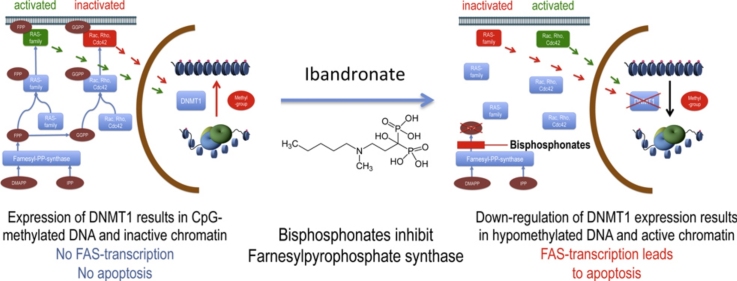
Graphical abstract
Keywords: Bisphosphonates, Ibandronate, Epigenetic DNA-methylation, Small GTPases, FAS apoptosis
Abstract
There is growing evidence that aminobisphosphonates like ibandronate show anticancer activity by an unknown mechanism. Biochemically, they prevent posttranslational isoprenylation of small GTPases, thus inhibiting their activity. In tumor cells, activated RAS-GTPase, the founding member of the gene family, down-regulates the expression of the pro-apoptotic gene FAS via epigenetic DNA-methylation by DNMT1. We compared ibandronate treatment in neoplastic human U-2 osteosarcoma and in mouse CCL-51 breast cancer cells as well as in the immortalized non-neoplastic MC3T3-E1 osteoblastic cells. Ibandronate attenuated cell proliferation in all cell lines tested. In the neoplastic cells we found up-regulation of caspases suggesting apoptosis. Further we found stimulation of FAS-expression as a result of epigenetic DNA demethylation that was due to down-regulation of DNMT1, which was rescued by re-isoprenylation by both geranylgeranyl-pyrophosphate and farnesylpyrophosphate. In contrast, ibandronate did not affect FAS and DNMT1 expression in MC3T3-E1 non-neoplastic cells. Data suggest that bisphosphonates via modulation of the activity of small-GTPases induce apoptosis in neoplastic cells by DNA-CpG-demethylation and stimulation of FAS-expression. In conclusion the shown epigenetic mechanism underlying the anti-neoplastic activity of farnesyl-transferase-inhibition, also explains the clinical success of other drugs, which target this pathway.
1. Introduction
The 3rd generation of aminobisphosphonates like alendronate, risedronate, zoledronic acid, ibandronate and other compounds affect bone resorption of osteoclasts by inhibiting isoprenylation of the small GTP-binding proteins and are therefore used as anti-resorptive treatment of osteoporosis. Additionally to their effects on bone, there is growing evidence for an anticancer activity of these drugs [1–8]. However, the mechanisms involved in these effects remain poorly understood.
Biochemically, aminobisphosphonates act principally by inhibiting farnesyl pyrophosphate (FPP) synthase – an enzyme of the mevalonate pathway – thereby preventing the post-translational modification (prenylation) of small guanosine triphosphate (GTP)-binding proteins (small-GTPases) that is essential for their function. Because small GTP-binding proteins modulate nearly every cellular activity, it is clear that functional inhibition from members of this protein family influences growth and differentiation of normal cells as well as of tumor cells [3,5,7,9–11].
The founding members of the large family of small-GTPases are the three RAS-proteins, HRAS, NRAS and KRAS. Activation of the RAS/RAF/MEK/ERK pathway, for instance by mutation of the involved GTPases or their regulating members, is responsible for the development of a plethora of cancers [12–14] and targeting this pathway seems to be a promising strategy in tumor therapy [15,16]. The RAS/RAF/MEK/ERK pathway activates DNA-methylation processes [17–19]. Epigenetic processes include histone modifications and methylation of CpGs (cytosine-guanosine dinucleotides) on the DNA, especially on gene promoters. Changes of the methylation state of gene promoters lead to alteration in gene expression patterns. This influences cellular differentiation and apoptosis and thus tumor formation (for review see Refs. [19–22]). DNA methylation dependent inactivation of tumor suppressor genes like cell cycle inhibitors (e.g. CDKN1A, CDKN1B, CDKN2A, CDKN2B) and LOX (lysyl oxidase) as well of the pro-apoptotic gene FAS (TNF receptor superfamily, member 6) is often observed during development of neoplastic diseases. Promoter CpG-hypermethylation of these genes was found in colon cancers [23], prostate carcinomas [24–26], breast cancers [27–29] or hematologic malignancies [30–33]. Consequently, several DNA demethylating agents were developed and are now in use as anti neoplastic drugs to reactivate genes such as FAS which plays a key role in immortality of cancer stem cells [34].
It has recently been shown that activated RAS prevents cellular apoptosis by epigenetic inhibition of Fas expression through stimulation of the RAF/MEK/MAPK1 pathway with subsequent Fas promoter methylation via DNMT1 (DNA-(cytosine-5-)-methyltransferase 1), an enzyme responsible for CpG methylation during cell replication [17]. Similarly, in osteoblasts, extracellular matrix (collagen type I) preserves CpG-methylation of the Fas promoter via MAPK1 and DNMT1, thus preventing apoptosis of proliferating osteoblasts [19]. Although, utmost efforts have been spent to clarify the relevance and the regulation of cytosine methylation for physiological and pathological development, only few progresses have been made until now. The involvement of RAS and other small GTP-binding proteins in bisphosphonates’ activity and the knowledge of apoptotic effects on bone cells, also of bisphosphonates of the 3rd generation [1,4,6,7,35–37], suggest that these drugs could modulate CpG-methylation of gene promoters.
Here, we demonstrate that the aminobisphosphonate ibandronate modulates the DNA methylation status of the FAS promoter by influencing the isoprenylate pathway in human U-2 osteosarcoma (OS) cells and CCL-51 cells, a murine mammary gland tumor cell line, but not in non-neoplastic immortalized MC3T3-E1 cells. Treatment with ibandronate leads to re-expression of FAS and to increased activity of apoptosis-associated caspases in the tumor cell lines. Knock down of FAS mRNA expression by siRNA technique largely re-establishes cell viability in ibandronate treated neoplastic U-2 OS cells. Our data suggest that epigenetic mechanisms play a key role in the apoptotic activity of bisphosphonates, and possibly many of their effects on cellular physiology including systemic changes within an organism.
2. Materials and methods
2.1. Cell culture
MC3T3-E1 cells, a clonal pre-osteoblastic cell line derived from newborn mouse calvaria (kindly donated by Dr. Kumegawa, Meikai University, Department of Oral Anatomy, Sakado, Japan) and the human osteosarcoma cell line U-2 OS were cultured in alpha-minimum essential medium (α-MEM; Biochrom, Berlin, Germany) supplemented with 50 μg/mL ascorbic acid (Sigma–Aldrich, St. Louis, MO), 5% fetal calf serum (Biochrom), and 10 μg/mL gentamycin (Sigma–Aldrich). CCL-51 cells, a murine mammary gland tumor cell line, were cultured in eagle minimum essential medium (EMEM, Sigma–Aldrich) supplemented with 292 μg/mL l-glutamine, 10% fetal calf serum and 10 μg/mL gentamycin. All cells were cultured in humidified air under 5% CO2 at 37 °C. For propagation, cells were subcultured twice a week using 0.001% pronase E (Roche, Mannheim, Germany) and 0.02% EDTA in Ca2+- and Mg2+-free phosphate-buffered saline (PBS) before achieving confluence. To prevent a potential phenotypic drift during repeated sub-cultures, the cells were not used for more than 4 weeks after thawing. For experiments, cells were seeded in culture dishes at a density of 20,000/cm2 as untreated controls or treated with the indicated compounds at times and concentrations specified. Ibandronate, geranylgeranyl-pyrophosphate (GGPP) and farnesyl-pyrophosphate (FPP) were purchased from Sigma-Aldrich. Ibandronate was dissolved in water and aliquots were frozen at −20 °C.
2.2. Cell viability/proliferation
To assess cell metabolic activity, a commercially available, MTT similar assay (EZ4U; Biomedica, Vienna, Austria) was used. For this purpose, the cell lines were incubated with increasing concentrations of ibandronate (1–50 μM for MC3T3-E1 and U-2 OS cells or 1–200 μM for CCL-51 cells). After a comparable doubling time for all three cell lines the assay was performed following the protocol of the supplier.
2.3. Cell counting
Cells were seeded in 24 multi-well culture dishes at a density of 20,000/cm2 and were either left untreated (controls) or treated with ibandronate, GGPP and FPP at the indicated concentrations for 72 h. Thereafter, cells were detached with 0.001% pronase E and the number of viable cells was assessed with Casy cell counter (Schaerfe Systems, Germany). Each experiment was performed in quadruplicate and experiments were carried out twice.
2.4. Measurement of caspase activity
Caspase 3/7 and caspase 8 activities were measured by using the Caspase-Glo 3/7 and Caspase-Glo 8 assay Kit (Promega, Corp., Madison, WI) following manufactures instructions. Briefly, after treatments, cells were lysed and substrate cleavage by caspases was measured by the generated luminescent signal with a 96 multi-well luminometer (Glomax, Promega). Each experiment was performed in quintuplicate and experiments were carried out twice.
2.5. Isolation of nucleic acids and expression analysis by qRT-PCR
DNA and RNA were extracted using a DNA/RNA Isolation Kit (Qiagen, Hilden, Germany) following manufacturers instructions. cDNA was synthesized from 0.5 μg RNA using the 1st Strand cDNA Synthesis Kit (Roche) as described by the supplier. The obtained cDNA was subjected to PCR amplification with a real-time cycler using FastStart SYBR-Green Master Mix (Roche) for the genes Fas and Dnmt1 (primers are shown in Table 1). The qRT-PCR was performed with 45 cycles composed of 30 s denaturation at 95 °C, 30 s annealing at the indicated temperature (Table 1) and 30 s extension at 72 °C after 10 min of initial denaturation at 95 °C. For normalization of the expression we used the 18S RNA TaqMan probe (4319413E, Applied Biosystems, Forster City, CA) in the respective master mix according to the suppliers suggested conditions (Applied Biosystems). All PCRs were performed in triplicate and expression was evaluated using the comparative quantitation method [38]. For each of the three time independent biological replicates, the triplicate results of the qRT-PCR were averaged and this mean value was treated as a single statistical unit.
Table 1.
Primer sequences.
