Abstract
Piscirickettsia salmonis is a fish bacterial pathogen that has severely challenged the sustainability of the Chilean salmon industry since its appearance in 1989. As this Gram-negative bacterium has been poorly characterized, relevant aspects of its life cycle, virulence and pathogenesis must be identified in order to properly design prophylactic procedures. This report provides evidence of the functional presence in P. salmonis of four genes homologous to those described for Dot/Icm Type IV Secretion Systems. The Dot/Icm System, the major virulence mechanism of phylogenetically related pathogens Legionella pneumophila and Coxiella burnetii, is responsible for their intracellular survival and multiplication, conditions that may also apply to P. salmonis. Our results demonstrate that the four P. salmonis dot/icm homologues (dotB, dotA, icmK and icmE) are expressed both during in vitro tissue culture cells infection and growing in cell-free media, suggestive of their putative constitutive expression. Additionally, as it happens in other referential bacterial systems, temporal acidification of cell-free media results in over expression of all four P. salmonis genes, a well-known strategy by which SSTIV-containing bacteria inhibit phagosome-lysosome fusion to survive. These findings are very important to understand the virulence mechanisms of P. salmonis in order to design new prophylactic alternatives to control the disease.
Introduction
Piscirickettsia salmonis is an aggressive facultative intracellular bacterium that severely threatens the sustainability of the salmon industry in Chile. The bacterium is known to be the etiological agent of Salmonid Rickettsial Septicaemia (SRS) or Piscirickettsiosis [1], a disease that produces systemic infection characterized by colonization of several organs including kidney, liver, spleen, intestine, brain, ovary and gills [1], [2]. Since it was first reported in Chile in 1989 [3], SRS has been reported in other latitudes but with significantly less impact [4], [5]. P. salmonis belongs to the group of Gamma proteobacteria [6], [7] and to date has been described as non-motile, non-encapsulated, pleomorphic but generally cocoid, with a diameter from 0.1 to 1.5 µm [8], [9], [10]. The bacterium was first described as an obligate intracellular pathogen and today is able to grow in cell-free media [11], [12], [13], [14]. P. salmonis productively infect salmonids and other economically important fish species [15], [16], [17]. This also confirms that the expected spread of the agent to other commercially relevant fish species has already started and is a clear indication of its potential threat [18]. Despite the high impact P. salmonis has had on the aquaculture industry, key aspects of its biology, pathogenesis and virulence are completely unknown, a situation that has significantly hampered control [19].
It was recently reported that P. salmonis infects macrophages by multiplying inside replicative vacuoles [10], [20], [21] and also induces apoptosis via caspase-3 activation in these cells [22]. Macrophage infection is a common strategy adopted by several intracellular pathogens to colonize and systemically spread into their hosts by using different molecular effectors to interfere with normal cell signaling and thus disable normal responses triggered to control and eliminate foreign invaders. One of the most important of these responses is lysosome-mediated degradation, which if impeded, allows the invader to succeed in its infection [23], [24]. After infection, phagosome acidification is an initial cell response that is essential to avoiding intracellular multiplication of many pathogenic organisms including Mycobacterium, Chlamydia spp., L. pneumophila, C. burnetii, and B.suis [25], [26]. The bacteria in turn react by isolating themselves in vacuoles inside the infected cell to avoid fusion of the acidified phagosome with a degrading lysosome [27]. In order to do that the bacteria inside the phagosome express different effectors molecules coded by different secretion system genes that promotes their replication [28].
In pathogenic bacteria, most of the seven secretion systems (SSs) described are conserved machineries involved in the secretion of virulence effectors as well as other molecules that tend to undermine key host-cell functions for allowing the pathogen to establish permissive niches to survive [29]. One of the most significant undermining targets of SSs is to impede phagosome-lysosome fusion. This distinctive feature is particularly enhanced by the different variants of Type IV Secretion Systems (TIVSSs). TIVSSs are one of many mechanisms widely used by various intracellular and non-intracellular pathogens [30], [31], [32]. In addition, TIVSSs are highly versatile as they are able to secrete not only proteins, such as the Helicobacter pylori Cag-type IVsecretion system [33], [34], but also DNA, as reported for Agrobacterium tumefaciens VirB/D4 system. Legionella pneumophila and Coxiella burnetii, two important human pathogens, use a particular TIVSS named Dot/Icm (Deficient in Organelle Trafficking/Intracellular Multiplication) to establish productive infections [35], [36]. The Dot/Icm system comprises around 20 different proteins including DotB and DotA as the most significant ones [35].The mimicking of Legionella pneumophila's system could represent one of the important components of the bacterial virulence for P. salmonis where the macrophage infections are preferred target for the multiplication of replicative vacuoles [37]. Furthermore, the L. pneumophila Dot/Icm system is also involved in phagocytosis, cytotoxicity, apoptosis and also in inhibition of phagosome-lysosome fusion which leads to the formation of a novel ribosome-lined phagosome [38], all of these may likewise be expected to occur in P. salmonis. We focus our attention on the experimental determination of whether P. salmonis possesses the Dot/Icm secretion system or an equivalent one for achieving productive infection, since P. salmonis is phylogenetically related to L. pneumophila and C. burnetti, assuming as well that they may share similar pathways to infect susceptible host cells. We have found four dot/icm homologues in the genome of P. salmonis by using PCR-based techniques. These putative P. salmonis dot/icm genes are all transcriptionally active in both tissue culture infected cells and cell-free media. Finally, it has also been demonstrated that the key phagosome-lysosome fusion event is hampered by P. salmonis, suggesting that the Dot/Icm system could participate in this process, supporting the relevancy of this secretion system in the potential infection of this novel pathogen.
Experimental Procedures
P. salmonis Growth Conditions
P. salmonis strain LF-89 (ATCC VR-1361) was routinely grown on sheep blood agar plates supplemented with 0.1% L-cysteine and 1% glucose [11], at 23C°. For liquid cultures a single colony of P. salmonis was used to inoculate 5 ml of MC1 broth [39], incubating the culture at 23°C with agitation of 100 rpm.
Degenerate Primer Design
In order to determine the presence of dot/icm genes within the P. salmonis genome, a set of PCR degenerate primers were designed by comparative analysis using 3 dot/icm genes (dotB, dotA and icmK) from phylogenetically related organisms such as L. pneumophila and C. burnetii. The retrieved sequences were aligned using CLUSTALW (http://www.ebi.ac.uk/clustalw/) [40] and primer properties were validated with the “Oligo Calculator” tool (http://www.basic.northwestern.edu/bio-tools/oligocalc.html). The sequences used in this study were the following: L. pneumophila icmK (Gen Bank: AAU26547.1), C. burnetii icmK (ABS77467.2), L. pneumophila dotB (AAU28734.1), C. burnetii dotB (ABX77916.1), L. pneumophila dotA (AAA79902.1) and C. burnetii dotA (YP_002306135.1). Primer sequences are shown in Table 1.
