Abstract
The ability of the opportunistic fungal pathogen Cryptococcus neoformans to resist oxidative stress is one of its most important virulence related traits. To cope with the deleterious effect of cellular damage caused by the oxidative burst inside the macrophages, C. neoformans has developed multilayered redundant molecular responses to neutralize the stress, to repair the damage and to eventually grow inside the hostile environment of the phagosome. We used microarray analysis of cells treated with hydrogen peroxide (H2O2) at multiple time points in a nutrient defined medium to identify a transcriptional signature associated with oxidative stress. We discovered that the composition of the medium in which fungal cells were grown and treated had a profound effect on their capacity to degrade exogenous H2O2. We determined the kinetics of H2O2 breakdown by growing yeast cells under different conditions and accordingly selected an appropriate media composition and range of time points for isolating RNA for hybridization. Microarray analysis revealed a robust transient transcriptional response and the intensity of the global response was consistent with the kinetics of H2O2 breakdown by treated cells. Gene ontology analysis of differentially expressed genes related to oxidation-reduction, metabolic process and protein catabolic processes identified potential roles of mitochondrial function and protein ubiquitination in oxidative stress resistance. Interestingly, the metabolic pathway adaptation of C. neoformans to H2O2 treatment was remarkably distinct from the response of other fungal organisms to oxidative stress. We also identified the induction of an antifungal drug resistance response upon the treatment of C. neoformans with H2O2. These results highlight the complexity of the oxidative stress response and offer possible new avenues for improving our understanding of mechanisms of oxidative stress resistance in C. neoformans.
Introduction
Cryptococcus neoformans is pathogenic basidiomycetous yeast with a ubiquitous worldwide distribution. It exists primarily as an environmental organism associated with soil and is known to have a particular association with bird guano [1]. However, C. neoformans is also an important opportunistic pathogen that causes invasive fungal infections and is responsible for about 1 million cases and 700,000 mortalities annually [2]. Most of the infections have been reported in patients with immune deficiencies, especially those with AIDS, but also in non-HIV immune compromised patients. The respiratory tract is the main portal of entry and the lung is the primary site of infection, though serious infections involving the central nervous system are common.
In the mammalian host, cell mediated immunity based on phagocytic cells is crucial to counteract fungal infections. Macrophages, neutrophils and other phagocytic cells generate potent reactive oxygen (ROS) and nitrogen (RNS) species that are toxic to most fungal and bacterial pathogens and cause damage to their DNA, protein and lipids. ROS and RNS are implicated in the killing of fungal pathogens such as Aspergillus fumigatus, Candida albicans, and C. neoformans by host immune cells [3], [4], [5]. Most of these conclusions are based on the positive correlation between resistance of the wild type and sensitivity of a specific deletion strain to oxidative stress in vitro and the corresponding respective virulence and avirulence phenotypes of these strains in a murine cryptococcosis model [6].
An important virulence related trait of C. neoformans is its ability to survive inside phagocytic cells. It not only resists killing by macrophages after phagocytosis, but can continue to replicate by budding within this environment and subsequently exit the macrophage without causing host cell lysis [7], [8], [9], [10], [11]. The ability of C. neoformans to survive and thrive inside this harsh environment suggests it must have mechanisms not only to neutralize the reactive molecular species to which it is exposed within the macrophages but also to repair the cellular damages caused by the oxidative and nitrosative stresses.
In C. neoformans, a number of genes coding for enzymes of antioxidant defense systems have been shown to be important for both in vitro oxidative stress resistance and also for in vivo pathogenesis [12]. Foremost among these is the highly conserved peroxiredoxin, Tsa1, which is induced under oxidative stress and is essential for C. neoformans resistance to peroxide stress [6]. The downstream components of the thioredoxin system, represented by the thioredoxins Trx1 and Trx2 and thioredoxin reductase Trr1, are involved in the reduction and recycling of the oxidized, inactive form of peroxiredoxin [13]. In C. neoformans, deletion of these genes renders them sensitive to oxidative stress, albeit with a less severe phenotype than that of a tsa1Δ strain, and decreased survival in macrophage culture conditions [14], [15]. C. neoformans also contains two glutathione peroxidases Gpx1 and Gpx2, both of which respond differently to various stressors; with only Gpx2 induced in response to H2O2 generated oxidative stress. Furthermore, both GPX1 and GPX2 deletion mutants were only mildly sensitive to oxidant killing by macrophages and exhibited no effect on virulence in a murine model [16]. Other enzymes that have been shown to play a role in oxidative stress resistance in C. neoformans include a cytosolic copper-zinc superoxide dismutase (Sod1) [17], the mitochondrial manganese superoxide dismutase (Sod2) [18], cytochrome c peroxidase (Ccp1) [19] and the alternative oxidase (Aox1) [20].
Whole genome microarray studies of C. neoformans experiencing oxidative stress have been reported in which the authors either used fungal cells engulfed by macrophages or grown in the nutrient rich YPD medium and treated with exogenous H2O2 [21], [22]. The magnitude of the transcriptional response was much weaker in the cells residing inside the macrophages compared to the robust transcriptional response observed in the cells treated with exogenous H2O2 in YPD medium, suggesting that the environment has a direct effect on the transcriptional response. This may also be due to the differences in the concentration of ROS released by the exogenous application of stress agent compared to that released inside the macrophages. Gene expression studies of oxidative stress resistance in a number of other fungal organisms such as S. cerevisiae, Schizosaccharomyces pombe and Candida sp have been published [23], [24], [25], [26]. They indicate that the sensitivity of the organisms to different concentrations of H2O2 and the magnitude of the elicited cellular response was highly dependent on the composition of the medium in which the cells were grown and treated. For example, S. pombe triggered different signaling networks mediated either by Pap1 or by Sty1 depending on the concentration of H2O2 used for the treatment [27]. Pap1 was more sensitive to H2O2 than the Sty1-mediated pathway and was responsible for inducing an adaptive response. This was shown by an induction of Pap1 with an extracellular concentration of 0.2 mM H2O2, whereas H2O2 concentrations above 0.2 mM failed to trigger this activation.
In a study of oxidative stress and aging in S. cerevisiae, the authors reported different degrees of sensitivity of yeast cells to external H2O2 when grown in different media [28]. In this study, there was a dramatic increase in the resistance of yeast cells growing in YNB in the presence of galactose and required the use of 10 mM of H2O2 for gene induction, even though the same strains grown in the presence of glucose exhibited 70% death when treated with 1.5 mM H2O2. The pathogenic fungi C. glabrata has shown to be resistant to up to 40 mM of H2O2, while C. albicans was found to be sensitive at this concentration of H2O2 [29]. Another pathogenic fungi, A. fumigatus, continued to grow during two-hour incubations with 5 mM H2O2 and tolerated up to 30 mM H2O2 [30], [31].
In the context of pathogenic fungi and phagocytosis, the concentration of H2O2 inside the phagosome during the oxidative burst is not precisely known, however multiple reports suggest that the effective H2O2 concentration may reach hundreds of micro molar [32], [33]. Additionally, the usable nutrient composition of the phagosome is essentially unknown, but it is likely to be a nutrient-limited environment with the lumen of the phagosomes reported to acidify to a final pH of ∼ 4.8 during maturation [34]. To more closely mimic in vivo conditions, we chose to treat C. neoformans cells with H2O2 in a nutrient limited YNB medium at pH 4.0. To stress the cells and avoid the induction of an overwhelming apoptotic death response, cells were treated with a concentration of H2O2 that resulted in the killing of ∼ 15% of initial cell population or lethal dose (LD∼15) which was determined via H2O2-generated oxidative stress death curves. These curves dictated the use of 1 mM H2O2 for treatment. By quantitatively measuring the concentration of H2O2 in the culture during the treatment, we discovered that within 30 minutes of incubation, all of the H2O2 was completely degraded from the medium. Therefore, we subjected RNA samples isolated from the cells at 5, 15, 30, 45 and 60 minutes post H2O2 treatment to microarray hybridization. We used a custom designed C. neoformans serotype A array using the predicted ORFs in the H99 (serotype A) C. neoformans genome identified by the Broad Institute genome sequencing project (http://www.broadinstitute.org/annotation/genome/cryptococcus_ neoformans/FeatureSearch.html). To facilitate the identification of underlying biological processes from the gene expression dataset, we generated a new gene ontology gene association file for C. neoformans genome. Herein, we report a global time-resolved genomic expression pattern of C. neoformans and identified over-represented biological processes which may point to potential mechanisms by which the fungus detects and destroys the oxidative stressor as well as repairs and recovers from the damage caused by the oxidative stress.
Materials and Methods
Strains, Media and Reagents
C. neoformans serotype A strain KN99α was used throughout this experiment. Cells were grown on rich medium, YPD (1% yeast extract, 2% Bacto peptone, and 2% dextrose), or minimal medium, YNB, pH 4.0 (6.7 g/liter yeast nitrogen base without amino acids plus 20 g/liter dextrose and 25 mM sodium succinate at pH 4.0). Solid media contained 2% Bacto agar. Antimycin-A from Streptomyces sp. (Cat No A8674), Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (Cat No -C2920), Salicylhydroxamic acid (SHAM) (Cat No -S607) and hydrogen peroxide solution (Cat No H1009) all were purchased from SIGMA-ALDRICH, St Louis, MO, USA. Estimation of H2O2 was performed using OxiSelect H2O2 assay kit from Cell Biolabs, San Diego, CA, USA following the protocol supplied with the reagent. Three independent experiments were carried out to calculate the standard error which is indicated by the error bars in the figures.
Determination of Stressor Concentration by Survival Curve
Exponentially growing cells (OD650 = 1.5) were treated with various concentrations of H2O2 in a shaking 30°C incubator. Aliquots were taken at various time points, cells pelleted by centrifugation at 4°C. Cell pellet was washed two times with cold PBS and finally resuspended in PBS for plating on solid YPD and incubated at 30°C. Colony forming units (CFU) were counted after 2 days.
Hydrogen Peroxide Sensitivity Tests
Overnight cultures were diluted to OD650 = 1.0. Ten times serially diluted cell suspension was spotted on various plates containing either the individual stressor or their combinations. Plates were incubated at 30°C and observed on different days.
Custom C. neoformans Serotype A Microarray Development
Of the 7239 predicted ORFs in the H99 (serotype A) C. neoformans genome identified through the Broad Institute C. neoformans genome sequencing project, 6,932 probes were designed. The remaining 307 genes were either identical paralogs or had primarily low complexity sequences. Single 60-mer oligonucleotide probes were designed for each of the open reading frames and were duplicated on each microarray to provide an estimate of intra array variance. The arrays were fabricated by Agilent Technologies, Santa Clara, CA.