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) | Tm (°C) |
|---|---|---|---|
| Primer for gene expression (mouse and human) | |||
| Dnmt1 | ACCGCTTCTACTTCCTCGAGGCCTA | GTTGCAGTCCTCTGTGAACACTGTGG | 62 |
| Fas | TATCAAGGAGGCCCATTTTGC | TGTTTCCACTTCTAAACCATGCT | 64 |
| Primer for Fas promoter methylation assessment | |||
| Mouse | CATACCCACAGGCAGTCTAGA | CAGCCCAGAGTAACTCACTTC | 62 |
| Human | CTGACTCCTTCCTCACCCT | CTTCCCCAACTCCGTACT | 64 |
| Primer for DNMT1 chromatin immune-precipitation of the Fas promoter | |||
| Mouse | CATACCCACAGGCAGTCTAGA | CAGCCCAGAGTAACTCACTTC | 62 |
| Human | CTGACTCCTTCCTCACCCT | CTTCCCCAACTCCGTACT | 62 |
2.6. FAS siRNA transfection
For FAS depletion by siRNA, U-2 OS cells were seeded at 20,000 cells/cm2 in a 48-multi-well plate and transfected with 100 nM of a mixture of two FAS siRNAs (SASI_Hs01 00079050, SASI_Hs01_00079052, Sigma–Aldrich) by electroporation using the Neon Transfection System (Invitrogen) following supplier's protocol. Next day cells were treated with 7.2 μM ibandronate (EC50). After 72 h of incubation, cell metabolic activity was measured with the EZ4U assay as described above. Furthermore to control FAS mRNA and protein knockdown, cells were seeded in a six-multi well plate or 5 cm petri dish at 20,000 cells/cm2 and transfected with the FAS siRNA as described above. 96 h after transfection mRNA or protein was isolated as described above and below and FAS mRNA and protein expression was measured by qRT-PCR and immune-blot, respectively.
2.7. Protein isolation and immune blotting
For whole cell protein extraction, cells were washed twice with PBS, scraped in SDS sample buffer (2% SDS, 100 mM β-mercaptoethanol, 125 mM Tris–HCl, pH 6.8) heated at 95 °C for 5 min and sonicated for further 5 min. To divide cell proteins into cytosolic and membrane fractions, cells were detached from culture vessels washed with PBS and incubated in hypotonic buffer (10 mM HEPES pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF) for 10 min on ice. Then, cells were broken by 20 strokes in a Dounce homogenizator and suspensions were centrifuged at 3300 × g for 20 min at 4 °C. Supernatants were centrifuged for another 30 min at 15,000 × g at 4 °C and the supernatants thus obtained were saved as cytosolic fraction. The pellets were dissolved in SDS sample buffer constituting the membrane fraction. Protein content was measured with Coomassie protein assay (Thermo Fisher Scientific, Waltham, MA) or with bicinchoninic acid assay (Sigma–Aldrich) for the SDS extracts. For immune-blotting 30 μg of protein extracts for RAS and FAS or 50 μg of protein extracts for DNMT1 were fractionated on 12%, 10% or 8% SDS-PAGE, respectively. Following SDS-gel electrophoresis, the proteins were transferred to nitrocellulose filters (Millipore) and blocked overnight with 10% blocking reagent (Roche Applied Science, Mannheim, Germany) in TN Buffer (50 mM Tris, 125 mM NaCl, pH 8). Subsequently, the filters were incubated for 1 h at room temperature with an antibody against DNMT1 (K-18, Santa Cruz Biotechnology, Santa Cruz, CA), against FAS and against RAS (610197 and 610002, BD Transduction Laboratories™) all diluted 1:200 in blocking buffer. Afterwards, filters were washed three times with immune blot wash buffer (TN buffer containing 0.01% Tween) and incubated for an additional hour with an anti-goat IgG HRP-labeled secondary antibody (Sigma–Aldrich) diluted 1:160,000 in blocking buffer or with an anti-mouse IgG/anti-rabbit IgG horseradish peroxidase (HRP)-labeled secondary antibody (Roche) diluted 1:10,000 in the blocking buffer, respectively. Finally, the blots were washed again three times with immune-blot washing buffer before detection of light emission with the BM chemiluminescence immune blotting kit (Roche Applied Science) as described by the supplier. Chemiluminescence was measured with an image acquisition system (Vilber-Lourmat, France).
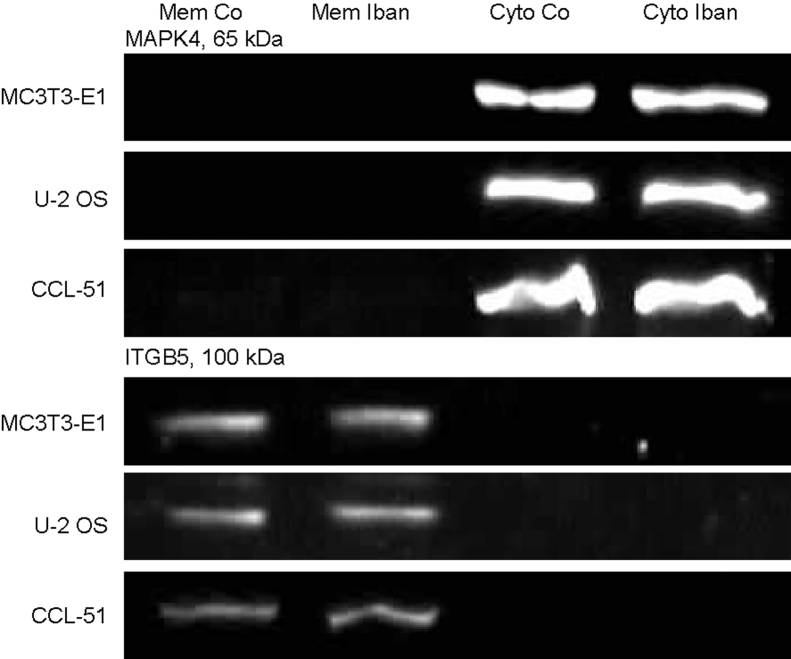
Cell fractionation was controlled by immunoblotting using antibodies against integrin-β5 (ITGB5, H-96, Santa Cruz Biotechnology, Santa Cruz, CA) for the membrane and mitogen-activated protein kinase 4 (MAPK4, Transduction Laboratories, BD, Franklin Lakes, NY) for the cytosolic fraction (Suppl. Fig. 3).
2.8. Specific promoter methylation
To analyze Fas promoter 5-methylcytosine levels, appropriate fragments of the targeted promoter regions were generated by digestion of 1 μg of genomic DNA with 20 U of the CpG methylation insensitive restriction enzymes MboII (Promega) or PstI (Fermentas, St. Leon-Rot, Germany) (for the murine or for the human Fas gene, respectively) for 1 h at 37 °C from cells cultured for 24, 48 and 72 h with 50 μM ibandronate in the medium. Subsequently, the enzyme was heat inactivated at 65 °C for 20 min. The Fas promoter region was selected as shown before [39]. In brief, in the murine cell lines two CpG rich regions were analyzed, the first at 2.6 kb and the second starting at 30 bp upstream from the Fas transcriptional start site (TSS) [17,19]. In the human osteosarcoma cell line, a CpG rich region of the proximal FAS promoter region was analyzed [19]. This fragment spanned from 79 bp upstream from the TSS into 359 bp of the first exon and showed an overall CpG content above 50%. PstI digestion generated a suitable fragment of 942 bp covering the CpG rich region. After digestion, DNA was purified using a commercially available PCR clean-up kit following the supplier's instructions. In the next step methylated DNA fragments were captured using the “MethylMiner Methylated DNA Enrichment Kit” (Invitrogen). In brief, methylated DNA was captured by methyl-CpG binding domain protein 2 (MBD2) coupled to magnetic beads to separate the modified from the non-modified DNA fractions. DNA was eluted from the magnetic beads with 200 μL of 2 M NaCl solution as single fraction independent from the CpG methylation density and concentrated by ethanol precipitation. Finally, the mean methylation status of the fragments was determined by amplifying the fragments by quantitative real-time PCR. Amplification ratios of the bound (methylated) DNA fraction to unbound (unmethylated) DNA fraction were calculated (for primer design see Table 1).
2.9. DNMT1 chromatin immune precipitation (ChIP) on Fas promoter
For this purpose, cells were treated with 50 μM ibandronate for 72 h. Subsequently, chromatin cross-linking, cell lysis, chromatin sharing, DNMT1 immune precipitation and DNA clean up were performed with the ChampionChip one-day kit (SABiosciences, Hilden, Germany) following manufacturer's instructions. Thereby, chromatin from untreated as well as from ibandronate treated cells was incubated overnight on a rotor at 4 °C with 4 μg of anti-DNMT1 antibody (K-18, Santa Cruz Biotechnology, Santa Cruz, CA) or with 4 μg of non-immune serum as negative control. Before immune precipitation, 1% (10 μL) of the chromatin was saved and stored at 4 °C for further use as reference. DNMT1 binding on appropriate FAS promoter regions was measured by qRT-PCR (for ChIP-FAS promoter primers see Table 1). For quantitation of the qRT-PCR values, for each sample, DNA signal of the DNMT1-precipitated chromatin was normalized to the unprecipitated chromatin.
2.10. Statistical analysis
Statistical analyses were performed either with ANOVA or with Student's t-test using Prism 4.03 (GraphPad Software, San Diego, CA), P ≤ 0.05 was considered as significant. The data of the experiments are presented as means ± standard deviation (SD).