Table 1. Degenerate primers used for the initial amplification of P. salmonis dotB, dotA and icmK genes.
| Primer | Sequence | gene |
| DotB-F1 | 5'-GCK TCA GAT ATW ACW ATY CAA AC-3' | dotB |
| DotB-R1 | 5′-TGT TTC AAK AAT RTC GAT RGT-3′ | dotB |
| IcmK-F1 | 5'-ATC GCC GAA AAR MGH RTT CCH CAR-3' | icmK |
| IcmK-R2 | 5′- GWT GAY ARS ACC ARR TGV CC-3′ | icmK |
| DotA-F3 | 5′-GAY CCR AAR ACY GWW GAA ATY-3′ | dotA |
| DotA-R4 | 5′-GGT CMG GSC GCA TWC KSA G-3′ | dotA |
Amplification and Sequencing Dot/icm Genes
For PCR analysis, P. salmonis DNA was extracted from 10 ml MC1 bacterial culture using the AxyPrep™ Multisource Genomic DNA Miniprep Kit (AxyGen Biosciences) in accordance with manufacturer’s instructions. The amplification of all target genes was done with GoTaq Flexi DNA Polymerase (Promega) in a 35-cycle PCR program, using the annealing the corresponding temperature for each primer set. PCR products were visualized on 1% agarose gels stained with GelRed™ (Biotium) and purified with the EZNA Gel Extraction Kit (Omega Biotek) in accordance with manufacturer’s instructions. Purified fragments were cloned into TOPO TA Cloning Kit (Invitrogen) and submitted for sequencing at Macrogen Inc, Korea. DNA sequences were analyzed with BLASTN and BLASTX software (http://blast.ncbi.nlm.nih.gov), limiting the query to bacterial sequences to determine their possible identities. Finally, P. salmonis sequences were aligned with analogous sequences obtained from BLAST analysis using CLUSTALW [40] and the alignments were processed using JALVIEW software [41]. Due to the L. pneumophila DotA has been reported as a transmembrane protein, an additional analysis using the SOSUI software (http://bp.nuap.nagoya-u.ac.jp/sosui/) was made for the putative P. salmonis DotA protein in order to determine transmembrane and hydrophobic domains.
Once the identities of the new P. salmonis sequences were obtained, specific primers against the P. salmonis dotB gene were designed for Long Range PCR (LR-PCR) purposes, in order to obtain a broader dot/icm gene array sequence. LR-PCR was performed using the combinations of the new specific dotB primers and the ITS primers (RTS1 and RTS4) [42],as described below: DotB-Forward/RTS1 (ITS Forward), DotB-Forward/RTS4 (ITS Reverse) and DotB-Reverse/RTS1 (ITS Forward). The PCR reactions were carried out with the Pfu Ultra™ high-fidelity DNA Polymerase (Stratagene) under the following conditions: 92°C for 5 minutes followed by 30 cycles of: 92° for 10 seconds, 51°C for 30 seconds and 68°C for 12 minutes. Amplification products were visualized in 1% agarose gel stained with GelRed™. The highest PCR product was selected for sequencing by creating a genome library. The amplicon was digested with the enzyme Sau3AI (New England Biolabs) for 15 minutes at 37°C and the digestion was then ligated to pBluescript SK+ vector (Fermentas) previously linearized with the enzyme BamHI (Promega) and treated with Alkaline Phosphatase (Promega). The ligation reaction was incubated at 22°C for 4 hours in the presence of T4 DNA Ligase (Promega) and the resultant product used to transform chemically-competent E. coli TOP10 cells (Invitrogen). Cells were plated on Luria-Bertani (LB) agar supplemented with Ampicillin 100 µg/ml and X-Gal (Promega) and incubated at 37°C overnight. Positive clones were submitted for sequencing at Macrogen Inc., Korea. The sequences were analyzed as described above.
P. salmonis Infection Kinetics in Two Cell Lines
In order to determine the transcriptional activity of the putative P. salmonis dot/icm genes during the host infection process, an infection kinetic assay was performed using qRT-PCR for two different cell lines of distinct origin.
The RTS11 cell line (Kindly donated by Dr. Niels Bols, University of Waterloo, Canada) was used as immune cell model since it presents two different cell morphologies: small, round, and non-adherent cells are monocyte-like and large adherent cells with typical macrophage morphology [43]. RTS11 cells were cultured at 20°C in cells in two 25 cm2 flasks with Leibovitz’s L-15 medium (Gibco) supplemented with 15% FBS (Gibco).
For a highly proliferative intracellular environment, it has been used the Sf21 insect cell line (Invitrogen) since it has been described for producing high titers of P. salmonis at 15 days post-infection [44]. Sf21 cells were maintained at 20°C in 25 cm2 flasks in Grace’s Insect Culture Medium (Biological Industries) supplemented with 20% FBS (Gibco).
Both cell lines kinetic infection were measured at earlier stages of the infection (24, 48 and 72 hours). A single colony of P. salmonis grown in BCG plates was used to inoculate 3 ml of MC1 medium, incubated at 23°C and 100 rpm until reaching an OD600 of 0.6 (12–16 hours approximately). Then, 200 µl of P. salmonis medium was used to infect each cell line using one cell flask for every kinetic time point including also a biological duplicate. For the RNA extraction the cells were scraped from the flask, centrifuged at 300 g for 10 minutes. Finally the cellular pellet was processed with Trizol® Reagent (Invitrogen) in accordance to the manufacturer’s instructions. The RNA concentration was measured with a Nanodrop-1000 spectrophotometer and kept al −80°C until use.
P. salmonis Growth Kinetics in Liquid Medium at Acidic pH Levels
The kinetic growth curves were measured for establishing the effect of the acidic pH levels over the transcriptional profile from the P. salmonis dot/icm genes. For this purpose, 3 ml of MC1 medium was inoculated with a single colony of P. salmonis grown in BCG plates, incubating the culture for 16 hours at 23°C and 100 rpm of agitation. Subsequently, this culture was used to inoculate 60 ml of MC1 broth and incubated for 24 hours under the same conditions described above. Six replicas of 50 ml each were started using 5 ml of the previous P. salmonis culture as inoculum, incubated at 23°C and 100 rpm. After 12 hours post-incubation the cultures were centrifuged at 6000 rpm for 20 minutes and resuspended in 50 ml MC1 at pH 4.0, 5.5 and 7.0, considering two 2 replica for each condition, and then incubated at 23°C and 100 rpm as usual. Samples from each culture were taken at 2, 4, 6 and 12 hours, centrifuging them at 6000 rpm for 20 minutes and processing the resultant pellets with the Trizol® Reagent (Invitrogen). The RNA was quantified in a Nanodrop-1000 spectrophotometer and kept at −80°C until use.
QRT-PCR of P. salmonis Dot/icm Genes
In order to quantify and compare the expression levels of the putative dot/icm genes under both cell lines used and growth in MC1 at different pH levels, relative quantification by qRT-PCR was made using as reference gene (housekeeping gene) the ITS (16 S–23 S internal transcribed spacer) for the normalization.
To proceed with the quantification, 2 µg RNA of each sample was pre-treated with 2 units of DNAse RQ1 (Promega) and incubatedat 37°C for 30 min in order to eliminate all putative DNA contamination. Subsequently, the DNAse treated samples were used for cDNA synthesis with M-MLV Reverse transcriptase (Promega) according to the manufacturer’s instructions in the presence of random primers (Promega). The qRT-PCR was performed using 20 µl reaction mixtures containing: 1X of Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Stratagene), 300 nM of each primer (see table 2) and 1 µl of template (standard curve or P. salmonis cDNA). As a host cell control, the elongation factor 1-alpha (EF1A) gene was used to normalize the qPCR and the RNA amount of each kinetic point. Samples were amplified and detected in a CFX96 Real Time PCR System (Biorad) using the following cycle profile for the dot/icm genes: 95°C for 3 minutes for initial denaturation; 95°C for 15 seconds followed by 40 cycles of: 58°C for 15 seconds and 60°C for 20 seconds. The primers RTS1 (5′-TGA TTT TAT TGT TTA GTG AGA ATG A-3′) and RTS4 (5′-ATG CAC TTA TTC ACT TGA TCA TA-3′) were used for ITS amplification with a cycling profile of: 95°C for 3 minutes for initial denaturation and 40 cycles of: 95°C for 15 seconds, 51°C for 15 seconds and 60°C for 20 seconds. Primers EF1A-For (5′-GTC TAC AAA ATC GGC GGT AT-3′) and EF1A-Rev (5′-CTT GAC GGA CAC GTT CTT GA-3′) were used for theEF1A amplification as described previously [21], using a cycling profile of: 95°C for 3 minutes for initial denaturation and 40 cycles of: 95°C for 15 seconds, 56°C for 15 seconds and 60°C for 20 seconds. In all cases, following the final cycle, melting curve analysis was performed to determine the specificity in each reaction tube (absence of primer dimers and other non-specific products) by heating the samples from 65 to 95°C in 0.5°C increments with a dwell time at each temperature of 5 seconds while continuously monitoring fluorescence. All Real-time PCR were assayed on every biological replicate and each sample was run in duplicate. Additionally, as a negative control qPCR reaction tube of DNAse treated RNA was run for all the experiments in order to confirm that samples were free of DNA contamination.
