations of antibodies and incubated for 30 min at 37 °C. TZM-bl cells were then added to the pseudovirus/antibody mixture and incubated for 48 h at 37 °C. Infection levels were determined with the Beta-Glo assay (Promega), and relative light units were read on a luminometer. Percent infection was calculated in relation to values from wells that received no antibody.
RT-PCR.
RT-PCR was performed with customized and commercially available primers, as detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Pamela Strauch and Jana M. Palaia for technical assistance, and members of the R.M.T. and R.P. laboratories for helpful discussions and comments on this study. This work was supported by the National Institutes of Health Grants AI052157 and AI078468 (to R.M.T.), AI052310 (to R.P.), and HD059527 (to E.N.J.); National Institute of Allergy and Infectious Disease Training Grant T32-AI07405 (to L.M.P.); and the Veterans Affairs Research Service (E.N.J.).
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213713110/-/DCSupplemental.
References
- 1.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33(4):542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston MI, Fauci AS. An HIV vaccine—evolving concepts. N Engl J Med. 2007;356(20):2073–2081. doi: 10.1056/NEJMra066267. [DOI] [PubMed] [Google Scholar]
- 3.Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455(7213):613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomaras GD, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: Virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verkoczy L, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci USA. 2010;107(1):181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 7.Ota T, et al. Anti-HIV B cell lines as candidate vaccine biosensors. J Immunol. 2012;189(10):4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13(9):1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1–infected individuals. PLoS Pathog. 2010;6(8):e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikell I, et al. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7(1):e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheid JF, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333(6049):1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329(5993):811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson CL, Pelanda R, Torres RM. Division of labor during primary humoral immunity. Immunol Res. 2012 doi: 10.1007/s12026-012-8372-9. 10.1007/s12026-012-8372-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Cerny J. Functional heterogeneity of marginal zone B cells revealed by their ability to generate both early antibody-forming cells and germinal centers with hypermutation and memory in response to a T-dependent antigen. J Exp Med. 2003;198(12):1923–1935. doi: 10.1084/jem.20031498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan TG, Gardam S, Basten A, Brink R. Altered migration, recruitment, and somatic hypermutation in the early response of marginal zone B cells to T cell-dependent antigen. J Immunol. 2005;174(8):4567–4578. doi: 10.4049/jimmunol.174.8.4567. [DOI] [PubMed] [Google Scholar]
- 18.Kruetzmann S, et al. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med. 2003;197(7):939–945. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weller S, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104(12):3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zandvoort A, Timens W. The dual function of the splenic marginal zone: Essential for initiation of anti-TI-2 responses but also vital in the general first-line defense against blood-borne antigens. Clin Exp Immunol. 2002;130(1):4–11. doi: 10.1046/j.1365-2249.2002.01953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxendale HE, et al. Immunogenetic analysis of the immune response to pneumococcal polysaccharide. Eur J Immunol. 2000;30(4):1214–1223. doi: 10.1002/(SICI)1521-4141(200004)30:4<1214::AID-IMMU1214>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Lottenbach KR, Barenkamp SJ, Reason DC. Somatic hypermutation and diverse immunoglobulin gene usage in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae Type 6B. Infect Immun. 2004;72(6):3505–3514. doi: 10.1128/IAI.72.6.3505-3514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott MG, Zachau HG, Nahm MH. The human antibody V region repertoire to the type B capsular polysaccharide of Haemophilus influenzae. Int Rev Immunol. 1992;9(1):45–55. doi: 10.3109/08830189209061782. [DOI] [PubMed] [Google Scholar]
- 24.Insel RA, Adderson EE, Carroll WL. The repertoire of human antibody to the Haemophilus influenzae type b capsular polysaccharide. Int Rev Immunol. 1992;9(1):25–43. doi: 10.3109/08830189209061781. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Martin F, Forbush KA, Perlmutter RM, Kearney JF. Evidence for selection of a population of multi-reactive B cells into the splenic marginal zone. Int Immunol. 1997;9(1):27–41. doi: 10.1093/intimm/9.1.27. [DOI] [PubMed] [Google Scholar]
- 26.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467(7315):591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: A hypothesis. Hum Antibodies. 2005;14(3-4):59–67. [PMC free article] [PubMed] [Google Scholar]
- 28.Snapper CM, et al. Comparative in vitro analysis of proliferation, Ig secretion, and Ig class switching by murine marginal zone and follicular B cells. J Immunol. 1993;150(7):2737–2745. [PubMed] [Google Scholar]