3. Results
3.1. Effects of ibandronate on cell multiplication and caspases activities
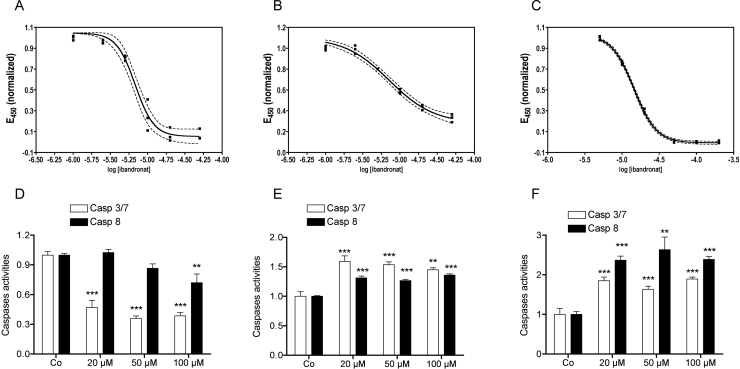
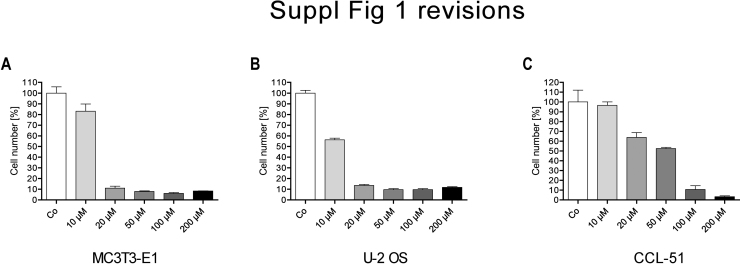
After 72 h of treatment, ibandronate attenuated cell multiplication in all cell lines tested, although with different half maximal effective concentrations (EC50, Fig. 1A–C). Using a MTT-like assay to assess cell viability/proliferation, in the bone related cells MC3T3-E1 (Fig. 1A) and U-2 OS (Fig. 1B), ibandronate showed a comparable half maximal effect (EC50) with 6.8 μM (95% c.i.: 6.00–7.67 μM) and 7.21 μM (95% c.i.: 5.90–8.79 μM), respectively, while the CCL-51 cells (Fig. 1C) were less sensitive with an EC50 of 14.4 μM (95% c.i.: 14.0–14.9 μM). Using cell counting only approximate similarities were found for the three cell-lines. This may be explained by the strong changes in cell morphology after ibandronate treatment, which makes it difficult to determine, if a cell is still viable (Suppl. Fig. 1A–C).
Fig. 1.
Ibandronate attenuated cell viability/proliferation and regulated caspases 3/7 and 8. After 3 days treatment ibandronate attenuated cell multiplication with a half maximal effect (EC50) at 6.8 μM (95% c.i.: 6.00–7.67 μM) in MC3T3-E1 cells (A) and 7.21 μM (95% c.i.: 5.90–8.79 μM) U-2 OS cells (B), while in CCL-51 a slightly higher concentration of 14.4 μM (95% c.i.: 14.0–14.9 μM) (C) was necessary to show the same effect. After the same treatment time, ibandronate down regulated caspase 3/7 in MC3T3-E1 cells significantly already at a concentration of 20 μM, while a significant down-regulation of 8 was found not until a concentration of 100 μM (D). In U-2 OS (E) and CCL-51 (F), however, ibandronate up regulated both caspases at all concentrations tested. Bars represent mean ± SD; **P ≤ 0.01; ***P ≤ 0.001; n = 4.
In MC3T3-E1 cells, caspase 8 and particularly caspase 3/7 activities were reduced after 72 h of ibandronate treatment in a concentration dependent manner showing significance at 20 μM for caspase 3/7 and at 100 μM for caspase 8 (Fig. 1D). An opposite effect was seen in the two tumor cell lines analyzed: caspase 8 as well as caspase 3/7 activities were significantly increased after 72 h of ibandronate treatment in human U-2 OS osteosarcoma cells (Fig. 1E) as well as in murine CCL-51 breast tumor cells (Fig. 1F).
3.2. Ibandronate up regulated FAS expression in a time and dose dependent manner, which was responsible for down regulation of cell viability/proliferation
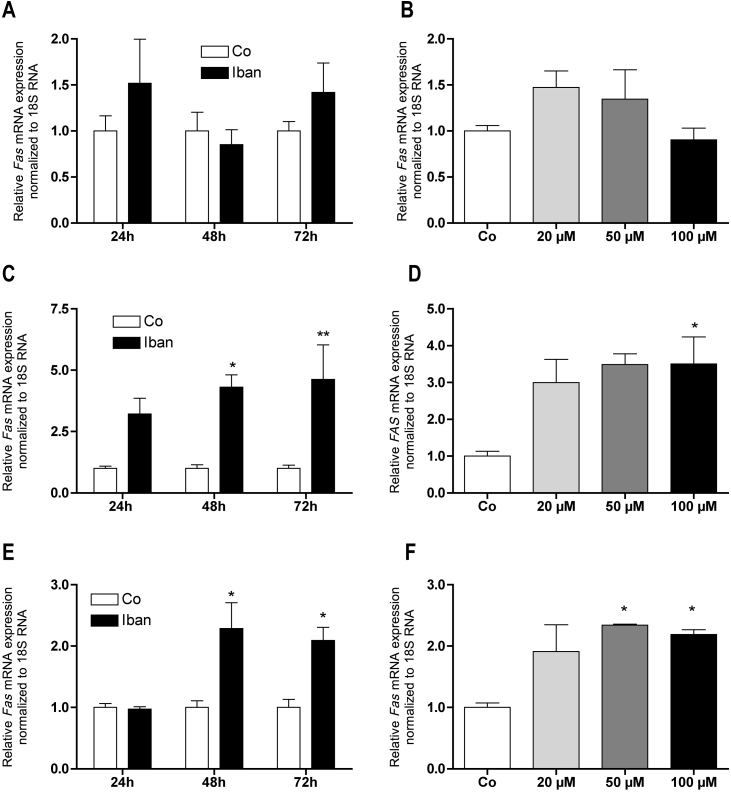
In line with the effects observed on caspase activities, ibandronate failed to significantly regulate the mRNA expression of the pro-apoptotic gene Fas in MC3T3-E1 cells at any time (Fig. 2A) and after 72 h at any concentration tested (Fig. 2B). In contrast, already after 24 h of treatment, FAS mRNA expression was strongly induced in U-2 OS cells (Fig. 2C) and after 48 h in CCL-51 cells (Fig. 2E). This stimulation was dose dependent but showed significance only at concentrations of 100 μM in U-2 OS cells (Fig. 2D) or higher than 20 μM in CCL-51 cells (Fig. 2F).
Fig. 2.
Ibandronate up regulated FAS expression in U-2 OS and CCL-51 but not in MC3T3-E1 cells (A and B). At 100 μM ibandronate up regulated FAS expression in U-2 OS already after 24 h (C) while in CCL-51 not until 48 h (E). In both cell lines the regulation was dose dependent, which reached significance at 100 μM in U-2 OS (D) and at 50 μM in CCL-51 (F). Bars represent mean ± SD; *P ≤ 0.05; **P ≤ 0.01; n = 3.
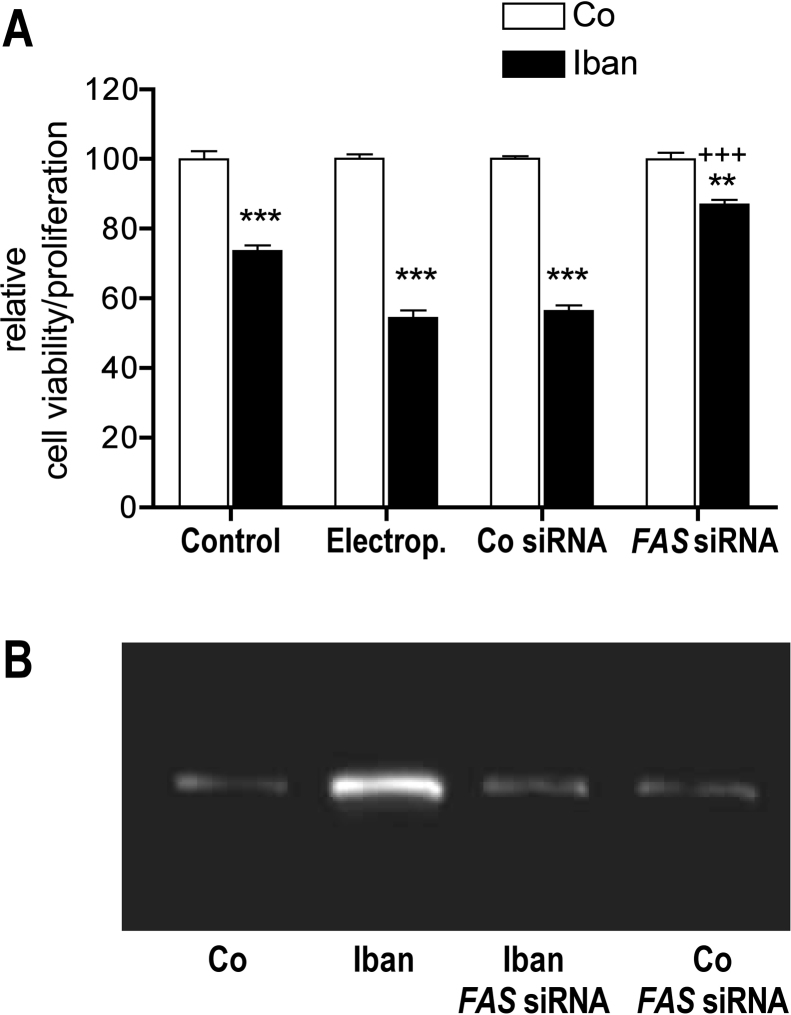
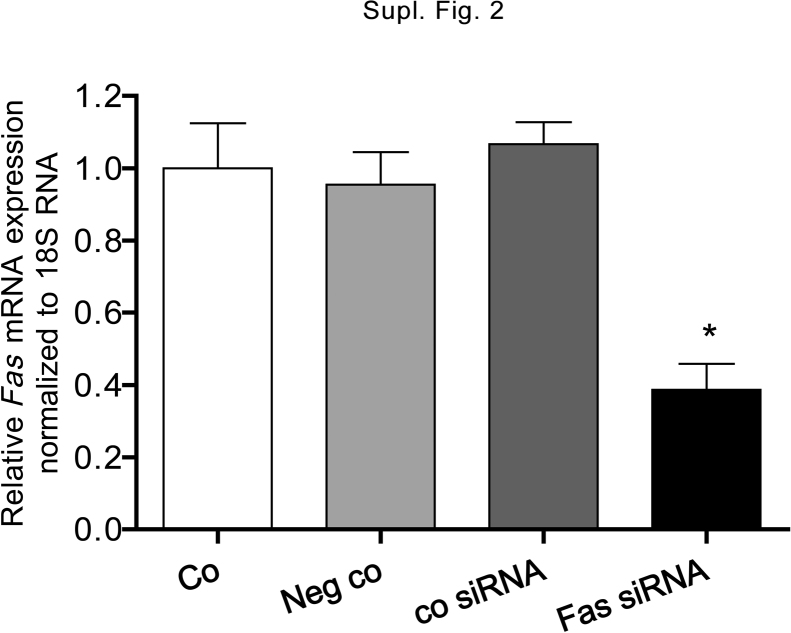
To demonstrate the role of FAS in ibandronate induced apoptosis, U-2 OS cells were transfected with FAS siRNA by electroporation and cell viability/proliferation was measured with the MTT-like assay (Fig. 3A). In ibandronate treated cells, a difference of 19.2% in down regulation of cell viability/proliferation was measured between electroporation of unexposed and electroporation of exposed cells, which suggest that electroporation makes cells more sensitive to ibandronate. This assumption is supported by a similar effect found by transfection of the control siRNA. Transfection of FAS siRNA recovered cell viability/proliferation to a large extent (87%), suggesting that a second, minor process is involved in down-regulation of cell viability/proliferation too. FAS knock down by siRNA was controlled by FAS mRNA expression (Suppl. Fig. 2).