Cell Preparation and Treatment
Two-day-old cultures of C. neoformans KN99α were used to inoculate three bioreplicates into YNB pH 4.0 media and incubated at 30°C with shaking (200 rpm) until the cells were in mid-log phase (OD650 ∼1.5–2.0). Each biological replicate culture was split into two cultures and H2O2 was added to one of each of these two cultures to a final concentration of 1 mM. Cultures were incubated for 60 minutes and sampled at 5, 15, 30, 45 and 60-minute intervals. Sodium citrate (50 mM final concentration) was added to control and test samples to rapidly halt the hydrogen peroxide stress. Cells were collected by centrifugation at 1800 g for 5 minutes and washed once with sterile phosphate buffered saline. The washed cell pellets were flash frozen and lyophilized. Lyophilized cells were stored at –80°C.
Isolation of Total RNA
RNA was isolated using the Agilent Total RNA Isolation (Protocol for Yeast) (Agilent Technologies, Palo Alto, CA.) according to the manufacturer’s instructions with the following modifications. The lyophilized pellet (approximately 1×108 cells) was vortexed with 0.5 g of 1 mm glass beads (Biospec, Inc.) for 5 min., followed by the addition of 600 uL lysis solution (kit supplied) and vortexed for a further 5 min. Disrupted cells were centrifuged at 16 000 g for 3 min. and the supernatant transferred to a pre-filtration column. Rest of the procedures was carried out as described in the Manufacturer’s protocol until the final step. Isolated RNA was treated with RNase free DNase. RNA was quantified using a Nanodrop ND-1000 (Nanodrop Technologies, Wilmington, DE.). The quality of purified RNA was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA.).
Labeling of Total RNA
Total RNA was directly fluorescently labeled with Cy3 or Cy5 using the Micromax ASAP RNA labeling kit (Perkin Elmer, Inc, Wellesley, MA.) according to manufacturer’s instructions with the following modifications. All reaction component volumes were doubled with the exception of RNA concentration resulting in a final reaction mix volume of 40 uL. After the addition of Stop buffer, to remove any unincorporated dye, the reaction mix volume was brought up to 100 uL with nuclease-free water followed by the addition of 3 volumes of 100% nuclease-free ethanol. This 400 uL reaction volume was then applied to an RNeasy Mini Kit column (Qiagen, Valencia, CA., USA) and processed according to manufacturer’s instructions (Yeast RNA isolation protocol, starting from RW1 wash). Dye incorporation was quantitated using a Nanodrop ND-1000 (Nanodrop Technologies) by measuring emission wavelength at 570 nm (Cy3) and 670 nm (Cy5). Labeled-RNA concentration and dye incorporation were used to determine the RNA labeling specific activity (pmol/ug). Labeled and unlabeled RNA were combined to adjust the sample specific activity to the empirically determined intensities of 40 and 45 pmol/ug for Cy3 and Cy5 labeled samples, respectively. Unstressed Cy3-labeled and 1 mM H2O2 stressed Cy5-labeled RNA were combined and brought to near-dryness in a vacuum centrifuge before being resuspended in 19 uL nuclease-free water and stored at –80°C.
Hybridization to Microarrays
Microarray hybridization and scanning were preformed according to Agilent Microarray processing specifications (MO gene LLC, St Louis, MO. USA).
Microarray Analysis
The LOESS balanced data from the Feature Extraction was used to assess replication consistency across biological and process replicates using a linear ANOVA model that considers all 13,864 probes on the array and data for all the replicates, to determine the significance of differential expression [35]. yrijk = α+Ai+Gk+(AG)ik+Dl+αrijk; where yrijk is the logarithm of the rth replicate model-based expression value of gene k at the ith time point with the jth treatment (i = 1, 2, 3, 4; j = 1, 2; k = 1,…, 41, 174). This model accounts for variance in genes (G), arrays (A), and array plus gene; α term accounts for error.
Quantitative Real Time PCR
Archive RNA extracted for the microarray experiments was used to make first-strand cDNA using the First-Strand cDNA synthesis kit for reverse transcription-PCR (Roche). This cDNA was used as template for real-time PCR analysis using SsoFast SYBR green PCR reagents (Biorad) according to the manufacturer’s recommendations. A BioRad CFX96 thermal cycler was programmed with the following two step PCR cycles: 5 s at 98°C, 5 s at 60°C, and a plate read was repeated in the second step for a total of 40 cycles. A melting curve was performed at the end of the reaction to confirm a single product. Standard curves were performed for each primer set and efficiencies calculated. The data were normalized to glucose-6-phosphate dehydrogenase cDNA expression included with each experiment.
Gene Ontology Annotation
The predicted protein sequences of H99 serotype A from the Broad assembly 2 (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/Info.html) were submitted to the GOAnno program of AgBase [36]. Each protein sequence was searched against the UniProt database, restricted to fungal sequences. The e-value cutoff for a significant hit was 10−10. The sequences and annotations from the Uniprot database were filtered to remove sequences that had only annotations with an evidence code of IEA (Inferred from Electronic Annotation) or ND (No biological Data available). Additional annotation was obtained by submitting those proteins without a match in the UniProt database to InterProScan [37]. Gene ontology assignments were made for domains with matches that had a e-value of <10−20. Gene ontology term enrichment was carried out using a hypergeometic test as implemented in the GOstats program, using a p-value of ≤0.01 as a criteria for significance [38].
Ubiquitination Analysis
C. neoformans cells grown in YNB, pH 4.0 were treated with varying concentrations of H2O2 (0–4 mM) for 30 min. At the end of the incubation, cells were rapidly chilled and collected by centrifugation. Cell pellet was lysed in 8 M urea containing buffer and equal amounts of protein per sample were used for immune blot analysis using antibodies specific to ubiquitin proteins (Rabbit polyclonal to ubiquitin cat # ab19247 from abcam at 4 K dilution). Uniform transfer of proteins across the membrane was verified by staining the membrane with Swift Membrane Stain (cat # 786-677, G BioSciences, MO, USA) before subjecting them to immuno blot analysis.
Results
Cells Grown in YNB Medium Exhibited Reduced Capacity to Breakdown Exogenous H2O2
To understand the fate of exogenous H2O2 once it has been added to C. neoformans culture, actively growing yeast cells in YNB, pH 4 medium were treated with 1 mM H2O2. At different time intervals aliquot of the cell culture was removed, cells were separated and the supernatant was used to measure the residual H2O2 in the medium. A concentration of 1 mM H2O2 was rapidly degraded by growing C. neoformans cells and was completely absent in the medium after 30 minutes (Figure 1A). The incubation of H2O2 either in the medium alone or in the spent medium did not cause significant H2O2 breakdown over time (Figure 1A). Incubation of heat killed C. neoformans cells did not affect the concentration of H2O2 over time clearly suggesting that actively growing cells are responsible for degradation of external H2O2.
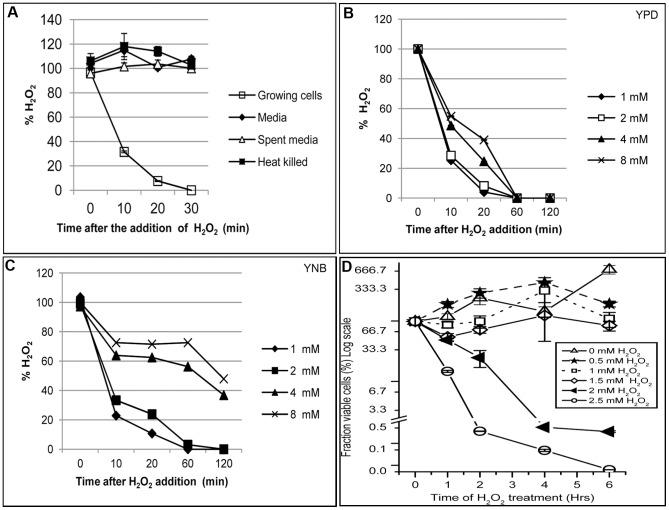
Figure 1. Kinetics of H2O2 degradation by actively growing C. neoformans cells and the effect of concentration of peroxide on fungal viability.
A: 1 mM H2O2 was added to KN99 cells growing in YNB, pH 4 at OD650 = 1.5 (□), YNB medium alone ( ), spent media (Δ) and to heat killed C. neoformans cells (▪). Various concentrations of H2O2 were added to the cells growing either in YPD (B) or in YNB (C) medium. At various time points samples were withdrawn, cells were separated by centrifugation and the supernatant was used for the quantitative estimation of H2O2. Percentage of residual H2O2 was plotted against treatment time. Error bars reflect standard error calculated from three independent experiments. D: Exponentially growing cells at OD650 = 1.5 were incubated with various concentrations of H2O2. At various time points aliquots were withdrawn, cells collected by centrifugation at 4°C, washed two times with cold PBS. Washed cells were serially diluted and plated on solid YPD media, then incubated at 30°C. After 3 days colony forming units (CFU) were counted. Fraction of viable cells was plotted against H2O2 treatment time.
C. neoformans cells are routinely grown in a nutrient rich YPD medium or a nutrient defined YNB medium for laboratory experiments. To investigate the influence of growth media on the capacity of the cells to breakdown exogenous H2O2, we treated C. neoformans cells either grown in YPD; a nutrient rich medium with a pH of 6.2–6.5 or YNB; a nutrient limited defined medium buffered to pH 4.0, at 30°C with various concentrations of H2O2. The kinetics of H2O2 breakdown was followed by measuring the residual H2O2 in the medium. When the cells were grown and treated in YPD, a concentration of H2O2 up to 8 mM was completely degraded in one hour at 30°C (Figure 1B). However, the cells grown and treated in YNB, pH 4.0, showed decreased capacity to breakdown exogenous H2O2, taking up to two hours to completely degrade even 2 mM of H2O2 (Figure 1C). Higher concentrations of H2O2 (4 and 8 mM) were decreased to ∼40–50% of the initial concentration by two hours, clearly indicating that unlike the cells grown in YPD, cells grown in nutrient limited YNB medium are much less efficient in degrading exogenous H2O2.
When the cells were grown to different densities (OD650 = 1–8) in either YPD or YNB medium and treated with various concentrations of H2O2 (2–8 mM), the rate of H2O2 breakdown increased as the culture density increased (data not shown). A cell culture grown in YPD to a density of OD650 = 2.7 was able to degrade 4 mM of H2O2 in just 20 min, while those with a density of OD650 ≥ 4.0 took only 10 minutes (Figure S1A). On the other hand cells grown in YNB, to a density of OD650 = 4.0 were unable to completely degrade 4 mM H2O2 in 2 hrs (Figure S1B). However, cell culture in YNB at a density of OD650 = 7.0 completely degraded 4 mM H2O2 in 10 minutes. Together these data indicate that capacity to degrade exogenous H2O2 depends not only on the medium but also on the density of the growing culture.