- 29.Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27(9):2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- 30.Guilbert B, Dighiero G, Avrameas S. Naturally occurring antibodies against nine common antigens in human sera, I: Detection, isolation and characterization. J Immunol. 1982;128(6):2779–2787. [PubMed] [Google Scholar]
- 31.Haspel MV, et al. Multiple organ-reactive monoclonal autoantibodies. Nature. 1983;304(5921):73–76. doi: 10.1038/304073a0. [DOI] [PubMed] [Google Scholar]
- 32.Fournier EM, et al. Dual-reactive B cells are autoreactive and highly enriched in the plasmablast and memory B cell subsets of autoimmune mice. J Exp Med. 2012;209(10):1797–1812. doi: 10.1084/jem.20120332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bono B, Madera M, Chothia C. VH gene segments in the mouse and human genomes. J Mol Biol. 2004;342(1):131–143. doi: 10.1016/j.jmb.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 35.Ehrenmann F, Giudicelli V, Duroux P, Lefranc MP. IMGT/Collier de Perles: IMGT standardized representation of domains (IG, TR, and IgSF variable and constant domains, MH and MhSF groove domains) Cold Spring Harb Protoc. 2011;2011(6):726–736. doi: 10.1101/pdb.prot5635. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carey JB, Moffatt-Blue CS, Watson LC, Gavin AL, Feeney AJ. Repertoire-based selection into the marginal zone compartment during B cell development. J Exp Med. 2008;205(9):2043–2052. doi: 10.1084/jem.20080559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reichard KK, McKenna RW, Kroft SH. Comparative analysis of light chain expression in germinal center cells and mantle cells of reactive lymphoid tissues: A four-color flow cytometric study. Am J Clin Pathol. 2003;119(1):130–136. doi: 10.1309/9MYM-D68F-U8YE-843D. [DOI] [PubMed] [Google Scholar]
- 39.Yount WJ, Dorner MM, Kunkel HG, Kabat EA. Studies on human antibodies, VI: Selective variations in subgroup composition and genetic markers. J Exp Med. 1968;127(3):633–646. doi: 10.1084/jem.127.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heng TS, Painter MW. Immunological Genome Project Consortium The Immunological Genome Project: Networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 41.Dosenovic P, et al. BLyS-mediated modulation of naive B cell subsets impacts HIV Env-induced antibody responses. J Immunol. 2012;188(12):6018–6026. doi: 10.4049/jimmunol.1200466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian C, et al. Evidence for preferential Ig gene usage and differential TdT and exonuclease activities in human naïve and memory B cells. Mol Immunol. 2007;44(9):2173–2183. doi: 10.1016/j.molimm.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahler DW, Pindzola JA, Swerdlow SH. Splenic marginal zone lymphomas appear to originate from different B cell types. Am J Pathol. 2002;161(1):81–88. doi: 10.1016/S0002-9440(10)64159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warsame AA, et al. Splenic marginal zone lymphoma with VH1-02 gene rearrangement expresses poly- and self-reactive antibodies with similar reactivity. Blood. 2011;118(12):3331–3339. doi: 10.1182/blood-2011-03-341651. [DOI] [PubMed] [Google Scholar]
- 45.Wu YC, et al. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116(7):1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toellner KM, et al. Low-level hypermutation in T cell-independent germinal centers compared with high mutation rates associated with T cell-dependent germinal centers. J Exp Med. 2002;195(3):383–389. doi: 10.1084/jem.20011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weller S, Reynaud CA, Weill JC. Vaccination against encapsulated bacteria in humans: Paradoxes. Trends Immunol. 2005;26(2):85–89. doi: 10.1016/j.it.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Guinamard R, Okigaki M, Schlessinger J, Ravetch JV. Absence of marginal zone B cells in Pyk-2-deficient mice defines their role in the humoral response. Nat Immunol. 2000;1(1):31–36. doi: 10.1038/76882. [DOI] [PubMed] [Google Scholar]
- 49.Rubtsov AV, et al. TLR agonists promote marginal zone B cell activation and facilitate T-dependent IgM responses. J Immunol. 2008;180(6):3882–3888. doi: 10.4049/jimmunol.180.6.3882. [DOI] [PubMed] [Google Scholar]
- 50.Genestier L, et al. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J Immunol. 2007;178(12):7779–7786. doi: 10.4049/jimmunol.178.12.7779. [DOI] [PubMed] [Google Scholar]
- 51.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: Up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101(11):4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 52.Agematsu K, et al. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur J Immunol. 1997;27(8):2073–2079. doi: 10.1002/eji.1830270835. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y, Agematsu K, Ochs HD, Sugane K. Functional analysis of human memory B-cell subpopulations: IgD+CD27+ B cells are crucial in secondary immune response by producing high-affinity IgM. Clin Immunol. 2003;108(2):128–137. doi: 10.1016/s1521-6616(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 54.Bryant VL, et al. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: Predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179(12):8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 55.Capolunghi F, et al. CpG drives human transitional B cells to terminal differentiation and production of natural antibodies. J Immunol. 2008;180(2):800–808. doi: 10.4049/jimmunol.180.2.800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.