Fig. 3.
Ibandronate down regulated cell viability/proliferation and up regulated FAS, which could be rescued by transfection of FAS siRNA. (A) After 3 days of treatment with ibandronate (7.2 μM, EC50) cell viability/proliferation was down regulated to 73.6% (control). After electroporation without siRNA ibandronate down regulated cell viability/proliferation to 54.4% (Electrop.) a comparable value, which was found after electroporation of a control siRNA (56.4%, Co siRNA). Electroporation of FAS siRNA resulted in attenuation of down-regulation of cell viability/proliferation to 87.0% demonstrating the FAS regulation on this parameter. The difference of 13.0% suggests a minor second mechanism on regulation of cell viability/proliferation. Bars represent mean ± SD; **P ≤ 0.01; ***P ≤ 0.001; untreated vs. Iban treated; +++P ≤ 0.001; Co siRNA vs. FAS siRNA; n = 3. (B) The immune-blot with anti-FAS antibody demonstrates that Ibandronate up regulated FAS protein expression as well. Transfection with FAS siRNA down regulated ibandronate stimulated FAS protein expression having no considerable effect on basal FAS expression.
Translation of the FAS mRNA expression to the protein level is demonstrated in Fig. 3B. Protein extracts isolated from FAS siRNA transfected cells, which were either left untreated or treated with ibandronate for 72 h, were subjected to immune-blotting and probed with an anti-FAS antibody. Similarly to mRNA regulation, when compared to controls, ibandronate up regulated FAS protein expression. FAS siRNA transfected cells, however, showed no significant difference to controls, neither untreated nor ibandronate treated.
3.3. Differential regulation of Dnmt1 expression by ibandronate
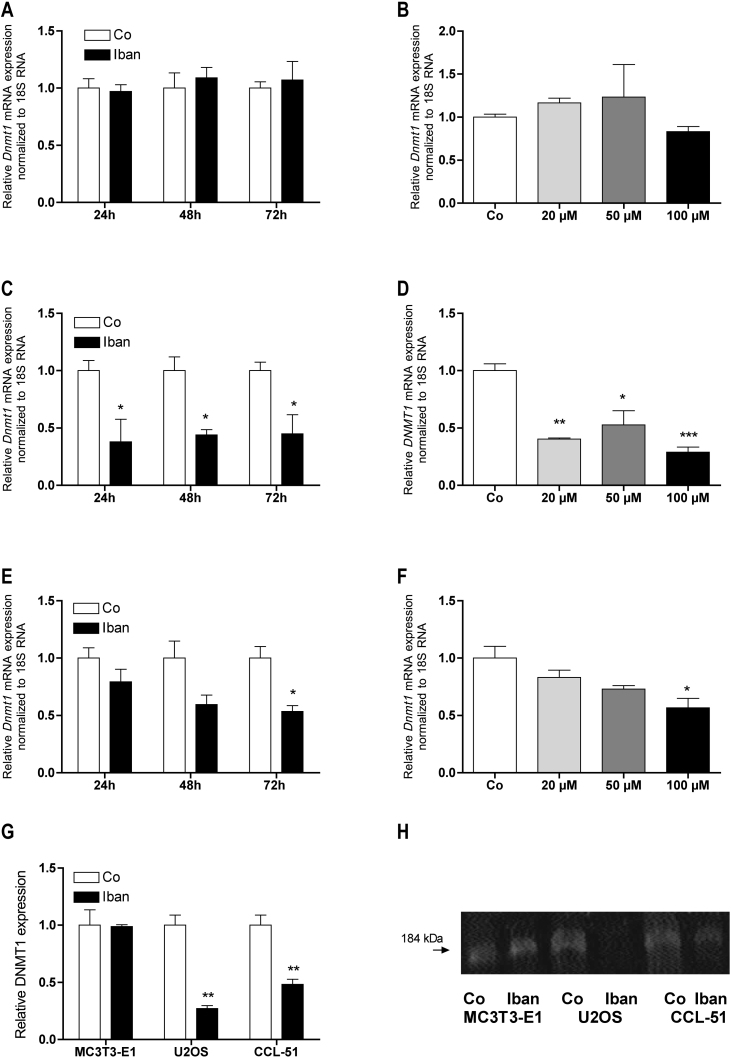
Similar to Fas expression, ibandronate failed to affect Dnmt1 expression in MC3T3-E1 cells at any time (Fig. 4A) and after 72 h at any concentration (Fig. 4B) tested. However, Dnmt1 expression was clearly down regulated in both tumor cell lines. In U-2 OS cells this effect was already visible after 24 h (Fig. 4C), while in CCL-51 cells significance was reached after 72 h of treatment with 50 μM ibandronate (Fig. 4E). At this time, in U-2 OS cells, DNMT1 expression was already strongly down regulated at 20 μM ibandronate in culture medium (Fig. 4D) whereby a comparable effect was reached at 100 μM ibandronate for CCL-51 cells (Fig. 4F). Fig. 6G and H compares the effect of 72 h of treatment with 50 μM ibandronate on DNMT1 protein expression by immune blot analysis in the three cell lines studied.
Fig. 4.
Ibandronate down regulated DNMT1 expression in U-2 OS and CCL-51 but not in MC3T3-E1 cells (A and B). At 100 μM ibandronate down regulated DNMT1 expression in U-2 OS already after 24 h (C) while in CCL-51 attenuation did not reach significance until 72 h (E). In both cell lines the regulation was dose dependent, which reached significance already at 20 μM in U-2 OS (D) and but not until 100 μM in CCL-51 (F). DNMT1 was also down regulated at the protein level as demonstrated by immuno-blot. Intensity measurements of 3 immune-blots revealed a significant down-regulation only for U-2 OS and CCL-51 cells but not for MC3T3-E1 cells (G). A representative blot is shown in (H). Bars represent mean ± SD; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n = 3.
Fig. 6.
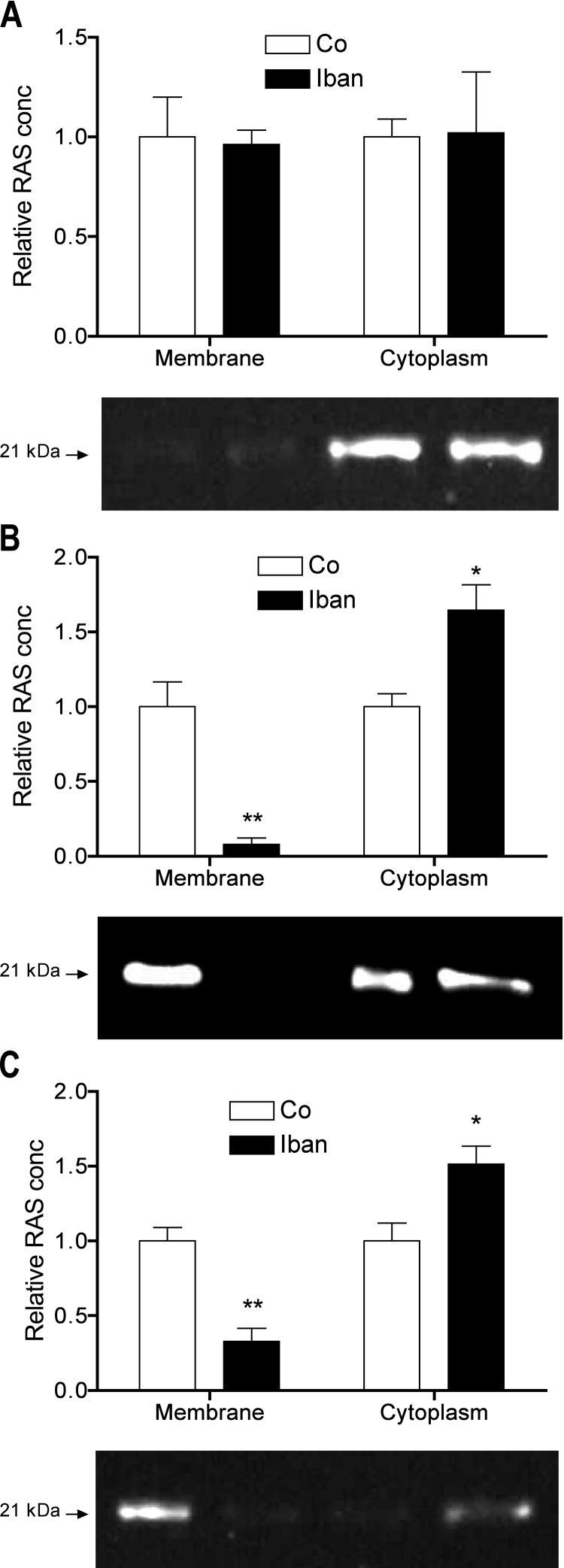
Ibandronate translocated RAS from the membranes to the cytoplasm in U-2 OS (B) and CCL-51 (C) but not in MC3T3-E1 cells (A). Intensity measurements of 3 immune-blots revealed a significant change in the localization of RAS in U-2 OS and CCL-51 cells but not in MC3T3-E1 cells (A). For each cell line a representative blot is shown. Bars represent means normalized to the signal of untreated cells ± SD; *P ≤ 0.05; **P ≤ 0.01; n = 3.