Because C. neoformans encounters oxidative stress in a nutrient limited environment inside the phagosome, we chose to use YNB, at pH 4.0 for all of our experiments. We were interested in inducing only oxidative stress response and supplemented YNB medium with 2% glucose and grew the cells at 30°C to prevent the non-specific induction of a transcriptional response due to either carbon source starvation or heat shock respectively. To determine the effect of exogenous H2O2 on the viability of C. neoformans cells, yeast cells grown in YNB, pH 4.0 at an OD650 of 1.0 were treated with various concentrations of H2O2. At various time points samples were withdrawn, cells collected by centrifugation, washed with cold PBS and serially diluted and plated onto YPD plates to count the colony forming units (CFU). A concentration of 0.5 mM H2O2 had no significant effect on cell viability at any time point (Figure 1D). Treatment of the cells with 1 mM H2O2 for one hour caused 15% cell death, but by two hours total number of viable cells had recovered to 100% of the initial cell population. Similarly, 1.5 mM H2O2 caused killing of ∼50% of cells by one hour, however by two hours, the fraction of viable cells increased to 70% of the cell population and by the four hour time point total viable cell population had reached 100%. This suggested that by two hours after addition, the majority of H2O2 had been degraded and the cell population had begun to recover from the H2O2 induced damage. Treatment of cells with 2 or 2.5 mM H2O2 caused a marked decrease in cell viability (Figure 1D) and prolonged incubation in the same medium did not increase their ability to degrade H2O2 and/or recover. From these results we concluded that sampling 1 mM H2O2 treated cells across a one-hour time-course should provide a detailed transcriptional response profile of this yeast specific to H2O2 induced oxidative stress.
C. neoformans Exhibited a Robust Transcriptional Response to Exogenous H2O2 Treatment
To analyze the transcriptional response of C. neoformans to H2O2, KN99α cells were treated with 1 mM H2O2 and samples were collected at 5, 15, 30, 45 and 60 minutes. Custom C. neoformans serotype A whole genome microarrays were used to measure the differential transcriptional response of cryptococcal cells to perturbation by 1 mM H2O2 relative to untreated cells. Statistical assessment of differential expression was obtained by fitting the balanced treated and untreated signal intensities to an analysis of variance (ANOVA) model as previously described [39]. We identified 2,930 differentially expressed (DE) probes (P<0.01) at one or more time points representing approximately 45% of the transcriptome represented on the array, indicating a very robust transcriptional response to H2O2 in C. neoformans (Tables 1 and S1).
Table 1. Differentially expressed probes at various time points during H2O2 treatment.
| 5 min | 15 min | 30 min | 45 min | 60 min | |
| Total (Differentially expressed) | 605 | 1272 | 2521 | 1289 | 835 |
| Up regulated | 264 | 661 | 1009 | 592 | 382 |
| Down regulated | 341 | 611 | 1512 | 697 | 453 |
We measured the transcript changes across a one-hour time-course and identified 605 probes differentially expressed (264 up-regulated, 341down-regulated) at the five minute time point (Table 1). At 15 minutes post-treatment, transcriptional activity increased to a total of 1272 differentially expressed probes (661 up- and 611 down-regulated). Differential expression peaked at the 30-minute sampling, to 2521 probes (1009 up- and 1512 down-regulated). Finally, at 45 and 60 minutes the number of differentially expressed probes decreased to 1289 (592 up- and 697 down-regulated) and 835 probes (382 up- and 453 down-regulated) respectively. Most extensive changes in gene expression were observed between 15 and 45 min after the initiation of the oxidative stress with a peak transcriptional response at 30 min post H2O2 treatment. The kinetics of transcriptional response (Figure 2A) is consistent with rate of H2O2 removal from the medium by actively growing yeast cells (Figure 1C).
Figure 2. Treatment of C. neoformans cells with 1 mM H2O2 elicited a robust transient transcriptional response.
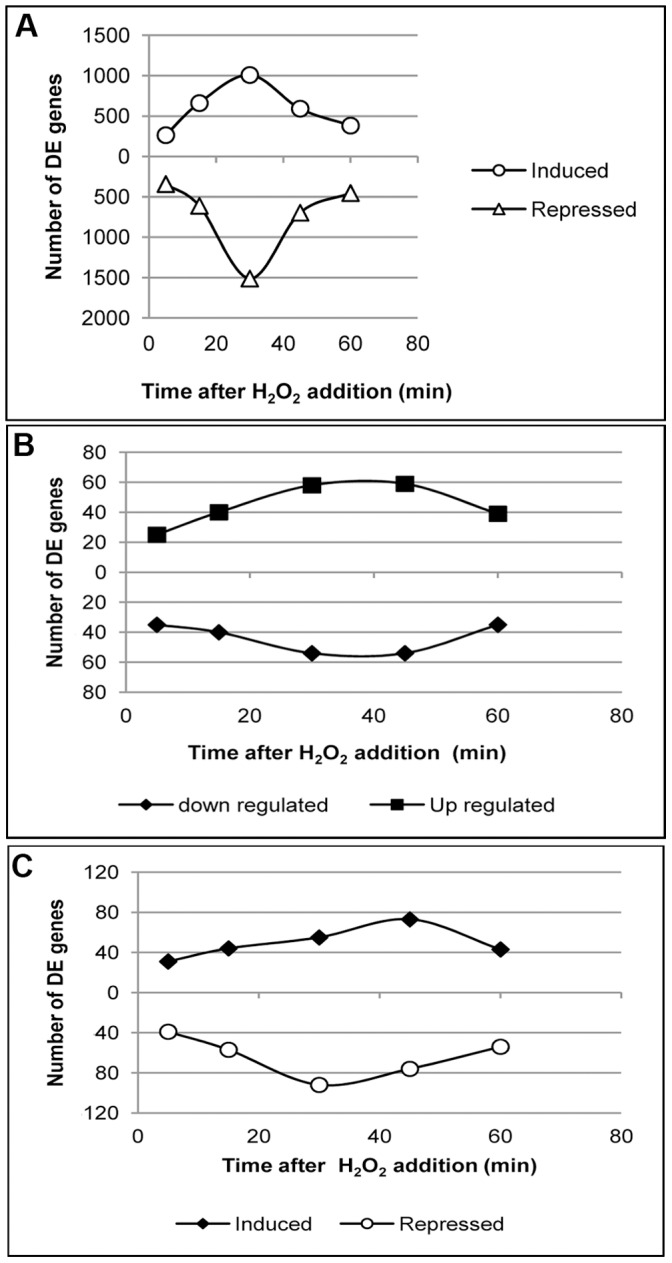
A:The number of differentially expressed (DE) genes (up-regulated above X axis and down regulated below X axis) were plotted at various time points during exposure to exogenous H2O2. B: Expression response of C. neoformans oxidation and reduction category to H2O2 treatment. C: Transcriptional pattern of differentially expressed genes belonging to metabolic process category during H2O2 stress.
We compared the sets of genes differentially expressed in response to H2O2 treatment in our experiment (treatment in YNB medium) to a previously published microarray dataset by Ko et al in which authors employed YPD medium for growth and treatment with H2O2. Consistent with our present findings on the influence of media composition and treatment conditions on the oxidative stress response we found considerable differences in the gene sets, but a high level of concordance for those genes identified as differentially expressed in both datasets [22]. We compared the identity of the DE transcripts at 30 and 60 minutes with the same two time points in KO et al., data. We observed 1582 DE transcripts compared to the 886 identified by Ko et al. There were 336 transcripts exhibiting overlap at the 30-minute time point on both arrays, with 306 (91%) of those showing the same direction in expression difference. At the 60-minute time point, we observed 836 differentially expressed transcripts compared to 1591 differentially expressed transcripts by Ko et al., with 333 genes overlapping and 276 (83%) of those showing the same direction in expression difference. The low percentage of probes that overlap at the same time point most likely reflects a combination of the differences in media conditions, the kinetics of H2O2 degradation, array probe differences, strains used and the intrinsic variation of experiments done in two different laboratories. We believe the high degree of concordance between the overlapping probes strongly validates both experimental approaches.
Quantitative Real Time PCR Experiments Confirmed the Microarray Dataset
Microarray results were validated by performing qRT-PCR analyses of six genes (CNAG_01211, CNAG_02132, CNAG_02288, CNAG_03238, CNAG_04001 and CNAG_06758) that encompassed a range of expression levels at 30 min time point. The mRNA abundance of these transcripts at 30 min after H2O2 treatment followed similar profile to microarray dataset validating the quality of our array. The linear regression analysis of the microarray and qRT-PCR measurements resulted in a correlation coefficient (R value) of 0.94, suggesting that array dataset correlated positively and closely with qRT quantification (data not shown).
A Large Number of Diverse Biological Processes are Enriched in the Transcriptional Response
Gene ontology (GO) enrichment analysis was performed and several biological processes were significantly enriched in our lists at all time points (Tables S2 and S3). As the number of differentially expressed genes increased at 30 and 45 minute time points (Figure 2A) so did the number of biological processes indicating that differentially expressed genes belong to diverse biological processes. A large number of biological processes perturbed at 30 and 45 minutes post H2O2 treatment and recovery of the viable cells at two hour time point (Figure 1D) emphasizes the potential importance and combined efforts of these biological processes in combating the H2O2 induced oxidative stress. The many GO process categories significantly affected by H2O2 induced stress at all time points include transport, metabolic process, oxidation-reduction, transmembrane transport, cellular response to stress, response to drug, transcription: DNA-dependent, ribosome biogenesis and assembly, pathogenesis, amino acid biosynthetic process, phosphorylation, protein transport, translation, filamentous growth, electron transport chain, signal transduction, cell cycle, ion transport, carbohydrate metabolic process, protein amino acid phosphorylation, fungal-type cell wall organization and biogenesis, lipid biosynthetic process, cell division and mRNA processing.
In addition to the above biological processes, a greater number of unique cellular processes significantly affected only at 30 and 45 min time points were identified (Table S2). They include response to DNA damage stimulus, proteosomal ubiquitin-dependent protein catabolic process, hyphal growth, DNA repair, RNA splicing, ER to Golgi vesicle-mediated transport, ion transport, protein folding, meiosis, chromatin modification, nuclear mRNA splicing via spliceosome, mitosis, tRNA processing, endocytosis, protein amino acid dephosphorylation, electron transport chain, small GTPase mediated signal transduction, vacuolar acidification, DNA replication, GTP catabolic process, carbohydrate metabolic process, sterol biosynthetic process, regulation of translation, and ribosomal large subunit assembly and maintenance. Identification of above GO terms clearly suggests that H2O2 causes substantial perturbation of cellular processes.