Table 2. Specific primers for P. salmonis dot/icm genes used for qRT-PCR.
| Primer | Sequence | gene |
| Ps-DotB-For | 5′-GCT ACA TCT CCA TTT CTT GAC CAT TTC-3 | dotB |
| Ps-DotB-Rev3 | 5'- GCA TTA GTG CCG AGC ATT ACA GG-3' | dotB |
| PS-IcmK-F | 5′-GCG CCA GAG CAG ATA CAT CAG TAT AAA G-3′ | icmK |
| PS-IcmK-R1 | 5′-GCC ACC GGA ACA TCT AAG CCT TTT AA-3′ | icmK |
| Ps-IcmE-For1 | 5′-GCC TTG GTT AAG TGT GAC CGT TG-3′ | icmE |
| PS-IcmE-Rev1 | 5′-GCT GTC ATT ACC TGC ATT AGA TCA TAG-3′ | icmE |
| Ps-DotA-For | 5′-GCT TAT GTC GCC ATT TCT GCA GCA CTT C-3′ | dotA |
| Ps-DotA-Rev2 | 5′-CCA CTC ACT CGG CAA ATT AAG CAG-3′ | dotA |
For all cases the real-time PCR efficiencies were calculated from the slope according to the established equation E = 10 (−1/slope) [45]. The threshold cycle (Ct) values of the CFX Manager Software (Biorad) were transformed to relative quantities as described by Peña et al [21].
For the conversion of the Ct values to relative quantities, reaction efficiencies were used. Relative gene expression for dotB, icmE, icmK and dotA were calculated using the values obtained to ITS of each assay as normalization factors. For the expression during infection kinetics the 24 hours post-infection was used as calibrator in both cell lines and for expression in MC1 at different pH the 2 hours of incubation at pH 7.0 was used as calibrator for all genes. Finally, a Mann-Whitney test was used to determine significant differences in gene expression between the biological and experimental replicates and significance was set at P<0.05.
In order to corroborate our results an additional analysis was made for relative quantification using the 2−ΔΔCt method [46]. The ITS data were used for normalization in all cases and the calibrator were the same used in the previous analyses.
Determination of Phagosome-lysosome Fusion
To determine that P. salmonis is able to fuse with lysosomes after bacterial phagocytosis, we performed immunofluorescence staining with three different infected-cell lines (RTS11, Sf21 and CHSE-214). RTS11 and Sf21 cells were cultured as described above and CHSE-214 cells (ATCC CRL 1681) were maintained in MEM medium (Gibco) supplemented with 15 mM HEPES, 10 mM sodium bicarbonate, and 10% FBS (Gibco) at 17°C [9].
For immunofluorescence, the cells were cultured in 6 well plates with glass coverslips previously treated with 0.1% of L-Polylysine (Sigma-Aldrich) diluted in sterile ultrapure water. After 7 days of incubation, the cells were infected with 50 µl of 12 hour-old P. salmonis culture and incubated for 5 days. As a control, 1 ml of12 hour P. salmonis culture was inactivated with 3% formaldehyde (Sigma-Aldrich) for 24 hours at 4°C, washed 5 times in PBS 1X after inactivation and finally resuspended in 1 ml of sterile PBS 1X. Finally, 50 µl of inactivated P. salmonis culture was used to infect one well of a CHSE-214 titer plate, and incubated for 48 hours. The infected cells with P. salmonis live and/or dead were subsequently incubated at 20°C for 3 hours in darkness with 75 nM of LysoTracker Red DND-99 (Invitrogen) diluted in the medium used for each cell line and then washed three times with PBS 1X sterile. The cells were fixed for 30 minutes in 3% paraformaldehyde in PBS 1X at pH 7.5 and immediately permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 20 minutes and washed 3 times with PBS 1X. The infected cells were incubated in the dark for 30 minutes at 20°C with a 1∶100 (v/v) dilution in BSA 3% in PBS 1X of the FITC-conjugated oligoclonal antibody anti-P. salmonis (SRS Flourotest, BiosChile). Afterwards, cells were washed three times with PBS 1X, mounted with Dako® mounting medium (Invitrogen). The samples were analyzed using a Leica TCS SP5 II Spectral Confocal Microscope (Leica Microsystems Inc.). The images were obtained with a Leica 40x/1.25 Oil HCX PL APO CS objective (Leica Microsystems Inc.).
Results and Discussion
P. salmonis Encoded ORFs dot/icm Homologue
Based upon on the available sequences TIVSS Dot/Icm genes of the pathogens L. pneumophila and C. burnetii, degenerate primers were designed over conserved regions of 10 out of 22 possible ORF counterparts. A preliminary screening yielded PCR-positive amplicons corresponding to three key genes: dotB, dotA and icmK, which were subsequently cloned into pCR 2.1 TOPO TA vector and submitted to sequence. A fourth putative gene, icmE, was obtained using LR-PCR. Interpretation and significance of the DNA sequences was analyzed using three different bioinformatics tools: BLASTN, which determines nucleotide sequence homologies; BLASTX, which interprets the protein-coding potential of the target sequence and ClustalW, which aligns and provides amino acid homologies of the presumed proteins.
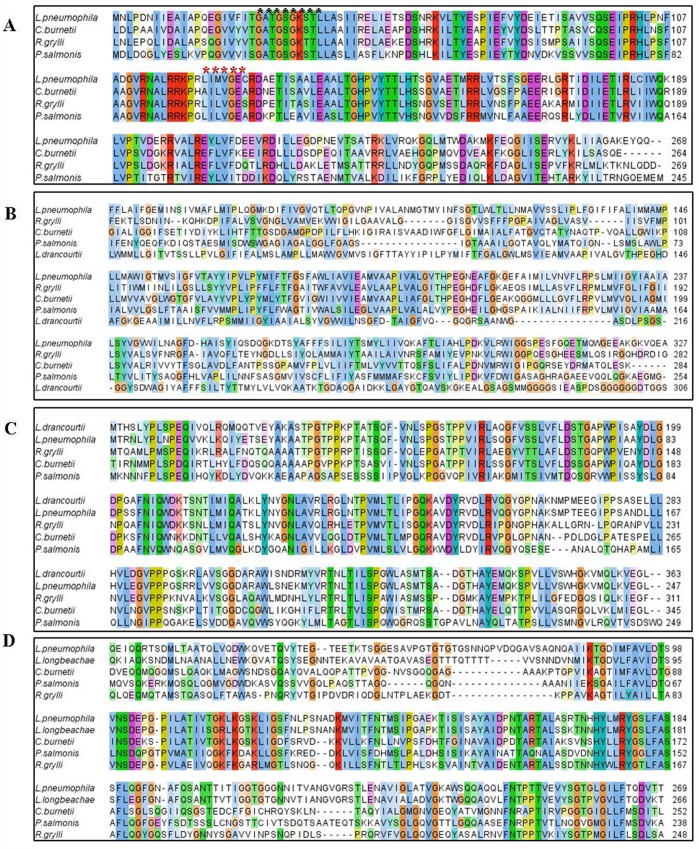
Regarding the putative P. salmonis dotB gene (Gene Bank: JX477678), BLASTN showed 69% and 68%of identity with L. pneumophila and C. burnetii, respectively. BLASTX analyses confirmed an in-frame ORF encoding a protein homologous to the DotB ATPase of the Dot/Icm secretion system of the two same reference pathogens mentioned above and also resemble to ATPase sequences from an array of different microorganisms (Table 3). About ClustalW alignment, this putative protein showed high degree of conservation with its counterparts (Figure 1–A). Furthermore, P. salmonis DotB contains two conserved distinctive motifs described for the P-loop NTPase superfamily: the first, consisting of a nucleotide phosphate-binding site, also known as the Walker A motif (GxxxxGK[S/T]); and the second, the Walker B motif (hhhh[D/E), where “h” is a hydrophobic residue [47]. The Walker A and B motifs bind the beta-gamma phosphate moiety of the donor molecule (either ATP or GTP) and the Mg2+ cation, respectively [48]. For the P. salmonis analogue, the putative Walker A motif involves the sequence GATGSGKS which fully matches the sequence of their reference counterparts, while the Walker B sequence (LILVGE) although not identical, does share the same physiochemical features as its counterparts (Figure 1–A). Nowadays, the L. pneumophila DotB is the best characterized protein, which fulfills three associated complementary functions: i) participation in the assembly of the TIVSS; ii) protein exportation; and iii) pilus retraction [49]. In addition, the prototype protein shares several structural features with type II and type IV secretion ATPases, which should also be present in the putative P. salmonis protein. One of such feature is an important hexameric ring structure that is believed to concede a loose association with the bacterial inner membrane despite its hydrophilic nature [49], [50], [51], [52].