3.4. Fas promoter methylation was altered in tumor cells after ibandronate treatment
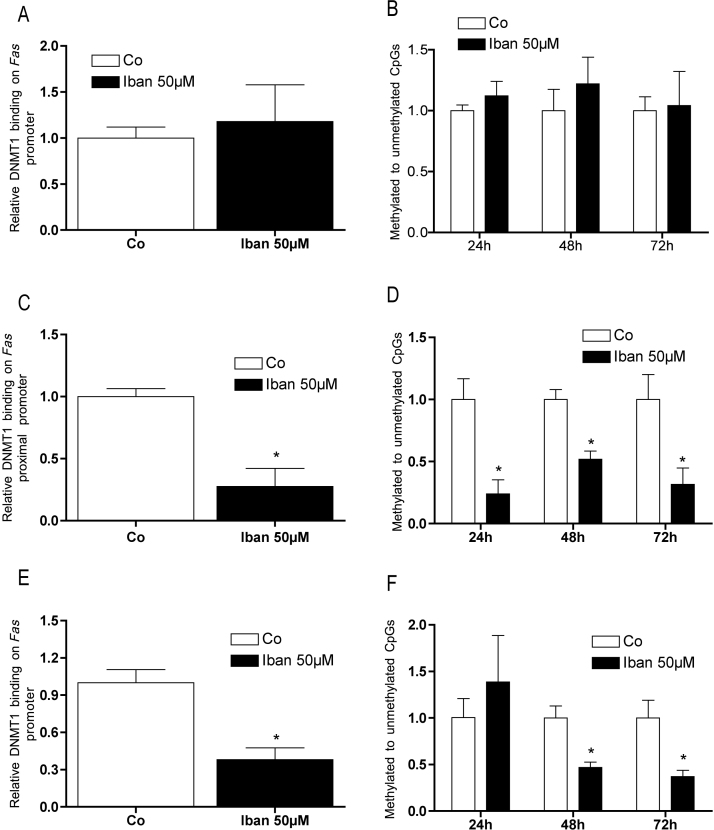
72 h after seeding, in MC3T3-E1 cells basal Fas promoter methylation was considerably lower as in the two neoplastic cell lines (Table 2). Furthermore, in MC3T3-E1 cells, 72 h of 50 μM ibandronate treatment failed to significantly affect DNMT1 binding on Fas promoter (Fig. 5A). After 72 h, in U-2 OS cells (Fig. 5C) as well as in CCL-51 cells (Fig. 5E) treatment with ibandronate significantly reduced binding of DNMT1 to the FAS promoter.
Table 2.
Methylation levels (ratio methylated vs. unmethylated) of the FAS promoters in the studied cell lines.
| Cell line | Methylation level |
|---|---|
| MC3T3-E1 | 0.0021 ± 0.012 |
| CCL-51 | 15.3 ± 4.2 |
| U2OS | 3.10 ± 0.63 |
Fig. 5.
DNMT1 bound to the FAS promoter, which resulted in change of the DNA methylation status in U-2 OS and CCL-51 but not in MC3T3-E1 cells (A and B). 50 μM ibandronate significantly reduced binding of DNMT1 to the promoter of FAS in U-2 OS cells (C) which resulted in a decreased ratio of methylated by unmethylated cytosines already after 24 h (D). In CCL-51 cells ibandronate attenuated binding of DNMT1 to the Fas promoter as well (E) but it decreased methylation not until 48 h treatment (F). Bars represent mean ± SD; *P ≤ 0.05; n = 3.
Reduced binding of DNMT1 to the Fas promoter suggested changes in DNA methylation, which was evaluated by comparing the concentration of methyl-cytosines vs. unmethylated cytosines in the promoter. While as expected, no change in the methylation status of the Fas promoter in MC3T3-E1 cells was found (Fig. 5B), ibandronate significantly reduced the methyl-cytosine concentration of the Fas promoter in U-2 OS already after 24 h (Fig. 5D) and after 48 h in CCL-51 cells (Fig. 5F).
3.5. Ibandronate altered RAS localization in the analyzed tumor cell-lines
Bisphosphonates inhibit the activities of the enzymes farnesyl- and geranylgeranyl-diphosphate synthase, which cause disruption of small-GTPases prenylation thus altering their sub-cellular distribution. Cells were either left untreated or treated with ibandronate. Thereafter, proteins of the membrane as well as of cytoplasmatic fraction were isolated and subjected to immune blotting. As shown in Fig. 6A, in MC3T3-E1 cells RAS was primarily localized in the cytoplasm and, therefore, ibandronate did not affect RAS localization in this cell-line. In U-2 OS cells (Fig. 6B) as well as in CCL-51 cells (Fig. 6C) RAS was primarily localized in cell membranes. After exposure to 50 μM ibandronate for 72 h, in both cell lines a clear shift in RAS localization from the cell membranes to cytoplasm was seen (Fig. 6B and C).
Cell fractionation was controlled by immune blotting of integrin-β5 for the membrane and mitogen-activated protein kinase 4 for the cytosolic fraction (Suppl. Fig. 3).
3.6. Bisphosphonate induced apoptosis of tumorigenic cells was differentially rescinded by GGPP and FPP
Inhibition of prenylation of small-GTPases by bisphosphonates can be rescued by parallel treatment with the substrate FPP for RAS or GGPP for the other modified small-GTPases.
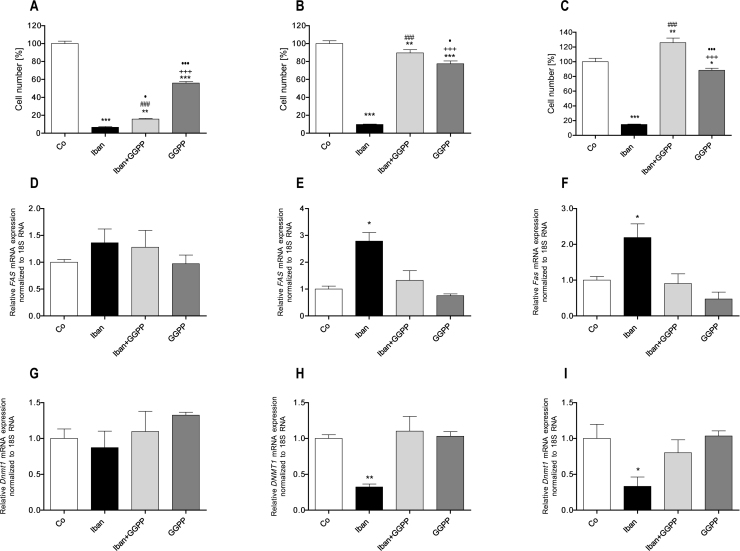
As already shown in Fig. 1 and Supplemental Fig. 1, treatment of MC3T3-E1 cells with 50 μM ibandronate reduced the cell number by about 90%. After the same culture time 10 μM GGPP by itself attenuated cell multiplication by about 50% and could only partially rescue the effect of ibandronate on these cells (Fig. 7A). In U-2 OS cells 50 μM ibandronate significantly reduced cell multiplication. Although GGPP itself reduced cell multiplication by about 25%, it could rescue the effect caused by ibandronate (Fig. 7B). Similarly, in CCL-51 cells GGPP could rescue ibandronate attenuated cell multiplication but, differentially, did only marginally influence cell multiplication (Fig. 7C). In MC3T3-E1 cells neither ibandronate nor GGPP influenced Fas expression (Fig. 7D). Again, in both other studied cell lines ibandronate induced FAS expression. GGPP alone had no significant effect on FAS expression, but down regulated ibandronate induced FAS expression in both U-2 OS (Fig. 7E) as well as in CCL-51 cell lines (Fig. 7F). Regarding DNMT1 expression, again all three treatments (ibandronate, GGPP or combination of both) showed no effect in MC3T3-E1 cells (Fig. 7G). However in both tumor cell lines, ibandronate, as expected, down regulated expression of DNMT1. GGPP counteracted this effect and had no effect when administered singularly (Fig. 7H and I). This supports our hypothesis that small-GTPases induce DNMT1 expression is inhibited by bisphosphonates leading to promoter demethylation and FAS expression.
Fig. 7.
Effects of GGPP on ibandronate regulated cell multiplication and expression of FAS and DNMT1. Ibandronate down regulated cell multiplication in all three cell lines tested. In MC3T3-E1 cells, GGPP itself attenuated cell multiplication but could not prevent ibandronate's down-regulation (A). In U-2 OS (B) and CCL-51 (C) cells, GGPP rescued down-regulation of cell multiplication. Moreover, the combination of both drugs increased cell multiplication in CCL-51 cells significantly (C). Neither ibandronate nor GGPP had an effect of Fas (D) and Dnmt1 (G) expression in MC3T3-E1 cells. In U-2 OS cells, GGPP rescued ibandronate's up-regulation of FAS (E) or down-regulation of DNMT1 (H), respectively. Equally, in CCL-51 cells, GGPP rescued ibandronate's up-regulation of Fas (F) or down-regulation of Dnmt1 (I), respectively. Bars represent mean ± SD; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 treatment vs. Co; +++P ≤ 0.001 GGPP vs. Ibn; •P ≤ 0.05; •••P ≤ 0.001 GGPP vs. Ibn + GGPP; ###P ≤ 0.001 Ibn vs. Ibn + GGPP; n = 3.
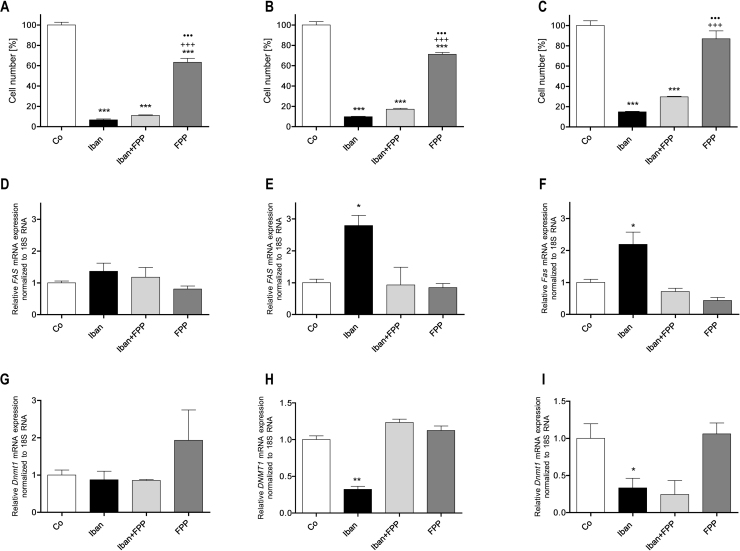
Next we tested the effects of FPP (40 μM) on cell multiplication (Fig. 8). In the immortalized MC3T3-E1 cell line (Fig. 8A), FPP reduced cell number to 63%, in U-2 OS cells to 71% (Fig. 8B) while it did not affect this parameter in CCL-51 cells (Fig. 8C). However, in contrast to GGPP, FPP could not rescue the down-regulation of cell multiplication by ibandronate (Fig. 8A–C). The effects of FPP on FAS expression were comparable to that of GGPP: no significant effects in MC3T3-E1 cells (Fig. 8D), while in both neoplastic cell lines (Fig. 8E and F) 40 μM FPP rescued the ibandronate mediated up-regulation of FAS. Comparable effects were also found on DNMT1 expression with no effects in MC3T3-E1 cells (Fig. 8G), while in U-2 OS FPP rescued the down-regulation of DNMT1 expression by ibandronate (Fig. 8H). Differentially, FPP did not change the attenuation of Dnmt1 expression in CCL-51 cells (Fig. 8I). FPP alone did not affect DNMT1 expression in any one of these cell lines.