Oxidation and Reduction Processes are Over-represented in the Transcriptional Response
As expected, our dataset identified oxidation-reduction process as one of the top ten enriched categories affected by H2O2 treatment (Table S3). Moreover, the number of genes belonging to oxidation and reduction category undergoing DE increased from 60 at the 5-minute time point to 80 at the 15-minute time point. At 30 and 45 minutes post-treatment, there were 112 genes with altered RNA levels that decreased to 74 at the 60-minute time point (Figure. 2B). Some of the genes identified in this category involve those that have previously been demonstrated to be important for oxidative stress resistance either in C. neoformans or in other fungal species. These include TSA1 coding for thiol specific antioxidase (CNAG_03482) [6], TRR1 coding for thioredoxin reductase (CNAG_05847) [15], cytochrome C peroxidase (CCP1; CNAG_01138) [19] and catalase genes CAT1 (CNAG_0498) and CAT3 (CNAG_00575) [40]. These genes exhibited consistent up-regulation at more than one time point (Table S4), suggesting their prolonged involvement in combating the H2O2-induced stress and further validating the quality of our array.
A more extensive analysis of the genes belonging to the oxidation-reduction functional category revealed potential novel mechanisms of oxidative stress resistance. Functional annotation of the C. neoformans genome using our GO database assigned a total of 514 genes to the oxidation-reduction functional category. We discovered that 205 genes exhibited differential expression at one or more time point with a maximum of 124 genes showing altered mRNA levels at 45 min post H2O2 treatment (Table S4). Reciprocal BLAST search analysis against the S. cerevisiae protein database identified potential orthologs for 145 genes (Table S4). Notably, 25 genes showed consistent up-regulation in at least three time points and 25 genes exhibited consistent down-regulation at a minimum of three time points as shown in Table S5. Sustained induction or repression of a gene may reflect its predominant role in oxidative stress resistance. In support of this hypothesis, genes such as CAT1, CAT3 and TRR1, known to be important for oxidative stress either in C. neoformans or in other fungal species were found to be up-regulated at three or more time points. The C. neoformans homologs of the S. cerevisiae genes ALD5, ZTA1, SCS7 and CIR2 were also induced at a minimum of three time points (Table S5) and deletion of these genes in S. cerevisiae has demonstrated their increased sensitivity to oxidative stress [41], [42], [43].
In addition to the core antioxidant response, our microarray data revealed the contribution of several additional gene products in oxidative stress resistance that may provide clues to novel mechanisms of stress resistance. The strong transcriptional regulation of 50 genes for a significant period of time during H2O2 treatment suggests an important role for these genes during oxidative stress (Table S4). For example the gene represented by CNAG_00654 was up-regulated at all time points after H2O2 treatment and is highly similar to sulfiredoxins (SRX1). The sulfiredoxins are critical for oxidative stress resistance in yeast and higher eukaryotes [44], [45]. The majority of the remaining gene products have not been characterized either in C. neoformans or in other fungal species. A preliminary bioinformatic analysis of a few select genes supported their potential roles during oxidative stress in C. neoformans and these are discussed below.
One of the genes that are under persistent transcriptional regulation in response to peroxide stress is CNAG_03238, predicted to belong to dioxygenase subfamily. It is similar to a putative dioxygenase gene from a saprophytic soil borne filamentous bacteria, and shows significant repression at four of the five time points post H2O2 treatment. Bacterial dioxygenases are reported to have either iron-sulphur center or non-heme mononuclear iron as cofactors. Importantly, these enzymes catalyze oxidation of the substrates at the expense of reduced NADPH [46]. Decreased expression of this gene in C. neoformans after H2O2 treatment suggests that this may be a part of protective mechanism of the yeast cells to inhibit NADPH-requiring reactions while cells are coping with oxidative stress.
Another uncharacterized gene, CNAG_01542, is predicted to have domains belonging to taurine catabolism dioxygenase super-family and exhibits increased expression at 30, 45 and 60 minutes. Proteins containing these domains have been demonstrated to be important for sulfonate metabolism, in the synthesis of Fe-S cluster protein family members and their expression in S. cerevisiae provides increased resistance to menadione-induced stress [47]. A gene represented by CNAG_02580, with a conserved 2OG- Fe (II) oxygenase superfamily domain also shows increased expression at 15, 30 and 60 minutes. The 2OG- Fe (II) oxygenase domains are found in enzymes belonging to prolyl hydroxylase (PHD) family and these enzymes play a role in hypoxia and oxidative stress response in Arabidopsis and other higher eukaryotes [48]. These preliminary results suggest that systematic characterization of the genes identified in our experiment may reveal the presence of novel mechanisms of oxidative stress resistance.
C. neoformans Elicits a Distinct Pattern of Transcriptional Regulation of Metabolic Process Genes during Oxidative Stress
Among the highly perturbed biological processes, we detected expression changes in numerous genes assigned to the metabolic process category. There are 608 genes assigned to the metabolic process category in the C. neoformans genome, with 272 (∼45%) showing differential expression at one or more time points during H2O2 treatment (Table S6). Of the differentially expressed genes, 98 were common with oxidation-reduction process. The extent of metabolic process perturbation by H2O2 treatment varied at different time points during the treatment. There were 70 genes exhibiting differential expression at 5 minutes, out of which 31 showed induction and 39 exhibited repression of transcription. The number of differentially expressed genes increased to 101 at 15 minutes with 57 showing repression (Figure 2C). The negative regulation of genes related to metabolic process increased at 30 minutes with 92 genes showing decreased levels of transcripts. However, samples at the 45-minute time point exhibited a significant increase in the number of up-regulated genes, suggesting that as the concentration of H2O2 in the medium decreases, there is a shift in the metabolic flux and reprogramming of metabolic processes. Interestingly even at the 60-minute (30 minutes after the complete absence of H2O2 from the medium) a majority of the metabolic process related genes were still under transcriptional regulation (Figure 2C). Out of the 272 metabolic process related genes displaying altered transcription, we detected 33 genes exhibiting consistent up-regulation at three or more time points and 45 genes had their transcript level decreased at three or more time points after H2O2 treatment (Table S6).
A subset of genes showing distinct differential expression pattern under metabolic process category are homologous to genes involved in ergosterol metabolism and antifungal drug resistance in other fungal species. Transient differential expression of a particular set of genes (related to ergosterol metabolism and antifungal drug resistance has been reported to contribute to the complex mechanisms of anti-fungal drug resistance [49]. Over expression of ERG11 has been reported in clinical isolates of drug resistant C. albicans, implicating its role in fungal drug resistance [50]. We observed persistent up-regulation of one of the identified homologs of S. cerevisiae ERG11 after H2O2 treatment (CNAG_ 05842, ERG110, and Figure S2). In S. cerevisiae ERG5 has been reported to bind to azole drugs by a similar mechanism as ERG11 and we observed increased expression of C. neoformans ERG5 (CNAG_06644) as well. Sterol analysis of the flucanazole resistant C. albicans showed defects in ERG2 and ERG3 [51]. Homologs of S. cerevisiae ERG2 (CNAG_00854) and ERG3 (CNAG_00519) showed altered expression levels in our microarray dataset (Figure S2). In addition to the above, we have observed altered regulation of 12 C. neoformans genes upon peroxide stress that are homologs of S. cerevisiae ergosterol biosynthesis genes (Figure S2) clearly suggesting a cross-talk between the mechanisms of oxidative stress and anti fungal drug resistance. Several families of ABC transporters have been characterized as efflux pumps in S. cerevisiae and C. albicans and are important for drug resistance [52]. The over expression of genes encoding two of the PDR5 family transporters in C. albicans CDR1 and CDR2 as well as that of a major facilitator CaMDR1 is one of the signature characteristics of resistant isolates [49], [53]. The C. neoformans genome contains two homologs of CDR1 (CNAG_04098, CNAG_04966) and a homolog of MDR1 (CNAG_00796). All three genes show differential transcriptional regulation, with CDR11 showing consistent up regulation at 15, 30 and 45 minutes and MDR1 exhibiting increased levels at 30, 45 and 60 minutes (Figure S2).
Cytochrome C Peroxidase Mediated Respiratory Chain is Essential for Oxidative Stress Resistance
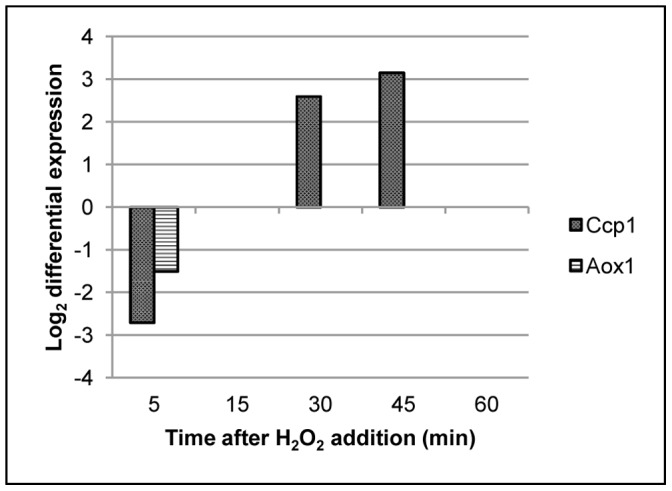
The mitochondrion is the site of most oxidation-reduction processes and we were particularly interested in the differential expression pattern of CCP1, which codes for cytochrome C peroxidase (CNAG_01138), and AOX1, encoding an alternative oxidase (CNAG_00162). C. neoformans CCP1 is known to protect against external oxidative stress inducing agents and consistent with that role, its transcription was increased at 30 and 45 minutes (Figure 3 and [19]). C. neoformans AOX1 is important for fungal pathogenesis and this phenotype has been attributed to its role in oxidative stress resistance [20]. However, transcription of AOX1 gene was not altered in our array after H2O2 treatment in the same manner as CCP1 (Figure 3). H2O2 treatment caused an initial repression of AOX1 mRNA levels with no further changes in its expression at subsequent time points. Therefore we decided to explore the potential functional consequences of the differential expression of these two genes in response to H2O2 induced oxidative stress.
Figure 3. Transcript abundance of C. neoformans CCP1 and AOX1 at various time points during H2O2 treatment.