Table 3. BLASTX results for P. salmonis dotB, dotA, icmK and icmE genes.
| GenBank N° | Organism | Protein | ID % | “e” Value |
| EDP46591.1 | Rickettsiella grylli | DotB | 69 | 2.0E–40 |
| CBJ10695.1 | Legionella longbeachae NSW150 | DotB | 69 | 5.0E–39 |
| ABX77916.1 | Coxiella burnetii RSA | DotB | 68 | 5.0E–38 |
| AAU28734.1 | Legionella pneumophila | DotB | 68 | 3.0E–38 |
| EHL29908.1 | Legionella drancourtii LLAP12 | DotA | 33 | 1.0E–24 |
| ABX79068.1 | Coxiella burnetii RSA | DotA | 31 | 1.0E–24 |
| AAP75479.1 | Legionella pneumophila | DotA | 29 | 2.0E–22 |
| EDP46828.1 | Rickettsiella grylli | DotA | 27 | 3.0E–20 |
| CAA75165.1 | Legionella pneumophila | IcmE | 45 | 5.0E–33 |
| CBJ13218.1 | Legionella longbeachae D-4968 | IcmE | 45 | 6.0E–32 |
| EDP45922.1 | Rickettsiella grylli | IcmE | 41 | 2.0E–30 |
| EDR35505.1 | Coxiella burnetii RSA | IcmE | 36 | 1.0E–30 |
| EHL29524.1 | Legionella drancourtii LLAP12 | IcmK | 49 | 1.0E–30 |
| AAS91991.1 | Legionella pneumophila | IcmK | 49 | 3.0E–30 |
| EDR35502.1 | Coxiella burnetii RSA | IcmK | 46 | 5.0E–29 |
| EDP46565.1 | Rickettsiella grylli | IcmK | 41 | 3.0E–28 |
The table shows proteins obtained with query coverage above 90 and e value of e−25.
Figure 1. ClustalW alignments among the most conserved regions of P. salmonis dot/icm protein products with their homologues. A:
DotB alignment, where the black and red asterisks show the Walker A and B, respectively (ATP binding site); B: DotA alignment; C: IcmK alignment; D: IcmE alignment. All figures were created using Jalview, where the color intensity shows the conservation degree of the aminoacids between the sequences.
Concerning to the dotA and icmK analogues, positive P. salmonis amplicons did not show sequence similarities via BLASTN analyses. Nevertheless, BLASTX analyses did reveal two putative proteins highly homologous to those expected for the DotA and the IcmK ORFs (Table 3). Additionally, Figures 1B and 1C show the ClustalW protein alignment of the putative P. salmonis DotA (JX477679) and IcmK (JX477681) sharing reasonable sequence similarity or identity percentage with their reference counterparts. DotA from L. pneumophila is an integral 113 kDa cytoplasmic membrane protein although it is exported by the Dot/Icm system during bacterial growth in liquid media [53]. It contains eight hydrophobic domains and is essential in regulating initial phagosome trafficking towards fusion to the lysosome, a pivotal early decision taken by the pathogen after macrophage uptake [54]. Similar to the L. pneumophila protein, the SOSUI analysis of the putative P. salmonis DotA reveals the presence of at least five hydrophobic domains with 23 residues each one, related with transmembrane regions (Table 4), indicating that this proteins could be associated with the bacterial membrane. The relevancy of L. pneumophila DotA is demonstrated by the fact that defective mutants are unable to inhibit the phagosome-lysosome fusion, which impedes productive infection [55].
Table 4. Hydrophobic domains of the P. salmonis DotA protein obtained using the SOSUI software.
| N terminal | Transmembrane region | C terminal | Type | Length |
| 275 | ALGGLFGAGSIGTAAAILGQTAV | 297 | SECONDARY | 23 |
| 313 | AWLPIALVVLGSLFTAAISFVVM | 335 | PRIMARY | 23 |
| 349 | IVWALSILEGLVAAPLVALALVY | 371 | PRIMARY | 23 |
| 389 | LNIIFRPVLMVIGVIAAMALTYV | 411 | PRIMARY | 23 |
| 435 | GMVNGIVSCFLIFIYASFMMMAF | 457 | PRIMARY | 23 |
Can be detected 5 putative transmembrane domains.
On the other hand, the L. pneumophila IcmK (DotH) protein is a periplasmatic and outer-membrane protein that possesses a peptide secretion signal and constitutes the scaffold core structure of the Dot/Icm complex in association with DotC, DotD, DotF and DotG [56], [57]. Its function is centered on pore formation and is crucial for macrophage killing [58].
The fourth putative gene, the icmE (JX477680) was obtained after sub cloning a 12 Kb LR-PCR amplicon. BLASTX analyses of a number of clones obtained from the LR-PCR yielded only one with the expected ORF, displaying high identity values with the IcmE protein of L. pneumophila, C. burnetii and R. gryllii (Table 3). ClustalW alignment of the P. salmonis IcmE protein confirmed the expected homology with other bacterial counterparts (Figure 1–D). In addition, the putative P. salmonis IcmE protein has conserved domains with the conjugational protein TrbI, similar to its counterpart in L. pneumophila, which also shares sequence homology with a plasmid-encoded protein TrbI from the IncP plasmid RK2 [59]. The main characteristic of the L. pneumophila IcmE or DotG (inner membrane protein) is that it interacts with other components of the complex TIVSS, triggering the assembly of the ATP-dependent export channel. The cascade involves DotF, which associates with outer membrane proteins IcmK, DotC and DotD to transfer energy from ATP hydrolysis to the outer membrane [57]. In general, the characterization of the IcmE analogue clearly suggests the existence of a functional Dot/Icm-like system in P. salmonis, which is sustained by the importance these genes have in other pathogenic bacteria.