Fig. 8.
Effects of FPP on ibandronate regulated cell multiplication and expression of FAS and DNMT1. Ibandronate down regulated cell multiplication in all three cell lines tested. In MC3T3-E1 and U-2 OS cells, FPP itself attenuated cell multiplication (A and B) but had no effect on CCL-51 cells (C). FPP could not rescue cell multiplication in these cell lines. Neither ibandronate nor FPP had an effect on FAS (D) or DNMT1 (G) expression in MC3T3-E1 cells. In U-2 OS cells, FPP rescued ibandronate's up-regulation of FAS (E) or down-regulation of DNMT1 (H), respectively. In CCL-51 cells, FPP rescued ibandronate's up-regulation of Fas (F) but not down-regulation of Dnmt1 (I), respectively. Bars represent mean ± SD; *P ≤ 0.05; ***P ≤ 0.001 treatment vs. Co; +++P ≤ 0.001 FPP vs. Ibn; •••P ≤ 0.001 FPP vs. Ibn + FPP; n = 3.
4. Discussion
A growing body of evidence attributes an anti-tumorigenic capacity to bisphosphonates. Several studies have shown that in patients affected by diverse malignances, bisphosphonate therapies show a positive outcome with regard to tumor growth and metastatic activity [40–46]. However, the mechanisms underlying those effects remain poorly understood.
By activation, the Ras superfamily of small GTPases attaches to cellular membranes and, among many other cell regulatory functions, promotes cell proliferation, differentiation and survival [12,16,47]. Aminobisphosphonates exert their pharmacologic activity by inhibiting the mevalonate pathway. By this way, these drugs prevent prenylation of small-GTPases as well as their attachment to the cellular membranes and thereby their function [5,7,48]. While the apoptotic impact of simple bisphosphonates (clodronate and etidronate) can be considered as secured, the mode of action of aminobisphosphonates on cells is not as clear [9,11,49,50]. Initially, J774 macrophages have been considered as a model of aminobisphosphonates’ action because this cell line matched the order of potency of the diverse bisphosphonates for inhibiting bone resorption and inducing apoptosis [7,51–53]. However using primary rabbit or human derived osteoclasts, it has been demonstrated that inhibition of bone resorption by bisphosphonates does not require osteoclast apoptosis [54–56]. Taking into account that J744 are lymphoma-derived tumorigenic cells [57] one can conclude that bisphosphonates act in tumor cells in a different way as compared to normal osteoclasts.
This is in accordance with the findings in this study, where in non neoplastic immortalized mouse preosteoblastic MC3T3-E1 cells, ibandronate attenuated cell multiplication by down-regulation of the cell cycle as suggested by decreased expression of cyclins Ccn2a and Ccn2d (not shown). Moreover, down-regulation of caspase 3/7 and caspase 8 in MC3T3-E1 cells assigned to aminobisphosphonates an anti-apoptotic activity as found in MLO-Y4 osteocytes [58] while in tumorigenic RAW 264.7 macrophages most bisphosphonates induced apoptosis [59]. In primary human osteoblasts at concentrations higher than 10−7 M zoledronate dose dependently reduced cell proliferation, although, at the highest concentration apoptosis was found as well [37]. Treatment of fetal immortalized osteoblasts with pamidronate and zoledronate resulted in a dose dependent down-regulation of cell proliferation with increased differentiation [60]. However, increased cell proliferation of human primary trabecular bone cells [61] and of human bone marrow stromal cells by alendronate and risedronate has been reported as well [62]. Summarizing these findings, in normal mesenchymal bone cells, bisphosphonates decrease proliferation but apoptosis was found only at very high concentrations, while in tumorigenic cells amino-bisphosphonates induce apoptosis.
Here we show that treatment of tumor cell lines with ibandronate down regulated genes of the DNA CpG-methylation apparatus (Dnmt1, Dnmt3b, Hells), which resulted in reduced binding of DNMT1 at the promoter of FAS in both tumor cell lines. Absence of the DNA-methyltransferases at the promoter results in reduced CpG-methylation of the promoter during further cell divisions, which is followed by increased expression of the gene [63–65]. Increased expression of FAS is followed by increased activity of caspase 3/7 and caspase 8, which is a classical sign of apoptosis. Interestingly, no change of FAS methylation status and of Fas expression was found in non-neoplastic MC3T3-E1 cells. The very low basal methylation level of Fas promoter in these cells can explain this fact. We have recently demonstrated that culturing these cells on culture dishes that are not covered by collagen results in down-regulation of DNMT1 and HELLS followed by demethylation of the Fas promoter and increased expression of the gene [19].
Small-GTPases play a central role in bisphosphonates’ activity. These drugs inhibit the activity of the enzymes, which are responsible for synthesis of FPP and GGPP, which are transferred to the amino acids motif CAAX anchoring of the small-GTPases to the cell membrane, which is a prerequisite for their action. Effects of bisphosphonates on small-GTPases mediated signal transduction can be rescued by co-treatment with GGPP or FPP, where in general FPP rescues RAS-activity and GGPP the activity of the RHO- and RAB-family [66,67]. Again, transformed cell lines behaved differentially; in CCL-51 and U-2 OS, GGPP rescued ibandronate attenuated cell multiplication, up regulated DNMT1 and, consequently, down regulated FAS expression. GGPP, however, could not rescue the immortalized non-neoplastic cell line MC3T3-E1 from the effects of ibandronate. We found no down-regulation of Dnmt1 and no up-regulation of Fas. Moreover, GGPP alone significantly decreased cell multiplication as well. GGPP is the posttranslational modifier of the RHO- and RAB-small-GTPases. Recently, it has been demonstrated that inhibition of prenylation by bisphosphonates rather activates than inactivates the RHO family of small-GTPases including RAC, CDC42 and RHO [9]. Interestingly, osteoblast-restricted Rac1 deletion leads to defective bone acquisition in vivo and knockdown impairs growth and induces apoptosis in the osteoblast cell line OP9 [68]. These data suggest that GGPP increases interaction of RAC1 with cellular membranes, which results in reduced activated RAC1 leading to decreased proliferation and increased apoptosis.
A down-regulation of cell multiplication was also found in mesenchymal MC3T3-E1 and U-2 OS cells by FPP but not in the neoplastic cell line resulting from an epithelial tumor. Down-regulation of cell multiplication by FPP suggests the involvement of RAS. After translation, FAS is farnesylated and transported to the endoplasmatic reticulum where it is prepared for palmitoylation and forwarded to the Golgi-membranes. This initiates a cycle of traffic to and from the plasma membranes, where it is activated [13,16]. Higher cellular concentrations of FPP could influence the equilibrium of this cycle and therefore alter differentiation, proliferation and apoptosis rate.
Unlike GGPP, FPP could not rescue down-regulation of cell multiplication by ibandronate in U-2 OS and CCL-51. This suggests that RHO- and RAB-small-GTPases are responsible for ibandronate's effect on this parameter. This is surprising because in U-2 OS, FPP canceled the effect on DNMT1 expression and inhibited up-regulation of FAS. An explanation is down-regulation of the cell cycle by ibandronate via down-regulation of cyclins and a possible up-regulation of cell cycle inhibitors [68–71]. From the rescue-experiments one can conclude that ibandronate inhibits cell multiplication by decreasing cell cycling and by increasing apoptosis. Apart from apoptosis a second process was also suggested by transfection of FAS siRNA, which could not completely recover ibandronate's down-regulation of cell viability/proliferation. However, this experiment strengthens our hypothesis that up-regulation of FAS is responsible for induction of apoptosis via caspases and confirms recent data obtained from HMC1 mast cell leukemia cells that were treated with demethylating drugs [34].
Although FPP and GGPP differentially influenced cell multiplication, unexpectedly, except for Dnmt1 expression in CCL-51, no differences were found in rescuing FAS and DNMT1 expression; both isoprenoids could recover the effects of ibandronate, which is in line with recent findings that both isoprenoids are able to recover bisphosphonates-induced apoptosis. There are several possible explanations for this finding [11]. Bisphosphonates are, with some exceptions [72,73], inhibitors of FPP-synthase but not of the GGPP-synthase. Therefore, when cells were treated with FPP some of the compounds will be converted to GGPP, which will modify and rescue RHO-family small-GTPases as well, at least partially [11]. Recently, it has been demonstrated that in presence of inhibition of farnesyltransferase, geranylgeranylation occurs instead of farnesylation of KRAS4A,B and NRAS but not for HRAS. Moreover, geranylgeranyltransferase can use FPP as substrate as well to modify proteins of the RHO-family [74–80]. As bisphosphonates deplete both FPP and GGPP, raising the concentration of one of the isoprenoids by addition, modification with the “wrong” modifier may occur. These facts may support a critical view on the implication of the rescue experiments, so that no unambiguous assignment of a small-GTPase can be made. Even though the presented results clearly demonstrate the involvement of small-GTPases in the epigenetic DNA methylation process and FAS mediated apoptosis, a detailed analysis of the large family of small-GTPases will be necessary to unravel the mechanism.
Taken together, our experiments support the common knowledge that more members of the group of small-GTPases are involved in the regulation of cell multiplication and apoptosis and suggest a different mode of action of bisphosphonates in non-transformed vs. neoplastic cell lines, as supported by previous findings [10,81].