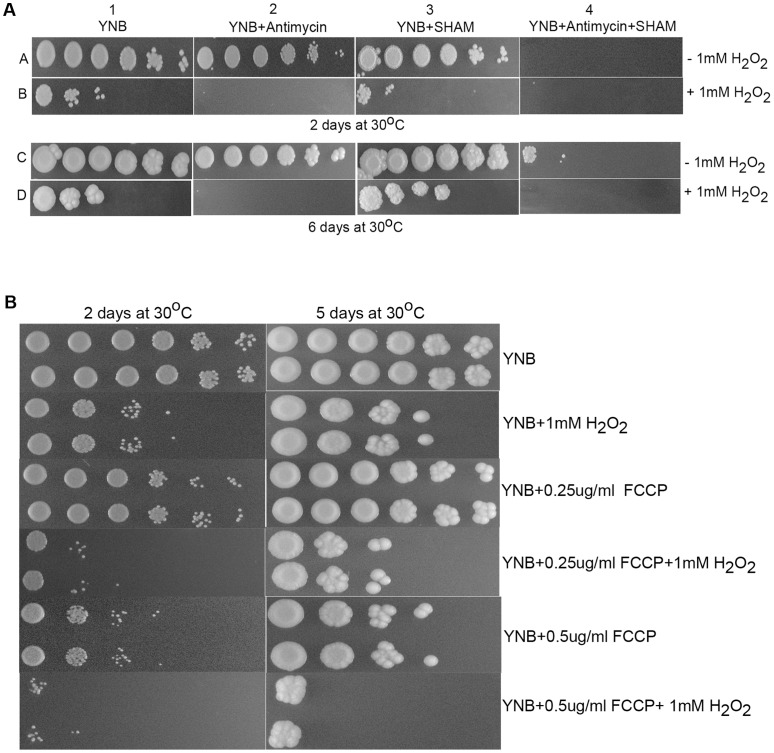
During mitochondrial respiration, C. neoformans utilizes both the classical electron transport chain, mediated by CCP1and an alternative oxidase chain mediated by AOX1 for the synthesis of ATP. These two pathways have been demonstrated to be functionally redundant in C. neoformans using specific respiratory chain inhibitors [20]. To elucidate the role of these two electron transport chains during H2O2 treatment, we treated cells with specific mitochondrial pathway inhibitors in conjunction with H2O2. The growth of KN99 cells in the presence of either antimycin, an inhibitor of the cytochrome c peroxidase pathway, or SHAM, an inhibitor of the alternate oxidase pathway, was found to be similar to that of the control plate (Figure 4A, A1–A3 and C1–C3). However, addition of both antimycin and SHAM resulted in severe impairment of growth of C. neoformans cells (Figure 4A, A4 and C4). Treatment of the cells with H2O2 in the presence of antimycin caused complete loss of growth when observed at 2 days (Figure 4A, B2) or at 6 days at 30°C (Figure 4A, D2). When cells were subjected to H2O2 stress in the presence of SHAM alone, slight growth was observed after two days (Figure 4A, B3) however, significant growth was restored after 6 days of incubation (Figure 4A, compare D1 to D3). Exposure of the cells to H2O2 in the presence of both antimycin and SHAM resulted in their complete loss of resistance to exogenous H2O2 (Figure 4A, compare panels B1 with B4 and D1 with D4). This provides evidence for a major role of CCP1 during H2O2 induced stress with a more limited role for the Aox1 protein, consistent with their differential expression pattern (Figure 3).
Figure 4. The affect of inhibitors of electron transport chain and mitochondrial function on C. neoformans oxidative stress resistance.
A: Logarithmically growing KN99 cells in YNB were 10 fold serially diluted and 5 ul of the cell suspension was spotted onto YNB agar containing either 0.5 ug/ml of antimycin (2) or 2 mM SHAM (3) or mixture of antimycin and SHAM (4) either in the absence (A and C) or in the presence (B and D) of 1 mM H2O2.B: Two independent cultures of C. neoformans cells were grown in YNB. At OD650 = 1.5, cells were collected, 10 fold serially diluted and 5 ul of the cell suspension was spotted onto solid YNB agar plates containing either 0.25 ug/ml or 0.5 ug/ml of FCCP in the presence or absence of 1 mM H2O2. Plates were incubated at 30°C for various days and photographed.
The preferential role of CCP1 during H2O2 induced oxidative stress was intriguing since the flow of electrons through the CCP1 mediated pathway generates more ROS than respiration through the AOX1 pathway [54]. It is plausible that utilization of the AOX1 mediated pathway during H2O2 induced stress may provide an additional benefit by minimizing the release of internal ROS. However, a potential advantage of using the CCP1 mediated pathway is its higher efficiency of ATP synthesis due to more proton pumping sites along the path of electron flow compared to the AOX1 mediated route. Therefore, we hypothesized that C. neoformans preferentially utilizes the CCP1 mediated electron transport chain to generate more ATP with which to carry out energy dependent processes including the repair of damage caused by H2O2 induced oxidative stress.
Functional Mitochondria are Essential for Protection Against Oxidative Stress
Additional roles for mitochondrial function during H2O2-induced oxidative stress were revealed by further analysis of the differential expression pattern of genes related to oxidation-reduction process. We identified 50 genes whose expression was either induced or repressed at a minimum of three time points during H2O2 (Table S5). Of the 50 oxidation-reduction genes altered at a minimum of three time points, 39 had potential homologs in S. cerevisiae. Out of those 39 genes, 22 are known or predicted to be located in the mitochondria (Table S5) according to their annotation in the Saccharomyces genome database [55]. The involvement of C. neoformans mitochondria during in vitro oxidative stress or while they are being phagocytosed is not well understood, though functional mitochondria are indispensable for growth inside the host [56]. The global transcriptome profile of C. neoformans at the site of central nervous system infection revealed higher expression of several respiratory genes demonstrating the importance of mitochondrial function for growth inside the host [57], [58]. Most fungal pathogens exhibit a distinct arrangement of electron transport chain compared to their host and a more flexible and complex pathways of electron flow that have not yet been fully elucidated [59]. The recent characterization of the hyper-virulent C. gatti responsible for the recent fatal fungal outbreak in Vancouver Island provides evidence for the potential role of mitochondria during oxidative stress [60]. In these studies the authors attributed the hyper-virulence to an increased intercellular proliferation rate of the fungal cells. In a phenotypic screen to identify fungal factors responsible for enhanced intracellular proliferation rate inside the host macrophage cell lines, Ma et al, identified a unique tubular mitochondrial morphology of yeast cells that positively correlated with enhanced intracellular parasitism, demonstrating a strong link between mitochondrial regulation and growth inside the macrophages [60]. Therefore we decided to explore the role of mitochondrial function in H2O2 induced oxidative stress.
To test the importance of mitochondrial function during H2O2 induced stress, we treated cells with a protonophore FCCP, known to disrupt mitochondrial function in other fungal organisms [61], [62]. C. neoformans cells were very sensitive to FCCP, with concentrations as low as 0.5 ug/ml affecting their growth in liquid culture (data not shown). By contrast, yeasts such as C. albicans and S. cerevisiae cells were able to grow normally in the presence of 2.5–5 ug/ml of FCCP [61], [62]. Because of this sensitivity, we used concentrations of 0.25 and 0.5 ug/ml of FCCP for our studies. Two independent cultures of KN99 cells were spotted on plates containing 1 mM H2O2 in the presence and absence of FCCP and incubated at 30°C. The capacity to withstand oxidative stress triggered by H2O2 challenge was markedly decreased by FCCP in a concentration dependent manner (Figure 4B), providing further support for the important role of functional mitochondria in oxidative stress resistance in C. neoformans.
Ubiquitin-dependent Protein Catabolic Genes are Sensitive to Oxidative Stress
The ubiquitin dependent proteosome pathway is one of the several cellular processes requiring ATP and is critical for maintaining cellular homeostasis. Protein ubiquitination is important during various stress conditions in yeast and higher eukaryotes and, more recently, protein ubiquitination has been shown to be important for stress resistance, adaptation and virulence of the human fungal pathogen C. albicans [63]. Moreover a previous microarray study on the oxidative stress resistance of C. neoformans found that a ubc8 deletion strain is more sensitive to H2O2 in vitro, demonstrating the importance of ubiquitin related processes for oxidative stress resistance [22].
We identified 186 genes that were assigned the ubiquitin related biological process terms protein polyubiquitinylation (GO: 0000209), deubiquitinating enzyme (GO: 0004843), ubiquitin-dependent protein catabolism (GO: 0006511), protein ubiquitinylation (GO: 0016567), and proteosomal ubiquitin-dependent protein catabolism (GO: 0043161) in the C. neoformans genome (Table S7). Of the 186 genes, 91 (49%) exhibited significant differential expression at one or more time points during H2O2 treatment (Table S7). In S. cerevisiae, proteins encoded by the ubiquitin conjugating (UBC) gene family have been implicated in a wide variety of cellular functions including those closely connected with oxidative stress resistance such as respiratory growth, glutathione homeostasis, UV and metal ion resistance and double strand DNA repair [64], [65]. Accordingly individual UBC gene deletion strains exhibit varying degree of sensitivities to diverse external stress conditions indicating the functional overlap between different UBC gene products [64], [66]. As it is likely that single ubiquitin related gene products are involved in multiple cellular stress conditions, we employed a biochemical strategy to directly address the role of ubiquitination during oxidative stress. One of the affects of H2O2 induced oxidative stress involves protein damage by irreversible oxidation and these oxidized proteins are recycled through the ubiquitin dependent proteasomal pathway in other systems [67]. To verify whether similar mechanisms operate in C. neoformans, we treated fungal cells with increasing concentration of H2O2 and subjected the cell lysates to immuno-blot analysis. The blots were probed with ubiquitin specific antibody to quantify the amount of ubiquitin tagged proteins in treated cells to their untreated counterpart. We observed an H2O2 concentration dependent increase in the amount of ubiquitin conjugated proteins in the total cell lysate (Figure 5A). Densitometric analysis of the immuno blot clearly showed direct correlation between the effective concentration of H2O2 used in the treatment and the number and intensity of ubiquitinated proteins in total cell lysate (Figure 5B).
Figure 5. H2O2 induced oxidative stress stimulates increased ubiquitination of C. neoformans proteins.
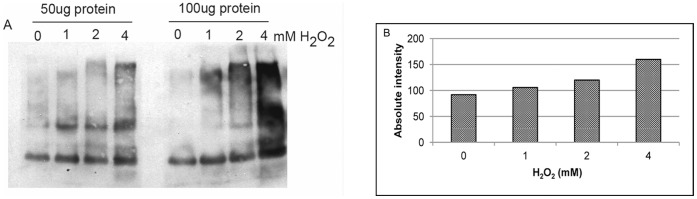
(A): Immuno blot analysis of the proteins from C. neoformans cells growing in YNB, pH 4 at OD650 = 1.5 and treated with various concentrations (0, 1, 2 and 4 mM) of H2O2. Cell lysates were prepared in urea containing denaturing buffer. Equal amount of protein was electrophoresed on SDS-polyacrylamide gel, transferred to nitrocellulose and ubiquitin conjugates were probed with polyclonal anti-ubiquitin antiserum. (B): Densitometric quantification of the signal from immunoblot A.