Dot/Icm Gene Expression during in vitro Infection
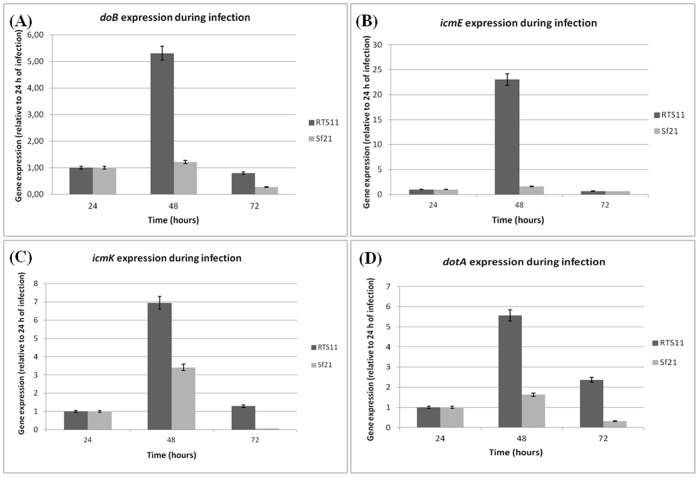
The evidences shown above suggest that P. salmonis comprises a putative Dot/Icm system including structural and proton-driving elements. If the putative P. salmonis dot/icm genes (TIVSS) were involved in modulating biogenesis of the vacuole, where the bacterium replicates inside the cells, these genes should be expressed during the infection process. Thus, the kinetic infection was evaluated at early stages of infection (24, 48 and 72 hours) in both cell lines: the phagocytic fish-derived RTS11 cell line and the non-phagocytic insect-derived Sf21 cell line. qRT-PCR analysis of dotB, icmK, icmE and dotA genes showed high levels of expression for at least the first three days after challenging (Figure 2). Our result shows a behavior is similar to that displayed by Brucella suis, where TIVSS genes are induced within the first few hours after bacterial uptake by macrophages [26]. In fact, the P. salmonis dot/icm gene expression was notoriously higher in the macrophage derived cell line (RTS11) with an evident overexpression at 48 hours with values of 5.31 for dotB, 23.04 for icmE, 6.96 for icmK and 5.57 for dotA (Figure 2). Nevertheless, expression of all dot/icm genes was higher at 48 hours post-infection in both cell lines, but the values obtained for the Sf21 cells were noticeably lower than RTS11 data (Figure 2), indicating that overexpression of these genes is preferentially triggered in macrophages. The high expression of dot/icm genes at 48 hours post-infection could be explained because P. salmonis grows at a slow rate in tissue culture cells and the major bacterial uptake by host cells may peak at 48 hours after infection and therefore expression of the dot/icm genes would peak at this time. A similar event has been observed in C. burnetti, which also displays low growth rates in vitro, and the expression of dotB and other Dot/Icm markers is not detected before 24 hours of infection, only at 48 hours post-infection the bacterium begin to appear in large vacuoles inside the cells [60]. Additionally, normalization with the 2−ΔΔCt method was made, obtaining similar results for all genes in both cell lines and the same tendency of expression was detected (Figure S1). While the dot/icm gene expression peaks at 48 hours post-infection, the ITS expression peaks at 72 hours in both cell lines and it was possible to detect an increasing ITS transcript in time (data not shown), possibly due to the multiplication of P. salmonis in both cell lines, which led to an increase in the number of transcripts. This result has validated our previous data, suggesting a differential expression in time of the secretion genes in earlier stages of infection. Negative RNA controls confirm a non-DNA contamination in the samples used for cDNA synthesis (data not shown). Furthermore, the EF1A gene expression on both cell lines demonstrate a constant amount of host cells during the kinetics (at least at earlier stages of infection) with Ct values of 30 and 32 for RTS11 and Sf21, respectively (Table S1) and consequently the eukaryotic mRNA is constant.
Figure 2. Expression profile of P. salmonis dot/icm genes during RTS11 and Sf21cell lines kinetic infection.
Gene expression was determined by qRT-PCR, using relative quantification. A: dotB gene expression number; B: icmE gene expression; C: icmK mRNA gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like a housekeeping gene. 24 hours post-infection in each cell line was used as calibrator (value = 1).
P. salmonis expresses dot/icm genes either in cell culture or in cell-free media (Figures 2 and 3). Comparatively, the Vir-like TIVSS genes in Brucella abortus and Brucella melitensis are constitutively expressed independent of the environment [61]. However, Brucella suisis unable to express the corresponding genes when growing in liquid media [62], suggestive that their expression might be responsive to limiting stress conditions.
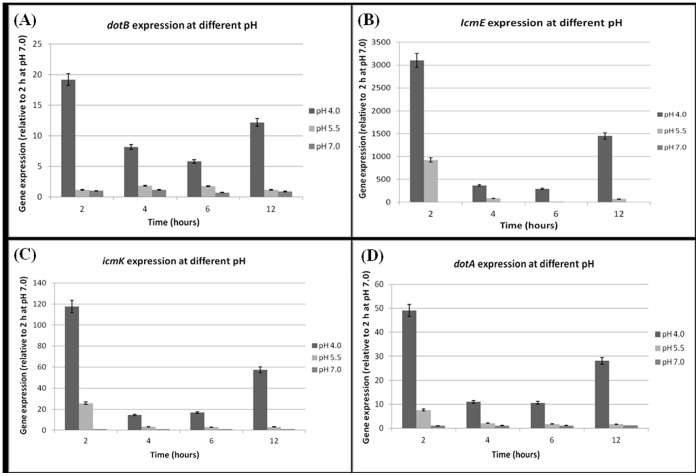
Figure 3. Expression profiles of dot/icm genes throughout P. salmonis growth kinetics at different pHs in MC1 medium.
Gene expression was determined by qRT-PCR, using relative quantification. A: dotB gene expression number; B: icmE gene expression; C: icmK gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like a housekeeping gene. Two hours of growth at pH 7.0 was used as calibrator (value = 1) for all genes. The figure shows that all genes were notably over-expressed at pH 4.0, particularly at 2 hours of incubation.
Acid pH Induces Overexpression of dot/icm Genes in P. salmonis
We hypothesized that phagosome acidification could be the signal that induces an intracellular over-expression of the P. salmonis dot/icm genes. So as to confirm this hypothesis, expressions of the dot/icm genes were evaluated by growth kinetics at pH 4.0, pH 5.5 and neutral pH.
qRT-PCR results show that dot/icm genes are clearly induced at acidic medium, specifically at pH 4.0 with values peaking at 2 hours of incubation (Figure 3). As seen in the figure, the lower the pH the higher the induction obtained. In comparison with expression at pH 7.0 at 2 hours of incubation, the induction at pH 4.0 increase 25-fold for dotB, 3000-fold for IcmE, 117-fold to icmK and 49-fold to dotA. At pH 5.5 the expression levels were higher than pH 7.0 but lower than pH 4.0, with values of 1.4, 900, 25 and 7.6 for dotB, icmE, icmK and dotA respectively. A similar expression tendency was observed for all genes using the 2−ΔΔCt normalization method (Figure S2), validating our previous data. As a control, ITS transcription (estimated by Ct values) remained stable at all times during the kinetics, except when the bacteria were grown at pH 4.0 with a decrease in 2 Ct units at 12 hours (Table S2), wherein the dot/icm expression is higher than at 4 and 6 hours (Figure 3), indicating that general protein expression is diminished at acidic pH, while dot/icm expression is promoted. A similar phenomenon is observed in B. suis, where 3 hours acid shock at pH 4.0 reduced the levels of protein synthesis while transcription of the TIVSS operon is strongly induced [63], [26]. Our data are consistent with the idea that acid pH induces over-expression of dot/icm genes, supporting our rationality where phagosome acidification could represents a crucial event in triggering expression of these genes during P. salmonis infection and could help as well to the intracellular survival along with other potential virulence mechanisms, at least initially at acid pH inside the cells.