As already mentioned above, most tumors or transformed neoplastic cells are characterized by an activated RAS/MAPK pathway [13–15], which results in methylation of cytosines in the promoter of tumor suppressor genes and of the pro-apoptotic FAS gene [12,24,30,32,33,82,83]. Both tumor cell lines, however, have a highly methylated FAS promoter, and therefore, an inhibited FAS expression. This suggests that RAS must be activated and attached to the cell membrane to enable bisphosphonates to display their epigenetic activity. Ibandronate, as demonstrated by immune-blotting, prevented RAS from attachment to membranes and translocated the protein into the cytoplasm; no change was found in MC3T3-E1 cells indicating that RAS is not attached to membranes, and therefore not or only marginally active, as generally found in normal, growth factor-unstimulated cells [47,84]. Our results could explain why tumor cells react differentially in response to bisphosphonates when compared with normal cells. It should be emphasized again that activation RAS/MAPK pathway seems to be a prerequisite for bisphosphonates’ action in tumor cells. Featuring the RAS/MAPK pathway is well founded by the fact that epigenetic silencing of the Fas promoter is a consequence from the signaling cascade of this pathway [17,39]. However, results of this study suggest that in addition to RAS, other members of small-GTPases must be involved in regulation of FAS promoter methylation as well. Differences in responses to ibandronate of the epithelial CCL-51 cells compared to the mesenchymal cells suggest involvement of RAC1, a gene that has been found important for osteoblastic proliferation, differentiation and apoptosis [68]. The use of specific inhibitors of small-GTPases will help to clarify the mechanism of the selective epigenetic activation of FAS and other epigenetic silenced tumor suppressor genes [8,15]. It should be emphasized that other inhibitors of small-GTPases may demonstrate epigenetic mechanisms as well.
A recent publication has demonstrated that bisphosphonate pulse treatment differentially affects mesenchymal stem cells [66]. Passive demethylation of a gene promoter needs at least one or two cell cycles. Short treatment of cells will activate promoter demethylation, which then will be inherited to the daughter cells, where apoptotic factors could further activate the demethylated FAS gene.
Methylation of tumor suppressor genes is a common mechanism in tumor development. Loss of hormone responsiveness in breast and prostate cancer as well as in leukemias may contribute to increased mortality that often happens by CpG-methylation of promoter DNA [25,26,28,29,85,86] and drugs decreasing CpG-methylation are promising for future treatment of such tumors. Our findings could possibly explain some contradictory results in clinical trials using bisphosphonates combined with an adjuvant endocrine therapy [40,87]; only a subgroup of patients with an epigenetic inactivated estrogen receptor ESR1 seems to respond to this therapy via RAS/MAPK pathway.
In summary, we demonstrated that ibandronate is an epigenetically active drug. In tumor cells it inhibits activated RAS and down-regulates DNMT1 expression. This leads to demethylation of CpGs in the FAS promoter resulting in activation of FAS transcription. Subsequently apoptosis is induced in the affected neoplastic cells. In contrast, ibandronate does not exert these effects in non-neoplastic (immortalized) cell lines.
Epigenetic mechanisms play an important role in multiple physiological and pathological processes [19]. Besides their use as antiresorptive treatment for osteoporosis, bisphosphonates as epigenetically active drugs with the potential to re-activate pro-apoptotic genes, could gain a consolidated role in antitumor treatment.
Conflicts of interest
The authors state that they have no conflicts of interest.
Acknowledgments
This study was supported by the Fonds zur Foerderung der Wissenschaftlichen Forschung (FWF; The Austrian Science Fund) Project P24370-B19, the Medical Scientific Fund of the Mayor of the City of Vienna (Project 2565), the WGKK (Social Health Insurance Vienna), and the AUVA (Austrian Social Insurance for Occupational Risks).
References
- 1.Caraglia M., Santini D., Marra M., Vincenzi B., Tonini G., Budillon A. Emerging anti-cancer molecular mechanisms of aminobisphosphonates. Endocr Relat Cancer. 2006;13:7–26. doi: 10.1677/erc.1.01094. [DOI] [PubMed] [Google Scholar]
- 2.Baron R., Ferrari S., Russell R.G. Denosumab and bisphosphonates: different mechanisms of action and effects. Bone. 2011;48:677–692. doi: 10.1016/j.bone.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Graaf M.R., Richel D.J., van Noorden C.J., Guchelaar H.J. Effects of statins and farnesyltransferase inhibitors on the development and progression of cancer. Cancer Treat Rev. 2004;30:609–641. doi: 10.1016/j.ctrv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Kucukzeybek Y., Gorumlu G., Cengiz E., Karabulut B., Sezgin C., Atmaca H. Apoptosis-mediated cytotoxic effects of ibandronic acid on hormone- and drug-refractory prostate cancer cells and human breast cancer cells. J Int Med Res. 2010;38:1663–1672. doi: 10.1177/147323001003800511. [DOI] [PubMed] [Google Scholar]
- 5.Luckman S.P., Hughes D.E., Coxon F.P., Graham R., Russell G., Rogers M.J. Nitrogen-containing bisphosphonates inhibit the mevalonate pathway and prevent post-translational prenylation of GTP-binding proteins, including Ras. J Bone Miner Res. 1998;13:581–589. doi: 10.1359/jbmr.1998.13.4.581. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson S., Huelsenbeck J., Fritz G. Mevalonate pathway inhibitors affect anticancer drug-induced cell death and DNA damage response of human sarcoma cells. Cancer Lett. 2011;304:60–69. doi: 10.1016/j.canlet.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 7.Rogers M.J., Crockett J.C., Coxon F.P., Monkkonen J. Biochemical and molecular mechanisms of action of bisphosphonates. Bone. 2011;49:34–41. doi: 10.1016/j.bone.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Walker K., Olson M.F. Targeting Ras Rho GTPases as opportunities for cancer therapeutics. Curr Opin Genet Dev. 2005;15:62–68. doi: 10.1016/j.gde.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Dunford J.E., Rogers M.J., Ebetino F.H., Phipps R.J., Coxon F.P. Inhibition of protein prenylation by bisphosphonates causes sustained activation of Rac, Cdc42, and Rho GTPases. J Bone Miner Res. 2006;21:684–694. doi: 10.1359/jbmr.060118. [DOI] [PubMed] [Google Scholar]
- 10.Reszka A.A., Halasy-Nagy J., Rodan G.A. Nitrogen-bisphosphonates block retinoblastoma phosphorylation and cell growth by inhibiting the cholesterol biosynthetic pathway in a keratinocyte model for esophageal irritation. Mol Pharmacol. 2001;59:193–202. doi: 10.1124/mol.59.2.193. [DOI] [PubMed] [Google Scholar]
- 11.Benford H.L., Frith J.C., Auriola S., Monkkonen J., Rogers M.J. Farnesol and geranylgeraniol prevent activation of caspases by aminobisphosphonates: biochemical evidence for two distinct pharmacological classes of bisphosphonate drugs. Mol Pharmacol. 1999;56:131–140. doi: 10.1124/mol.56.1.131. [DOI] [PubMed] [Google Scholar]
- 12.Vigil D., Cherfils J., Rossman K.L., Der C.J. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy. Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo F., Agudo-Ibanez L., Crespo P. The Ras-ERK pathway: understanding site-specific signaling provides hope of new anti-tumor therapies. Bioessays. 2010;32:412–421. doi: 10.1002/bies.200900155. [DOI] [PubMed] [Google Scholar]
- 14.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montagut C., Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Ahearn I.M., Haigis K., Bar-Sagi D., Philips M.R. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazin C., Wajapeyee N., Gobeil S., Virbasius C.M., Green M.R. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patra S.K., Szyf M. DNA methylation-mediated nucleosome dynamics and oncogenic Ras signaling: insights from FAS, FAS ligand and RASSF1A. FEBS J. 2008;275:5217–5235. doi: 10.1111/j.1742-4658.2008.06658.x. [DOI] [PubMed] [Google Scholar]
- 19.Thaler R., Agsten M., Spitzer S., Paschalis E.P., Karlic H., Klaushofer K. Homocysteine suppresses the expression of the collagen cross-linker lysyl oxidase involving IL-6, Fli1, and epigenetic DNA methylation. J Biol Chem. 2011;286:5578–5588. doi: 10.1074/jbc.M110.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delcuve G.P., Rastegar M., Davie J.R. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- 21.Huang K., Fan G. DNA methylation in cell differentiation and reprogramming: an emerging systematic view. Regen Med. 2010;5:531–544. doi: 10.2217/rme.10.35. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 23.Petak I., Danam R.P., Tillman D.M., Vernes R., Howell S.R., Berczi L. Hypermethylation of the gene promoter and enhancer region can regulate Fas expression and sensitivity in colon carcinoma. Cell Death Differ. 2003;10:211–217. doi: 10.1038/sj.cdd.4401132. [DOI] [PubMed] [Google Scholar]
- 24.Santourlidis S., Warskulat U., Florl A.R., Maas S., Pulte T., Fischer J. Hypermethylation of the tumor necrosis factor receptor superfamily 6 (APT1, Fas, CD95/Apo-1) gene promoter at rel/nuclear factor kappaB sites in prostatic carcinoma. Mol Carcinog. 2001;32:36–43. doi: 10.1002/mc.1062. [DOI] [PubMed] [Google Scholar]
- 25.Reibenwein J., Pils D., Horak P., Tomicek B., Goldner G., Worel N. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67:427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 26.Sasaki M., Tanaka Y., Perinchery G., Dharia A., Kotcherguina I., Fujimoto S. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J Natl Cancer Inst. 2002;94:384–390. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 27.Brinkman J.A., El-Ashry D. ER re-expression and re-sensitization to endocrine therapies in ER-negative breast cancers. J Mammary Gland Biol Neoplasia. 2009;14:67–78. doi: 10.1007/s10911-009-9113-0. [DOI] [PubMed] [Google Scholar]