Discussion
The transcriptional regulation of protective mechanisms against oxidative stress is critical for C. neoformans both for establishing an infection in the host and also for surviving extended periods of time in the environment. The kinetics of H2O2 degradation by cells growing in YNB indicates that actively growing C. neoformans cells have an efficient mechanism to degrade exogenous H2O2. This may involve the activity of functional catalase enzymes reported to be present in C. neoformans [40]. The absence of H2O2 degrading activity in the culture supernatant is consistent with an earlier study reporting the absence of secreted catalase family proteins in C. neoformans genome [40]. The higher capacity of H2O2 breakdown by cells growing in YPD compared to those grown in YNB was rather surprising and clearly suggests that growth media composition may significantly affects cell’s constitutive redox potential. Moreover, we also demonstrated that cells grown to a higher optical density were able to degrade exogenous H2O2 more rapidly compared to the culture at lower optical density, again emphasizing that the cell culture and treatment conditions may significantly influence the magnitude and intensity of the induced cellular transcriptional response to oxidative stress. Therefore these important experimental parameters need to be considered when comparing different datasets generated for H2O2 mediated stress induced either in the same organism or between different organisms.
At every time point, both induced and repressed genes displayed reciprocal transient profiles with negative regulation of transcription more predominant at all 4 of the 5 time points post H2O2 treatment (Figure 2A and Table 1). It took almost 30 minutes for the cells to exhibit maximum differential expression, even though the vast majority of the H2O2 is degraded by that time. The amount of time it takes for cells to maximally respond transcriptionally may depend on the growth history of the cells prior to H2O2 challenge. During the initial stages of encounter with H2O2 cells may use their preexisting pool of antioxidant defenses to neutralize H2O2, and the concentration of cellular antioxidant molecules may likely vary depending on the environment in which cells are grown and treated. Accordingly, the duration, intensity and the complexity of the elicited transcriptional response will differ depending on the environment in which C. neoformans exist prior to H2O2 treatment. It is plausible that the antioxidant capacity of the cells in YPD is significantly higher than those grown in YNB and thus cells grown in YPD may require a higher concentration of H2O2 to exhibit similar magnitude of transcriptional response at 30 min after treatment, compared to those grown in YNB. This observed influence of growth conditions on the sensitivity of yeast cells to peroxide stress may be of special importance in C. neoformans pathogenesis since various species of Cryptococcus has been discovered to be associated with diverse ecological niches such as avian guano, vegetables, wood, dairy products, and soil [68], [69]. The composition of these environmental conditions may therefore determine the level of yeast cells’ constitutive redox potential. The ability to cause infection in a mammalian host will depend on their capacity to resist initial oxidative burst inside the macrophages. Therefore, increased resistance to oxidative stress as a result of environmental factors may play a role in enhanced virulence during infection.
In the oxidation-reduction category we found interesting gene regulation patterns of thioredoxin family genes. Among the genes of thioredoxin family, we found increased expression of TSA1 (CNAG_03482) at the 15 and 30 minute time points consistent with its established role in oxidative stress and virulence [6]. C. neoformans TSA1 is a typical 2-cys peroxiredoxin and its homolog in S. cerevisiae is located in the cytoplasm [70]. C. neoformans TSA3 (CNAG_06917) and DOT5 (CNAG_02854) belong to 1-cys peroxidase family and have shown increased expression at both protein and mRNA levels in response to oxidative stress in a previous study [6]. The homolog of C. neoformans TSA3 and DOT5 in S. cerevisiae are located to mitochondria and nucleus respectively and are known to play a major role in oxidative stress resistance [70]. The sub-cellular location of Tsa3 and Dot5 in C. neoformans is not known and their deletion in C. neoformans did not affect oxidative stress resistance in either in vitro or in vivo conditions [6]. The strong negative regulation of both TSA3 and DOT5 shown in our dataset is similar to the pattern of expression of the majority of genes of potential mitochondrial origin suggesting that both TSA3 and DOT5 may contribute additionally to oxidative stress resistance in C. neoformans by unknown mechanisms.
Three of the four known catalase genes in C. neoformans showed differential expression. Increased expression of CAT1 is consistent it being the only functional catalase as determined by the in-gel activity staining of C. neoformans cell extracts [40]. The similar profile of CAT1 and CAT3 suggests these genes may be regulated by similar factors, consistent with both belonging to the same phylogenetic clade [40]. However, computational analysis of the promoter regions of these two genes in the past has not identified any conserved cis-acting elements [40]. Interestingly both these genes show increased amounts of mRNA even at 60 minutes while by 30 minutes all the H2O2 in the medium had been degraded. This suggests that the CAT1 and CAT3 gene products may have additional novel roles during in vitro oxidative stress induced by H2O2.
In addition to their initial exposure to oxidative stress inside the macrophages, C. neoformans undergoes major metabolic adaptation during growth inside the host [57], [71]. The majority of the metabolic changes discovered during infection so far have been attributed to the task of growing in a limited nutrient environment. We observed significant differential expression of genes assigned to the metabolic process functional category, despite the presence of 2% glucose in our study (Table S6). Widespread differential expression of the genes related to metabolic processes was also observed during nitrosative stress in the presence of 2% glucose, suggesting the presence of overlapping mechanisms of oxidative and nitrosative stress protection [72]. A unique feature of the metabolic process genes induced by H2O2 involves the genes encoding pentose phosphate pathway (PPP) enzymes. A major product of PPP is the production of NADPH that is critical for the function of proteins required for repairing oxidative protein damage. Accordingly the components and function of PPP have been shown to be important for resistance and adaptation to oxidative stress in yeast and higher eukaryotes. In both S. cerevisiae and C. glabrata, H2O2 treatment increased the expression of genes belonging to the pentose phosphate pathway [73], [74]. Moreover, expression of glucose 6-phosphate dehydrogenase (ZWF1) is increased during exposure of C. albicans to nitrosative stress [75]. In S. cerevisiae, independent deletion strains of pentose phosphate pathway genes such as 6-phospho gluconate dehydrogenase (GND1), D-ribulose-5-phosphate 3-epimerase (RPE1), transketolase 1 and transketolase 2 (TKL1and TKL2), glucose-6 phosphate dehydrogenase (ZWF1) and transaldolase (TAL1) all exhibit increased sensitivity to oxidative stress [76]. Interestingly, we observed no differential expression of PPP genes, suggesting a more limited role of this metabolic pathway during oxidative stress in C. neoformans. Previous studies demonstrated that neither the ZWF1 mRNA nor protein levels were altered during nitrosative stress in C. neoformans. Consistent with this deletion of ZWF1 gene in C. neoformans did not increase their sensitivity to either oxidative or nitrosative stress [72], [77]. These are consistent with our present observations that pentose phosphate pathway does not participate in peroxide induced oxidative stress resistance in C. neoformans.
A major mechanism of adaptation to carbon source limitation during growth inside the host is increased expression of genes belonging to carbon metabolism [71]. We observed increased expression of three genes from the TCA cycle, aconitase, succinate dehdrogenase and malic enzyme (Table S6). Increased TCA cycle activity may drive the rate of electron flow through the CCP1-mediated mitochondrial electron transport chain to meet the sudden increase in demand for ATP necessary for repairing damaged proteins. Previous serial analysis of gene expression (SAGE) revealed a high abundance of tags corresponding to phopsphophenol pyruvate carboxykinase (PCK1), a main control enzyme for the regulation of gluconeogenesis in the lung-exposed cryptococcal library suggesting that gluconeogenesis is important for fungal survival in the host lung due to the limited availability of glucose [71]. Interestingly, in spite of exposing C. neoformans cells to H2O2 in the presence of glucose we also observed significant and consistent up-regulation of PCK1 (CNAG_04217) after H2O2 treatment (Table S6). The genes encoding glycolytic functions including glucose 6-phosphate isomerase (CNAG_03916) and phosphoglycerate mutase (CNAG_05892) showed decreased expression in our dataset similar to their low abundance in the lung-exposed cryptococcal SAGE library [71]. Decreased glycolysis with concomitant increase in gluconeogenesis may be critical for the regeneration of sugar phosphates that are the substrates of nucleotide biosynthesis, glycosylation and cell wall biosynthesis processes. The role of these processes during peroxide stress is supported by their identification after the GO analysis of the differentially expressed genes (Table S3). Further, the importance of C. neoformans cell wall biogenesis during oxidative stress through protein kinase C mediated cell integrity pathway has already been documented [78].
Unlike the response of the TCA cycle and the gluconeogenic pathway, genes of the glyoxylate cycle that converts acetyl-CoA to succinate for the synthesis of carbohydrates responded completely differently in our dataset compared to the previously published lung exposed cryptococcal SAGE library [71]. We observed no elevated expression of acetyl-CoA synthase (ACS1), malate synthase (MLS1), pyruvate decarboxylase (PDC1) or alcohol dehydrogenase (ADH1) during oxidative stress in contrast to their increased expression during growth conditions in the host lung. The up-regulation of the glyoxylate pathway during pulmonary infection conditions has been attributed to the higher amount of acetate present in lung tissues [71]. This is consistent with the absence of the differential expression of genes related to glyoxylate pathway in our treatment conditions and also confirms the specificity of the differential expression response we obtained to the oxidative stress induced by H2O2.
Peroxide-induced oxidative stress also caused significant perturbation of genes of amino acid biosynthesis pathway (Table S6). Increased amino acid biosynthetic gene expression has also been reported in C. neoformans during nitrosative stress [72]. It is possible that amino acid biosynthesis pathway has important overlapping functions in oxidative and nitrosative stress resistance.
In S. cerevisiae transcriptional up-regulation of CCP1 after H2O2 treatment and its role in stress signaling has been reported to contribute to oxidative stress resistance [79], [80]. While C. neoformans CCP1 was initially identified by bioinformatic analysis, the AOX1gene was identified by its significant up-regulation in C. neoformans in response to exposure to 37°C temperature [19], [20]. However, in a separate study employing SAGE during early murine pulmonary infection studies, authors identified tags for AOX1 as being enriched in the mouse lung library, but not CCP1 [71]. These results indicate that transcriptional regulation of C. neoformans CCP1 and AOX1are potentially subject to different mechanisms. Moreover, the AOX1 deleted strains exhibited a slight virulence phenotype in a mouse model while CCP1 deleted strains were avirulent. However, both CCP1 and AOX1 independent gene deletion strains exhibited in vitro oxidative stress sensitivity towards H2O2 and tert-butyl hydroperoxide respectively [20], [58]. The differential expression pattern of CCP1 and AOX1 observed in our dataset in response to peroxide stress further supports that their transcription is under the control of different signaling mechanisms. The absence of AOX1 up regulation seen in our dataset is similar to the one reported in the previously published microarray analysis [22]. Moreover transcriptional response of AOX1observed in our microarray analysis was in agreement with the results obtained in our laboratory when we subjected RNA samples to RNA seq analysis (unpublished results). The functional consequence of differential expression of CCP1 during H2O2 treatment is dramatic and is reflected in the ability of its specific inhibitor antimycin to completely abolish oxidative stress resistance to H2O2, whereas specific inhibition of AOX1 had little effect on oxidative stress resistance.