P. salmonis Inhibition of Phagosome-lysosome Fusion
Phagocytosis is a process mediated by binding of organisms or large particles to plasma membrane receptors on phagocytes followed by internalization in newly formed phagosomes, organelles which mature into acidic and protease-rich phagolysosomes, where phagocytosed microorganisms or materials are killed and/or degraded [64]. In macrophages, phagosome-lysosome membrane fusion is a tightly regulated event essential for intracellular microorganism killing [65]. For this reason, many bacterial pathogens have developed strategies, which allow them to evade phagosome-lysosome fusion to survive and multiply within the intracellular environment [66]. For example, B. abortus is able to replicate in a compartment segregated from the endocytic pathway and the maturation of the Brucella-containing vacuole involves sustained interactions and fusion with the endoplasmic reticulum (ER), which creates a replicative compartment with ER-like properties, in a TIVSS-dependent process [67]. Bartonella henselae and S. typhimurium have a specific competence to actively avoid the host endocytic pathway after entry into macrophages and epithelial cells, from within a specialized non-endocytic membrane-bound vacuole is formed in a TIVSS- and TIIISSS-dependent event, respectively [66], [68], [69].
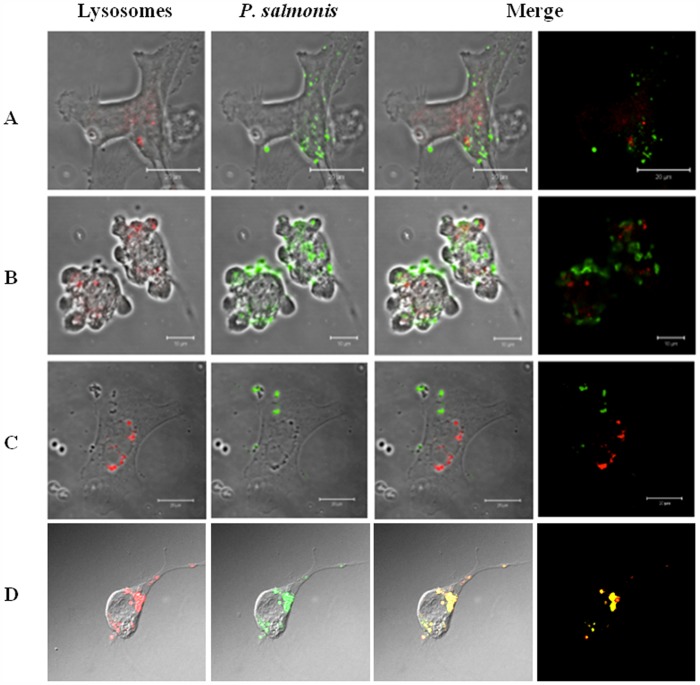
As our data support the possibility that P salmonis dot/icm genes could play a pivotal role in the biogenesis of the replicative vacuole, we decided to determine whether a phagosome-lysosome escape mechanism exists in P. salmonis infected cells. Three different cell lines, the non-phagocytic insect (Sf21), a second non-phagocytic salmonid embryo kidney (CHSE-214) and the phagocytic trout macrophage/monocyte (RTS11), were infected for 5 days because at this time the typical P. salmonis-induced Cytopathic Effect (CPE) is fully evident in all cell lines used, confirming the viability of the bacterium and therefore is an evidence of productive infection. After day five, the infected cells were stained with a lysosomal-specific probe (LysoTracker(™) Red) to detect the organelle and also with aspecific FITC-labeled anti-P salmonis to evaluate possible co-localization with the phagosome-containing bacteria. Samples were then analyzed under a Laser Scanning Confocal Microscope. As seen in Figure 4, phagosome-lysosome fusion does not occur in any of the three cell lines, indicating that P. salmonis could use this strategy to promote its replication inside isolated vacuole. In contrast, the control where formaldehyde inactivated bacterium was used to infect the RTS11 cell line, showed clearly that the inactive P. salmonis is unable to evade the phagosome-lysosome fusion and consequently is derived directly to the endocytic pathway for being degraded. This final result is consistent to sustain that P. salmonis is able to evade the phagosome-lysosome fusion event that could be targeted by the expression and effectors secretion of the Dot/Icm system and/or by another virulence mechanism to ensure their multiplication in the infected cells.
Figure 4. Confocal Laser Scanning Microscopy of P. salmonis infection on three cell lines showing the escape of phagosome-lysosome fusion.
The immunofluorescence was made 5 days post-infection. Lysosomes were stained in red with LysoTracker Red reagent and P. salmonis was detected with a FITC conjugated antibody. A: CHSE-214 cell line infected with P. salmonis. B: Sf21 cell line infected with P. salmonis. C: RTS11 cell line infected with P. salmonis. D: CHSE-214 cell line infected with formaldehyde-inactivated P. salmonis, the immunofluorescence stain was made at 48 hours after infection.
Comparatively, L.pneumophila and C. Burnetiiare known to depend on a similar pathway in which an initial generation of an organelle-like structure inside the host cell helps them support bacterial replication [70], [60]. Therefore, it is feasible to think that due to the similarities shared by these two pathogens with P. salmonis, the latter could use the same strategy to avoid phagosome-lysosome fusion, and if so, dot/icm gene products should hold this responsibility.
Final Remarks
Most intracellular and facultative pathogens employ TIVSS as a preferential mechanism to direct biogenesis of a vacuolar replicative niche that circumvents default maturation through the endolysosomal cascade and favors their intracellular multiplication [71]. This also seems to be the case for Piscirickettsia salmonis. Out of the two existent ancestral lineages of the TIVSS: the VirB/D4 system of A. tumefaciens and the Dot/Icm system of L. pneumophila, sometimes referred as the TIVSS-A and TIVSS-B systems, respectively [72], P. salmonis clearly uses the latter. Existing evidence suggests that proteins translocated by the Dot/Icm system are critical for successful parasitism of macrophages by either C. burnetii and/or L. pneumophila [73]. In this report, we have demonstrated the functional existence of four essential components of Dot/Icm secretion system (dotB, dotA, icmK and icmE) in the genome of fish pathogen P. salmonis. Moreover, the putative corresponding polypeptide products also share distinctive features with their counterparts, corroborating our interpretation that the system exists, in spite of the fact that not all presumptive components have been characterized. Our results also indicate that these genes are expressed during the infection as well as in growing cell-free medium, suggestive of a constitutive expression in response to the stress signaling compatible with the rough environment the bacteria faces in vivo. In addition, qRT-PCR experiments show that the dot/icm genes are over expressed in acid pH, indicating possibly that phagosome acidification is the triggering event that produces dot/icm gene expression and therefore protein secretion via the Dot/Icm system to favor intracellular bacterial replication. Finally, we have also demonstrated that P. salmonis-containing vacuoles do not fuse with lysosomes, indicating that there is a bacterium-driven interference in the endosomal maturation process that ensures bacterial survival, of which the Dot/Icm secretion system should be responsible by delivering effectors proteins inside the host cell.
Hence, we may conclude that the P. salmonis Dot/Icm secretion system represents one of the mechanisms associated with its virulence and pathogenesis, similar to what happen with the closely related pathogens L. pneumophila and C. burnetii. In order to demonstrate this action, knock out gene experiments could be carried out once P. salmonis be efficiently transformed.
Supporting Information
P. salmonis dot/icm gene expression during an infection kinetic in RTS11 and Sf21 cell lines. The normalization was made using the 2−ΔΔCt method. A: dotB gene expression number; B: icmE gene expression; C: icmK mRNA gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like housekeeping gene. 24 hours post-infection in each cell line was used as calibrator (value = 1).
(DOC)
P. salmonis dot/icm gene expression during a growth kinetic at different pH. The normalization was made using the 2−ΔΔCt method. A: dotB gene expression number; B: icmE gene expression; C: icmK mRNA gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like a housekeeping gene. Two hours of growth at pH 7.0 was used as calibrator (value = 1).
(DOC)
EF1A gene Ct values of RTS11 and Sf21 cell lines during infection kinetics determined by qRT-PCR.
(DOC)
ITS (16 S-23 S internal transcribed spacer) Ct values obtained during P. salmonis growth kinetic at different pH.
(DOC)
Acknowledgments
We thank Dr. Constanza Cárdenas for figures preparation.
Funding Statement
This work was supported by FONDECYT grant 1120584 for Sergio H. Marshall and by a Conicyt Doctoral Scholarship for Fernando A. Gómez. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fryer JL, Lannan CN, Garcés HL, Larenas JL, Smith PA (1990) Isolation of Rickettsiales-like organism from diseased coho salmon (Oncorhynchuskisutch) in Chile. Fish Pathology 25: 107–114. [Google Scholar]
- 2. Tobar JA, Jerez S, Caruffo M, Bravo C, Contreras F, et al. (2011) Oral vaccination of Atlantic salmon (Salmo salar) against salmonid rickettsialsepticaemia. Vaccine317: 83–92. [DOI] [PubMed] [Google Scholar]
- 3. Bravo S, Campos M (1989) Síndrome del salmón Coho. Chile Pesquero 54: 47–48. [Google Scholar]
- 4. Olsen AB, Evensen O, Speilberg I, Melby HP, Hastein T (1993) Nylaksesykdomforarsaketavrickettsie. NorskFiskeoppdrett 12: 40–J1. [Google Scholar]
- 5. Almendras FF, Jones SRM, Fuentealva C, Wrigth GM (1997) In vitro infection of a cell line from Ictalurusnebulosus with Piscirickettsia salmonis . Can J Vet Res 61: 66–68. [PMC free article] [PubMed] [Google Scholar]
- 6. Mauel MJ, Giovannoni SJ, Fryer JL (1999) Phylogenetic analysis of Piscirikettsia salmonis by 16S, internal transcribed spacer (ITS) and 23S ribosomal DNA sequencing. Dis Aquat Organ 35: 115–123. [DOI] [PubMed] [Google Scholar]
- 7. Fryer JL, Hedrick RP (2003) Piscirickettsia salmonis: a Gram-negative intracellular bacterial pathogen of fish. J Fish Dis 26: 251–262. [DOI] [PubMed] [Google Scholar]
- 8. Fryer JL, Mauel MJ (1997) The Rickettsia: an emerging group of pathogens in fish. Emerg Infect Dis 3: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rojas V, Olivares J, del Río R, Marshall SH (2008) Characterization of a novel and genetically different small infective variant of Piscirickettsia salmonis . MicrobPathog 44: 370–378. [DOI] [PubMed] [Google Scholar]
- 10. Rojas V, Galanti N, Bols NC, Marshall SH (2009) Productive infection of Piscirickettsia salmonis in macrophages and monocyte-like cells from rainbow trout, a possible survival strategy. J Cell Biochem 108: 631–637. [DOI] [PubMed] [Google Scholar]
- 11. Mauel MJ, Ware C, Smith PA (2008) Culture of Piscirickettsia salmonis on enriched blood agar. J Vet Diagn Invest 20: 213–214. [DOI] [PubMed] [Google Scholar]
- 12. Mikalsen J, Skjaervik O, Wiik-Nielsen J, Wasmuth MA, Colquhoun DJ (2008) Agar culture of Piscirickettsia salmonis, a serious pathogen of farmed salmonid and marine fish. FEMS Microbiol Lett 278: 43–47. [DOI] [PubMed] [Google Scholar]
- 13. Gómez F, Henríquez V, Marshall SH (2009) Additional evidence of the facultative intracellular nature of the fish bacterial pathogen Piscirickettsia salmonis . Arch Med Vet 41: 261–267. [Google Scholar]
- 14. Yáñez AJ, Valenzuela K, Silva H, Retamales J, Romero A, et al. (2012) Broth medium for the successful culture of the fish pathogen Piscirickettsia salmonis . Dis Aquat Organ 97: 197–205. [DOI] [PubMed] [Google Scholar]
- 15. Mauel MJ, Miller DL, Frazier K, Liggett AD, Styer L, et al. (2003) Characterization of a piscirickettsiosis-like disease in Hawaiian tilapia. Dis Aquat Organ 53: 249–255. [DOI] [PubMed] [Google Scholar]
- 16. Athanassopoulou F, Groman D, Prapas Th, Sabatakou O (2004) Pathological and epidemiological observations on rickettsiosis in cultured sea bass (Dicentrarchuslabrax L.) from Greece. J ApplIchth 20: 525–529. [Google Scholar]
- 17. Arkush KD, McBride AM, Mendonca HL, Okihiro M, Andree KB, et al. (2005) Genetic characterization and experimental pathogenesis of Piscirickettsia salmonis isolated from white seabassAtractoscionnobilis . Dis Aquat Organ 63: 139–149. [DOI] [PubMed] [Google Scholar]
- 18. Marshall SH, Henríquez V, Gómez FA, Cárdenas C (2011) ISPsa2, the first mobile genetic element to be described and characterized in the bacterial facultative intracellular pathogen Piscirickettsia salmonis . FEMS Microbiol Lett 314: 18–24. [DOI] [PubMed] [Google Scholar]
- 19. Gómez FA, Cárdenas C, Henríquez V, Marshall SH (2011) Characterization of a functional toxin-antitoxin module in the genome of the fish pathogen Piscirickettsia salmonis . FEMS Microbiol Lett 317: 83–92. [DOI] [PubMed] [Google Scholar]
- 20. McCarthy UM, Bron JE, Brown L, Pourahmad F, Bricknell IR, et al. (2008) Survival and replication of Piscirickettsia salmonis in rainbow trout head kidney macrophages. Fish Shellfish Immunol 25: 477–484. [DOI] [PubMed] [Google Scholar]
- 21. Peña AA, Bols NC, Marshall SH (2010) An evaluation of potential reference genes for stability of expression in two salmonid cell lines after infection with either Piscirickettsia salmonis or IPNV. BMC Res Notes 3: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rojas V, Galanti N, Bols NC, Jiménez V, Paredes R, et al. (2010) Piscirickettsia salmonis induces apoptosis in macrophages and monocyte-like cells from rainbow trout. J Cell Biochem110: 468–476. [DOI] [PubMed] [Google Scholar]
- 23. Rosenberger CM, Finlay BB (2003) Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat Rev Mol Cell Biol 4: 385–396. [DOI] [PubMed] [Google Scholar]
- 24. Tobar JA, González PA, Kalergis AM (2004) Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fc gamma receptors on dendritic cells. J Immunol173: 4058–4065. [DOI] [PubMed] [Google Scholar]
- 25. Joiner KA (1997) Membrane-protein traffic in pathogen-infected cells. J Clin Invest 99: 1814–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, et al. (2002) The Brucellasuis virB operon is induced intracellularly in macrophages. Proc Natl Acad Sci USA 99: 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sinai AP, Joiner KA (1997) Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu Rev Microbiol 51: 415–462. [DOI] [PubMed] [Google Scholar]
- 28.Flores-Herrera O, Riveros H, Sosa A, Vázquez E (2003) Sistemas de secreción de proteínas en las bacterias Gram negativas: biogénesisflagelar y translocación de factores de virulencia. Mensaje BioquímicoVol XXVII.