The different arrangement of fungal proteins in the electron transport chain compared to their hosts points to the special importance of mitochondrial function in pathogenesis. This is supported by recent reports showing an active role for functional mitochondria in fungal virulence and drug resistance [81]. The significant perturbation of expression of genes related to mitochondrial function in our microarray is not surprising since earlier gene expression studies in C. neoformans in the presence of various stress conditions such as nitrosative stress, heat shock and growth inside the host, all identified genes related to the mitochondria and respiratory chain to be differentially expressed [57], [72]. FCCP has previously been used to investigate mitochondrial function in other yeasts such as S. cerevisiae and C. albicans [62], [82]. The inhibition of ATP synthesis by FCCP and the subsequent increased sensitivity of yeast cells to H2O2 provides evidence that C. neoformans cells undergoes substantial damage by H2O2 and energy dependent repair mechanisms are critical for recovering from oxidative stress. The increased concentration of ubiquitin tagged proteins observed by us during H2O2 treatment (Figure 5) further supports the requirement of ATP for damage repair after oxidative stress exposure.
Permanent oxidation of proteins disrupts their structure and debilitates their function. The ubiquitin-dependent proteasomal system is part of the protein degrading mechanism that helps maintain cellular homeostasis. The covalent attachment of ubiquitin to proteins as a selection for degradation is the hallmark of the ubiquitin-dependent pathway. ATP-dependent degradation of ubiquitinated proteins is catalysed by the 26S proteasome complex that is composed of a 20S core particle and a 19S regulatory particle. The observation that exposure of C. neoformans to H2O2 causes increased ubiquitination of cellular proteins (Figure 5) is consistent with the significant induction of UBI4 (CNAG_01920). UBI4 dependent ubiquitination has been shown to play a major role in the ubiquitin dependent protein degradation pathway during thermal, cell wall and oxidative stress conditions in S. cerevisiae and C. albicans [63], [83]. We observed up-regulation of homologs of S. cerevisiae RSP5 (CNAG_05355) (Table S7). RSP5 encodes an essential E3 ubiquitin ligase in S. cerevisiae, and its transcription pattern is similar to UBI4, indicating that it may be required for ligating potential substrates with ubiquitin.
Deletion mutants of ubiquitin conjugating enzymes UBC4 and UBC5 in S. cerevisiae were exceedingly sensitive to stress conditions [66]. The increased interaction of Ubc4p with 26S proteasome has been shown in S. cerevisae upon heat stress [84]. The homolog of S. cerevisiae UBC4 in C. neoformans (CNAG_01084, Table S7) exhibits persistent expression at four time points, suggesting its major role in recognizing oxidatively damaged substrates. A slight induction of (at only one time point, Table S7) homologs of S. cerevisiae UBC6 and UBC8 in C. neoformans suggests that they may have a weaker affinity towards the oxidized protein substrates. Proteins encoded by homologs of S. cerevisiae UBC1, UBC2, UBC13, UBC5, and UBC12 all showed decreased expression, further supporting the major role of only Ubc4 enzyme in recognizing and diverting damaged proteins to proteasome pathway for their degradation during oxidative stress. C. neoformans RAD4, RAD16 and RAD7 were continuously induced for the majority of H2O2 treatment time (Table S7). S. cerevisiae Rad7p has a functional ubiquitin ligase activity and its interaction with Rad16p and Rad4p are critical for the proteasome dependent nucleotide excision repair [85]. Increased amount of C. neoformans Rad4, Rad16 and Rad7 suggests that they may be involved in the proteasomal dependent repair of DNA damage caused by H2O2 induced oxidative stress. We observed repression of the majority of genes coding for the 19S regulatory particle of the proteasome RPN1, RPN2, RPN5, RPN6, RPN7, RPN8, RPN9, RPN11 and RPN12) and RPT gene (RPT3, RPT4, RPT5 and RPT6) families. Several recent reports in yeast (S. cerevisiae) and mammalian systems indicate that degradation of oxidized proteins may occur either by ubiquitin/ATP-dependent (catalysed by 26S) or ubiquitin/ATP-independent (catalysed by 20S) mechanisms [67], [86], [87]. Moreover the 20S proteasome complex alone has been shown to possess the capacity to recognize and degrade oxidatively damaged proteins in vitro [67], [86], [87]. This was also supported by observations that oxidative stress stimulates the separation of the 19S regulatory particle from the 26S core particle to yield the 20S proteasome complex [88]. The RPN and RPT family of genes belong to the 19S proteasome complex. The repression of these genes during H2O2 treatment indicates that the 19S regulatory particle may play a minor role in maintaining cellular homeostasis during oxidative stress in C. neoformans.
By examining gene expression differences over a time course that paralleled the kinetics of H2O2 removal, we identified many more genes affected by oxidative stress than were identified in previous studies of oxidative stress in C. neoformans. The pattern of the transcriptional response mirrored the kinetics of peroxide removal and allowed us to infer potential mechanisms for the response as well as the recovery from oxidative stress. We found potential novel mechanisms for the role of mitochondria and expanded our understanding of the role of ubiquitin-dependent proteolysis in recovery from oxidative e stress.
Supporting Information
The affect culture density on H2O2 breakdown by C. neoformans cells. A 4 mM of H2O2 was added to cultures at various densities (OD650) growing in YPD (A) and YNB (B). At various time points samples were withdrawn, cells separated by centrifugation and the supernatant was used for H2O2 estimation. The percentage of residual H2O2 was plotted against H2O2 treatment time. Standard bars reflect standard error calculated from three independent experiments.
(TIF)
Differential expression pattern of C. neoformans genes related to anti-fungal drug resistance upon peroxide stress.
(TIF)
Global transcriptional profile of C. neoformans genome during H2O2 induced oxidative stress.
(XLSX)
Gene ontology enrichment analysis of the differentially expressed probes at various time points during H2O2 treatment.
(PDF)
Master list of gene ontology annotation of the differentially expressed probes at various time points.
(XLSX)
Differential expression pattern of C. neoformans genes assigned to oxidation-reduction functional category at various time points during exposure to oxidative stress.
(XLSX)
List of C. neoformans differentially expressed genes assigned to oxidation-reduction functional category that exhibited persistent induction or repression for a minimum of three time points during H2O2 induced oxidative stress.
(PDF)
Differential gene expression profile of the genes belonging to metabolic process functional category with proposed gene names.
(XLSX)
Transcript abundance at multiple time points of the genes belonging to C. neoformans ubiquitin dependent protein catabolic processes during H2O2 induced oxidative stress.
(XLSX)
Acknowledgments
We acknowledge Kimberly Gerik and Roger Herr for helpful discussions and comments on the manuscript. We thank Cathy Grisham and Fiona McCarthy for their assistance in setting up a fungal database to use with GOAnna.
Funding Statement
This work was funded by NIH NIAID R01AI050184 and NIH NHLBI R01HL088905 grants to JKL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nielsen K, De Obaldia AL, Heitman J (2007) Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell 6: 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530. [DOI] [PubMed] [Google Scholar]
- 3. Hasenberg M, Behnsen J, Krappmann S, Brakhage A, Gunzer M (2011) Phagocyte responses towards Aspergillus fumigatus. Int J Med Microbiol 301: 436–444. [DOI] [PubMed] [Google Scholar]
- 4. Del Poeta M (2004) Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot Cell 3: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seider K, Heyken A, Luttich A, Miramon P, Hube B (2010) Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr Opin Microbiol 13: 392–400. [DOI] [PubMed] [Google Scholar]
- 6. Missall TA, Pusateri ME, Lodge JK (2004) Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol Microbiol 51: 1447–1458. [DOI] [PubMed] [Google Scholar]
- 7. Feldmesser M, Tucker S, Casadevall A (2001) Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol 9: 273–278. [DOI] [PubMed] [Google Scholar]
- 8. Tucker SC, Casadevall A (2002) Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc Natl Acad Sci U S A 99: 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alvarez M, Casadevall A (2007) Cell-to-cell spread and massive vacuole formation after Cryptococcus neoformans infection of murine macrophages. BMC Immunol 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alvarez M, Casadevall A (2006) Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16: 2161–2165. [DOI] [PubMed] [Google Scholar]
- 11. Ma H, Croudace JE, Lammas DA, May RC (2007) Direct cell-to-cell spread of a pathogenic yeast. BMC Immunol 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Missall TA, Lodge JK, McEwen JE (2004) Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell 3: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wood PM, Hollomon DW (2003) A critical evaluation of the role of alternative oxidase in the performance of strobilurin and related fungicides acting at the Qo site of complex III. Pest Manag Sci 59: 499–511. [DOI] [PubMed] [Google Scholar]
- 14. Missall TA, Lodge JK (2005) Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol Microbiol 57: 847–858. [DOI] [PubMed] [Google Scholar]
- 15. Missall TA, Lodge JK (2005) Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans. Eukaryot Cell 4: 487–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Missall TA, Cherry-Harris JF, Lodge JK (2005) Two glutathione peroxidases in the fungal pathogen Cryptococcus neoformans are expressed in the presence of specific substrates. Microbiology 151: 2573–2581. [DOI] [PubMed] [Google Scholar]
- 17. Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, et al. (2003) Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun 71: 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giles SS, Batinic-Haberle I, Perfect JR, Cox GM (2005) Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot Cell 4: 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giles SS, Perfect JR, Cox GM (2005) Cytochrome c peroxidase contributes to the antioxidant defense of Cryptococcus neoformans. Fungal Genet Biol 42: 20–29. [DOI] [PubMed] [Google Scholar]
- 20. Akhter S, McDade HC, Gorlach JM, Heinrich G, Cox GM, et al. (2003) Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect Immun 71: 5794–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan W, Kraus PR, Boily MJ, Heitman J (2005) Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 4: 1420–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ko YJ, Yu YM, Kim GB, Lee GW, Maeng PJ, et al. (2009) Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 8: 1197–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen D, Toone WM, Mata J, Lyne R, Burns G, et al. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 14: 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ikner A, Shiozaki K (2005) Yeast signaling pathways in the oxidative stress response. Mutat Res 569: 13–27. [DOI] [PubMed] [Google Scholar]
- 26. Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, et al. (2006) Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 40: 1201–1209. [DOI] [PubMed] [Google Scholar]
- 27. Vivancos AP, Jara M, Zuin A, Sanso M, Hidalgo E (2006) Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Genet Genomics 276: 495–502. [DOI] [PubMed] [Google Scholar]
- 28. Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, et al. (2008) Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol Biol Cell 19: 865–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuellar-Cruz M, Briones-Martin-del-Campo M, Canas-Villamar I, Montalvo-Arredondo J, Riego-Ruiz L, et al. (2008) High resistance to oxidative stress in the fungal pathogen Candida glabrata is mediated by a single catalase, Cta1p, and is controlled by the transcription factors Yap1p, Skn7p, Msn2p, and Msn4p. Eukaryot Cell 7: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chauhan N, Inglis D, Roman E, Pla J, Li D, et al. (2003) Candida albicans response regulator gene SSK1 regulates a subset of genes whose functions are associated with cell wall biosynthesis and adaptation to oxidative stress. Eukaryot Cell 2: 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Enjalbert B, MacCallum DM, Odds FC, Brown AJ (2007) Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun 75: 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim SH, Kim B, Yadavalli VK, Pishko MV (2005) Encapsulation of enzymes within polymer spheres to create optical nanosensors for oxidative stress. Anal Chem 77: 6828–6833. [DOI] [PubMed] [Google Scholar]
- 33. Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, et al. (2012) Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med 52: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yates RM, Hermetter A, Russell DG (2005) The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic 6: 413–420. [DOI] [PubMed] [Google Scholar]
- 35. Cui X, Churchill GA (2003) Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, et al. (2006) AgBase: a functional genomics resource for agriculture. BMC Genomics 7: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Falcon S, Gentleman R (2007) Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258. [DOI] [PubMed] [Google Scholar]
- 39. Kerr MK, Martin M, Churchill GA (2000) Analysis of variance for gene expression microarray data. J Comput Biol 7: 819–837. [DOI] [PubMed] [Google Scholar]
- 40. Giles SS, Stajich JE, Nichols C, Gerrald QD, Alspaugh JA, et al. (2006) The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot Cell 5: 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lopes J, Pinto MJ, Rodrigues A, Vasconcelos F, Oliveira R (2010) The Saccharomyces cerevisiae Genes, AIM45, YGR207c/CIR1 and YOR356w/CIR2, Are Involved in Cellular Redox State Under Stress Conditions. Open Microbiol J 4: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM (2005) A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol 1: 2005 0001. [DOI] [PMC free article] [PubMed]
- 43. Fernandez MR, Porte S, Crosas E, Barbera N, Farres J, et al. (2007) Human and yeast zeta-crystallins bind AU-rich elements in RNA. Cell Mol Life Sci 64: 1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Biteau B, Labarre J, Toledano MB (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984. [DOI] [PubMed] [Google Scholar]
- 45. Soriano FX BP, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE (2009) Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Molecules and Cells 27: 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harayama S, Kok M, Neidle EL (1992) Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol 46: 565–601. [DOI] [PubMed] [Google Scholar]
- 47. Honjoh K, Matsuura K, Machida T, Nishi K, Nakao M, et al. (2010) Enhancement of menadione stress tolerance in yeast by accumulation of hypotaurine and taurine: co-expression of cDNA clones, from Cyprinus carpio, for cysteine dioxygenase and cysteine sulfinate decarboxylase in Saccharomyces cerevisiae. Amino Acids 38: 1173–1183. [DOI] [PubMed] [Google Scholar]
- 48. Hieta R, Myllyharju J (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J Biol Chem 277: 23965–23971. [DOI] [PubMed] [Google Scholar]
- 49. White TC, Marr KA, Bowden RA (1998) Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11: 382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. White TC (1997) Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41: 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Venkateswarlu K, Taylor M, Manning NJ, Rinaldi MG, Kelly SL (1997) Fluconazole tolerance in clinical isolates of Cryptococcus neoformans. Antimicrob Agents Chemother 41: 748–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, et al. (2009) Efflux-mediated antifungal drug resistance. Clin Microbiol Rev 22: 291–321, Table of Contents. [DOI] [PMC free article] [PubMed]
- 53. Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, et al. (1995) Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother 39: 2378–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turrens JF, Alexandre A, Lehninger AL (1985) Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch Biochem Biophys 237: 408–414. [DOI] [PubMed] [Google Scholar]
- 55. Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40: D700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Toffaletti DL, Nielsen K, Dietrich F, Heitman J, Perfect JR (2004) Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr Genet 46: 193–204. [DOI] [PubMed] [Google Scholar]
- 57. Steen BR, Zuyderduyn S, Toffaletti DL, Marra M, Jones SJ, et al. (2003) Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot Cell 2: 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toffaletti DL, Del Poeta M, Rude TH, Dietrich F, Perfect JR (2003) Regulation of cytochrome c oxidase subunit 1 (COX1) expression in Cryptococcus neoformans by temperature and host environment. Microbiology 149: 1041–1049. [DOI] [PubMed] [Google Scholar]
- 59. Guerrero-Castillo S, Araiza-Olivera D, Cabrera-Orefice A, Espinasa-Jaramillo J, Gutierrez-Aguilar M, et al. (2011) Physiological uncoupling of mitochondrial oxidative phosphorylation. Studies in different yeast species. J Bioenerg Biomembr 43: 323–331. [DOI] [PubMed] [Google Scholar]
- 60. Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, et al. (2009) The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A 106: 12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jin JH, Seyfang A (2003) High-affinity myo-inositol transport in Candida albicans: substrate specificity and pharmacology. Microbiology 149: 3371–3381. [DOI] [PubMed] [Google Scholar]
- 62. Demasi AP, Pereira GA, Netto LE (2006) Yeast oxidative stress response. Influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. FEBS J 273: 805–816. [DOI] [PubMed] [Google Scholar]
- 63. Leach MD, Stead DA, Argo E, MacCallum DM, Brown AJ (2011) Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol 79: 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hiraishi H, Mochizuki M, Takagi H (2006) Enhancement of stress tolerance in Saccharomyces cerevisiae by overexpression of ubiquitin ligase Rsp5 and ubiquitin-conjugating enzymes. Biosci Biotechnol Biochem 70: 2762–2765. [DOI] [PubMed] [Google Scholar]
- 65. Hwang GW, Furuchi T, Naganuma A (2008) The ubiquitin-conjugating enzymes, Ubc4 and Cdc34, mediate cadmium resistance in budding yeast through different mechanisms. Life Sci 82: 1182–1185. [DOI] [PubMed] [Google Scholar]
- 66. Seufert W, Jentsch S (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jung T, Grune T (2008) The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 60: 743–752. [DOI] [PubMed] [Google Scholar]
- 68. Sorrell TC, Ellis DH (1997) Ecology of Cryptococcus neoformans. Rev Iberoam Micol 14: 42–43. [PubMed] [Google Scholar]
- 69. Mak S, Klinkenberg B, Bartlett K, Fyfe M (2010) Ecological niche modeling of Cryptococcus gattii in British Columbia, Canada. Environ Health Perspect 118: 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morgan BA, Veal EA (2007) Functions of typical 2-Cys peroxiredoxins in yeast. Subcell Biochem 44: 253–265. [DOI] [PubMed] [Google Scholar]
- 71. Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW (2008) Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol 69: 1456–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Missall TA, Pusateri ME, Donlin MJ, Chambers KT, Corbett JA, et al. (2006) Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: implications for virulence. Eukaryot Cell 5: 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, et al. (1998) The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem 273: 22480–22489. [DOI] [PubMed] [Google Scholar]
- 74. Roetzer A, Klopf E, Gratz N, Marcet-Houben M, Hiller E, et al. (2011) Regulation of Candida glabrata oxidative stress resistance is adapted to host environment. FEBS Lett 585: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hromatka BS, Noble SM, Johnson AD (2005) Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell 16: 4814–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Juhnke H, Krems B, Kotter P, Entian KD (1996) Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet 252: 456–464. [DOI] [PubMed] [Google Scholar]
- 77. Brown SM, Upadhya R, Shoemaker JD, Lodge JK (2010) Isocitrate dehydrogenase is important for nitrosative stress resistance in Cryptococcus neoformans, but oxidative stress resistance is not dependent on glucose-6-phosphate dehydrogenase. Eukaryot Cell 9: 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK (2008) PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell 7: 1685–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kwon M, Chong S, Han S, Kim K (2003) Oxidative stresses elevate the expression of cytochrome c peroxidase in Saccharomyces cerevisiae. Biochim Biophys Acta 1623: 1–5. [DOI] [PubMed] [Google Scholar]
- 80. Charizanis C, Juhnke H, Krems B, Entian KD (1999) The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7). Mol Gen Genet 262: 437–447. [DOI] [PubMed] [Google Scholar]
- 81. Shingu-Vazquez M, Traven A (2011) Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell 10: 1376–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J (2009) The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology 155: 413–423. [DOI] [PubMed] [Google Scholar]
- 83. Finley D, Ozkaynak E, Varshavsky A (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48: 1035–1046. [DOI] [PubMed] [Google Scholar]
- 84. Tongaonkar P, Chen L, Lambertson D, Ko B, Madura K (2000) Evidence for an interaction between ubiquitin-conjugating enzymes and the 26S proteasome. Mol Cell Biol 20: 4691–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Reed SH, Gillette TG (2007) Nucleotide excision repair and the ubiquitin proteasome pathway–do all roads lead to Rome? DNA Repair (Amst) 6: 149–156. [DOI] [PubMed] [Google Scholar]
- 86. Davies KJ (2001) Degradation of oxidized proteins by the 20S proteasome. Biochimie 83: 301–310. [DOI] [PubMed] [Google Scholar]
- 87. Breusing N, Grune T (2008) Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem 389: 203–209. [DOI] [PubMed] [Google Scholar]
- 88. Wang X, Yen J, Kaiser P, Huang L (2010) Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3: ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The affect culture density on H2O2 breakdown by C. neoformans cells. A 4 mM of H2O2 was added to cultures at various densities (OD650) growing in YPD (A) and YNB (B). At various time points samples were withdrawn, cells separated by centrifugation and the supernatant was used for H2O2 estimation. The percentage of residual H2O2 was plotted against H2O2 treatment time. Standard bars reflect standard error calculated from three independent experiments.
(TIF)
Differential expression pattern of C. neoformans genes related to anti-fungal drug resistance upon peroxide stress.
(TIF)
Global transcriptional profile of C. neoformans genome during H2O2 induced oxidative stress.
(XLSX)
Gene ontology enrichment analysis of the differentially expressed probes at various time points during H2O2 treatment.
(PDF)
Master list of gene ontology annotation of the differentially expressed probes at various time points.
(XLSX)
Differential expression pattern of C. neoformans genes assigned to oxidation-reduction functional category at various time points during exposure to oxidative stress.
(XLSX)
List of C. neoformans differentially expressed genes assigned to oxidation-reduction functional category that exhibited persistent induction or repression for a minimum of three time points during H2O2 induced oxidative stress.
(PDF)
Differential gene expression profile of the genes belonging to metabolic process functional category with proposed gene names.
(XLSX)
Transcript abundance at multiple time points of the genes belonging to C. neoformans ubiquitin dependent protein catabolic processes during H2O2 induced oxidative stress.
(XLSX)