- 29. Tseng TT, Tyler BM, Setubal JC (2009) Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9 Suppl 1S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thanassi DG, Hultgren SJ (2000) Multiple pathways allow protein secretion across the bacterial outer membrane. Curr Op Cell Biol 12: 420–430. [DOI] [PubMed] [Google Scholar]
- 31. Schulein R, Dehio C (2002) The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intra-erythrocytic infection. Mol Microbiol 46: 1053–1067. [DOI] [PubMed] [Google Scholar]
- 32. Jones KM, Lloret J, Daniele JR, Walker GC (2007) The type IV secretion system of Sinorhizobiummeliloti strain 1021 is required for conjugation but not for intracellular symbiosis. J Bacteriol 189: 2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeo HJ, Waksman G (2004) Unveiling molecular scaffolds of the type IV secretion system. J Bacteriol 186: 1919–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Angelini A, Cendron L, Goncalves S, Zanotti G, Terradot L (2008) Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori . Proteins 72: 1212–1221. [DOI] [PubMed] [Google Scholar]
- 35. Segal G, Feldman M, Zusman T (2005) The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii . FEMS Microbiol Rev 29: 65–81. [DOI] [PubMed] [Google Scholar]
- 36. Sauer JD, Shannon JG, Howe D, Hayes SF, Swanson MS, et al. (2005) Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by co-infection. Infect Immun 73: 4494–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zink SD, Pedersen L, Cianciotto NP, Abu-Kwaik Y (2002) The Dot/Icm type IV secretion system of Legionella pneumophila is essential for the induction of apoptosis in human macrophages. Infect Immun70: 1657–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zusman T, Yerushalmi G, Segal G (2003) Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. . Infect Immun 71: 3714–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marshall SH, Gómez FA, Ramírez R, Nilo L, Henríquez V (2012) Biofilm generation by Piscirickettsia salmonis under growth stress conditions: A putative in vivo survival/persistence strategy in marine environments. Res Microbiol 163(8): 557–566. [DOI] [PubMed] [Google Scholar]
- 40. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 41. Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427. [DOI] [PubMed] [Google Scholar]
- 42. Marshall S, Heath S, Henríquez V, Orrego C (1998) Minimally invasive detection of Piscirickettsia salmonis in cultivated salmonids via the PCR. Appl Environ Microbiol 64: 3066–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ganassin RC, Bols NC (1998) Development of a monocyte/macrophage-like cell line, RTS11, from rainbow trout spleen. Fish Shellfish Immunol 8: 457–476. [Google Scholar]
- 44. Birkbeck TH, Griffen AA, Reid HI, Laidler LA, Wadsworth S (2004) Growth of Piscirickettsia salmonis to high titers in insect tissue culture cells. Infect Immun 72: 3693–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Let 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 46. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using Real Time Quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 47. Gangwar D, Kalita MK, Gupta D, Chauhan VS, Mohmmed A (2009) A systematic classification of Plasmodium falciparum P-loop NTPases: structural and functional correlation. Malar J 18: 8–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hubbard PA, Padovani D, Labunska T, Mahlstedt SA, Banerjee R, et al. (2007) Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonicaciduria. J BiolChem282: 31308–31316. [DOI] [PubMed] [Google Scholar]
- 49. Sexton JA, Yeo HJ, Vogel JP (2005) Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol Microbiol 57: 70–84. [DOI] [PubMed] [Google Scholar]
- 50. Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ (1995) Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae . EMBO J 14: 1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rashkova S, Spudich GM, Christie PJ (1997) Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol 179: 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krause S, Barcena M, Pansegrau W, Lurz R, Carazo JM, et al. (2000) Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc Natl Acad Sci USA 97: 3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagai H, Roy CR (2001) The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J 20: 5962–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roy CR, Berger KH, Isberg RR (1998) Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol 28: 663–674. [DOI] [PubMed] [Google Scholar]
- 55. Scaturro M, Meschini S, Arancia G, Stefano F, Ricci ML (2009) Characterization of a spontaneous avirulent mutant of Legionella pneumophila Serogroup 6: evidence of DotA and flagellin involvement in the loss of virulence. J Microbiol 47: 768–773. [DOI] [PubMed] [Google Scholar]
- 56. Andrews HL, Vogel JP, Isberg RR (1998) Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect Immun 66: 950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vincent CD, Buscher BA, Friedman JR, Williams LA, Bardill P, et al. (2006) Identification of non-dot/icm suppressors of the Legionella pneumophilaΔdotL lethality phenotype. J Bacteriol 188: 8231–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morozova I, Qu X, Shi S, Asamani G, Greenberg JE, et al. (2004) Comparative sequence analysis of the icm/dot genes in Legionella . Plasmid 51: 127–147. [DOI] [PubMed] [Google Scholar]
- 59. Segal G, Shuman HA (1999) Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii . Mol Microbiol 33: 669–670. [DOI] [PubMed] [Google Scholar]
- 60. Zamboni DS, McGrath S, Rabinovitch M, Roy CR (2003) Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol49: 965–976. [DOI] [PubMed] [Google Scholar]
- 61. Rouot B, Alvarez-Martinez MT, Marius C, Menanteau P, Guilloteau L, et al. (2003) Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect Immun 71: 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carle A, Höppner C, Ahmed Aly K, Yuan Q, den Dulk-Ras A, et al. (2006) The Brucella suis type IV secretion system assembles in the cell envelope of the heterologous host Agrobacterium tumefaciens and increases IncQ plasmid pLS1 recipient competence. Infect Immun 74: 108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin J, Ficht TA (1995) Protein synthesis in Brucellaabortus induced during macrophage infection. Infect Immun 63: 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Funato K, Beron W, Yang CZ, Mukhopadhyay A, Stahl PD (1997) Reconstitution of phagosome-lysosome fusion in streptolysin O-permeabilized cells. J BiolChem 272: 16147–16151. [DOI] [PubMed] [Google Scholar]
- 65. Zimmerli S, Majeed M, Gustavsson M, Stendahl O, Sanan DA, et al. (1996) Phagosome-lysosome fusion is a calcium-independent event in macrophages. J Cell Biol 132: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buchmeier NA, Heffron F (1991) Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium . Infect Immun 59: 2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, et al. (2006) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kyme PA, Haas A, Schaller M, Peschel A, Iredell J, et al. (2005) Unusual trafficking pattern of Bartonellahenselae -containing vacuoles in macrophages and endothelial cells. Cell Microbiol 7: 1019–34. [DOI] [PubMed] [Google Scholar]
- 69. AlpucheAranda CM, Swanson JA, Loomis WP, Miller SI (1992) Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA 89: 10079–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gal-Mor O, Zusman T, Segal G (2002) Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J Bacteriol184: 3823–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Voth DE, Heinzen RA (2007) Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii . Cell Microbiol 9: 829–840. [DOI] [PubMed] [Google Scholar]
- 72. Rikihisa Y, Lin M, Niu H (2010) Type IV secretion in the obligatory intracellular bacterium Anaplasmaphagocytophilum . Cell Microbiol 12: 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, et al. (2011) Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio 2: e00175–11 doi:10.1128/mBio.00175–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P. salmonis dot/icm gene expression during an infection kinetic in RTS11 and Sf21 cell lines. The normalization was made using the 2−ΔΔCt method. A: dotB gene expression number; B: icmE gene expression; C: icmK mRNA gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like housekeeping gene. 24 hours post-infection in each cell line was used as calibrator (value = 1).
(DOC)
P. salmonis dot/icm gene expression during a growth kinetic at different pH. The normalization was made using the 2−ΔΔCt method. A: dotB gene expression number; B: icmE gene expression; C: icmK mRNA gene expression; D: dotA gene expression. Gene expression was normalized by the use of ITS like a housekeeping gene. Two hours of growth at pH 7.0 was used as calibrator (value = 1).
(DOC)
EF1A gene Ct values of RTS11 and Sf21 cell lines during infection kinetics determined by qRT-PCR.
(DOC)
ITS (16 S-23 S internal transcribed spacer) Ct values obtained during P. salmonis growth kinetic at different pH.
(DOC)