- 28.Fackler M.J., Umbricht C.B., Williams D., Argani P., Cruz L.A., Merino V.F. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res. 2011;71:6195–6207. doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vesuna F., Lisok A., Kimble B., Domek J., Kato Y., van der Groep P. Twist contributes to hormone resistance in breast cancer by downregulating estrogen receptor-alpha. Oncogene. 2011;31(27):3223–3234. doi: 10.1038/onc.2011.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oki Y., Issa J.P. Epigenetic mechanisms in AML – a target for therapy. Cancer Treat Res. 2010;145:19–40. doi: 10.1007/978-0-387-69259-3_2. [DOI] [PubMed] [Google Scholar]
- 31.Wu J., Wood G.S. Reduction of Fas/CD95 promoter methylation, upregulation of Fas protein, and enhancement of sensitivity to apoptosis in cutaneous T-cell lymphoma. Arch Dermatol. 2011;147:443–449. doi: 10.1001/archdermatol.2010.376. [DOI] [PubMed] [Google Scholar]
- 32.Hopfer O., Komor M., Koehler I.S., Freitag C., Schulze M., Hoelzer D. Aberrant promotor methylation in MDS hematopoietic cells during in vitro lineage specific differentiation is differently associated with DNMT isoforms. Leuk Res. 2009;33:434–442. doi: 10.1016/j.leukres.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Karlic H., Reitermaier R., Ghanim V., Hermann H., Thaler R., Spitzer S. 5-Azacytidine and decitabine induce demethylation and re-expression of FAS (CD95) and promoter apoptosis in neoplastic cells in acute myeloid leukemia (AML) Blood. 2011;118:3463. AHS Annual Meeting Abstracts. [Google Scholar]
- 34.Ghanim V., Herrmann H., Heller G., Peter B., Hadzijusufovic E., Blatt K. 5-Azacytidine and decitabine exert proapoptotic effects on neoplastic mast cells: role of FAS-demethylation and FAS re-expression, and synergism with FAS-ligand. Blood. 2012;119:4242–4252. doi: 10.1182/blood-2011-09-382770. [DOI] [PubMed] [Google Scholar]
- 35.Fujita H., Kurokawa K., Ogino T., Ono M., Yamamoto M., Oka T. Effect of risedronate on osteoblast differentiation, expression of receptor activator of NF-kappaB ligand and apoptosis in mesenchymal stem cells. Basic Clin Pharmacol Toxicol. 2011;109:78–84. doi: 10.1111/j.1742-7843.2011.00685.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang J., Stern P.H. Dose-dependent differential effects of risedronate on gene expression in osteoblasts. Biochem Pharmacol. 2011;81:1036–1042. doi: 10.1016/j.bcp.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corrado A., Neve A., Maruotti N., Gaudio A., Marucci A., Cantatore F.P. Dose-dependent metabolic effect of zoledronate on primary human osteoblastic cell cultures. Clin Exp Rheumatol. 2010;28:873–879. [PubMed] [Google Scholar]
- 38.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thaler R., Karlic H., Spitzer S., Klaushofer K., Varga F. Extra-cellular matrix suppresses expression of the apoptosis mediator Fas by epigenetic DNA methylation. Apoptosis. 2010;15:728–737. doi: 10.1007/s10495-010-0462-3. [DOI] [PubMed] [Google Scholar]
- 40.Gnant M., Mlineritsch B., Schippinger W., Luschin-Ebengreuth G., Postlberger S., Menzel C. Endocrine therapy plus zoledronic acid in premenopausal breast cancer. N Engl J Med. 2009;360:679–691. doi: 10.1056/NEJMoa0806285. [DOI] [PubMed] [Google Scholar]
- 41.Pazianas M., Abrahamsen B., Eiken P.A., Eastell R., Russell R.G. Reduced colon cancer incidence and mortality in postmenopausal women treated with an oral bisphosphonate – Danish National Register Based Cohort Study. Osteoporos Int. 2012;23(11):2693–2701. doi: 10.1007/s00198-012-1902-4. [DOI] [PubMed] [Google Scholar]
- 42.Bundred N. Antiresorptive therapies in oncology and their effects on cancer progression. Cancer Treat Rev. 2012;38(6):776–786. doi: 10.1016/j.ctrv.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Pazianas M., Russell R.G. Potential therapeutic effects of oral bisphosphonates on the intestine. Ann N Y Acad Sci. 2011;1240:E19–E25. doi: 10.1111/j.1749-6632.2011.06372.x. [DOI] [PubMed] [Google Scholar]
- 44.Rodrigues P., Hering F.O., Meller A. Adjuvant effect of IV clodronate on the delay of bone metastasis in high-risk prostate cancer patients: a prospective study. Cancer Res Treat. 2011;43:231–235. doi: 10.4143/crt.2011.43.4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daubine F.R.L.E.B., Marquez M., Nilsson S., Schroder T., Holmberg A.R. Treatment of bone metastasis in prostate cancer: efficacy of a novel polybisphosphonate. Anticancer Res. 2011;31:4141–4145. [PubMed] [Google Scholar]
- 46.Cardwell C.R., Abnet C.C., Veal P., Hughes C.M., Cantwell M.M., Murray L.J. Exposure to oral bisphosphonates and risk of cancer. Int J Cancer. 2012;131(5):E717–E725. doi: 10.1002/ijc.27389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prior I.A., Hancock J.F. Ras trafficking, localization and compartmentalized signalling. Semin Cell Dev Biol. 2012;23(2):145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher J.E., Rogers M.J., Halasy J.M., Luckman S.P., Hughes D.E., Masarachia P.J. Alendronate mechanism of action: geranylgeraniol, an intermediate in the mevalonate pathway, prevents inhibition of osteoclast formation, bone resorption, and kinase activation in vitro. Proc Natl Acad Sci U S A. 1999;96:133–138. doi: 10.1073/pnas.96.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreau M.F., Guillet C., Massin P., Chevalier S., Gascan H., Basle M.F. Comparative effects of five bisphosphonates on apoptosis of macrophage cells in vitro. Biochem Pharmacol. 2007;73:718–723. doi: 10.1016/j.bcp.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 50.Monkkonen H., Auriola S., Lehenkari P., Kellinsalmi M., Hassinen I.E., Vepsalainen J. A new endogenous ATP analog (ApppI) inhibits the mitochondrial adenine nucleotide translocase (ANT) and is responsible for the apoptosis induced by nitrogen-containing bisphosphonates. Br J Pharmacol. 2006;147:437–445. doi: 10.1038/sj.bjp.0706628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogers M.J., Chilton K.M., Coxon F.P., Lawry J., Smith M.O., Suri S. Bisphosphonates induce apoptosis in mouse macrophage-like cells in vitro by a nitric oxide-independent mechanism. J Bone Miner Res. 1996;11:1482–1491. doi: 10.1002/jbmr.5650111015. [DOI] [PubMed] [Google Scholar]
- 52.Luckman S.P., Coxon F.P., Ebetino F.H., Russell R.G., Rogers M.J. Heterocycle-containing bisphosphonates cause apoptosis and inhibit bone resorption by preventing protein prenylation: evidence from structure-activity relationships in J774 macrophages. J Bone Miner Res. 1998;13:1668–1678. doi: 10.1359/jbmr.1998.13.11.1668. [DOI] [PubMed] [Google Scholar]
- 53.Coxon F.P., Benford H.L., Russell R.G., Rogers M.J. Protein synthesis is required for caspase activation and induction of apoptosis by bisphosphonate drugs. Mol Pharmacol. 1998;54:631–638. [PubMed] [Google Scholar]
- 54.Halasy-Nagy J.M., Rodan G.A., Reszka A.A. Inhibition of bone resorption by alendronate and risedronate does not require osteoclast apoptosis. Bone. 2001;29:553–559. doi: 10.1016/s8756-3282(01)00615-9. [DOI] [PubMed] [Google Scholar]
- 55.Sato M., Grasser W. Effects of bisphosphonates on isolated rat osteoclasts as examined by reflected light microscopy. J Bone Miner Res. 1990;5:31–40. doi: 10.1002/jbmr.5650050107. [DOI] [PubMed] [Google Scholar]
- 56.Breuil V., Cosman F., Stein L., Horbert W., Nieves J., Shen V. Human osteoclast formation and activity in vitro: effects of alendronate. J Bone Miner Res. 1998;13:1721–1729. doi: 10.1359/jbmr.1998.13.11.1721. [DOI] [PubMed] [Google Scholar]
- 57.Ralph P., Moore M.A., Nilsson K. Lysozyme synthesis by established human and murine histiocytic lymphoma cell lines. J Exp Med. 1976;143:1528–1533. doi: 10.1084/jem.143.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plotkin L.I., Weinstein R.S., Parfitt A.M., Roberson P.K., Manolagas S.C., Bellido T. Prevention of osteocyte and osteoblast apoptosis by bisphosphonates and calcitonin. J Clin Invest. 1999;104:1363–1374. doi: 10.1172/JCI6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plotkin L.I., Manolagas S.C., Bellido T. Dissociation of the pro-apoptotic effects of bisphosphonates on osteoclasts from their anti-apoptotic effects on osteoblasts/osteocytes with novel analogs. Bone. 2006;39:443–452. doi: 10.1016/j.bone.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 60.Reinholz G.G., Getz B., Pederson L., Sanders E.S., Subramaniam M., Ingle J.N. Bisphosphonates directly regulate cell proliferation, differentiation, and gene expression in human osteoblasts. Cancer Res. 2000;60:6001–6007. [PubMed] [Google Scholar]
- 61.Im G.I., Qureshi S.A., Kenney J., Rubash H.E., Shanbhag A.S. Osteoblast proliferation and maturation by bisphosphonates. Biomaterials. 2004;25:4105–4115. doi: 10.1016/j.biomaterials.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 62.von Knoch F., Jaquiery C., Kowalsky M., Schaeren S., Alabre C., Martin I. Effects of bisphosphonates on proliferation and osteoblast differentiation of human bone marrow stromal cells. Biomaterials. 2005;26:6941–6949. doi: 10.1016/j.biomaterials.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 63.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 64.Fraga M.F., Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Wilson A.S., Power B.E., Molloy P.L. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Ebert R., Zeck S., Krug R., Meissner-Weigl J., Schneider D., Seefried L. Pulse treatment with zoledronic acid causes sustained commitment of bone marrow derived mesenchymal stem cells for osteogenic differentiation. Bone. 2009;44:858–864. doi: 10.1016/j.bone.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 67.Coxon F.P., Helfrich M.H., Van’t Hof R., Sebti S., Ralston S.H., Hamilton A. Protein geranylgeranylation is required for osteoclast formation, function, and survival: inhibition by bisphosphonates and GGTI-298. J Bone Miner Res. 2000;15:1467–1476. doi: 10.1359/jbmr.2000.15.8.1467. [DOI] [PubMed] [Google Scholar]
- 68.Lane S.W., De Vita S., Alexander K.A., Karaman R., Milsom M.D., Dorrance A.M. Rac signaling in osteoblastic cells is required for normal bone development but is dispensable for hematopoietic development. Blood. 2012;119:736–744. doi: 10.1182/blood-2011-07-368753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubista B., Trieb K., Sevelda F., Toma C., Arrich F., Heffeter P. Anticancer effects of zoledronic acid against human osteosarcoma cells. J Orthop Res. 2006;24:1145–1152. doi: 10.1002/jor.20129. [DOI] [PubMed] [Google Scholar]
- 70.Forsea A.M., Muller C., Riebeling C., Orfanos C.E., Geilen C.C. Nitrogen-containing bisphosphonates inhibit cell cycle progression in human melanoma cells. Br J Cancer. 2004;91:803–810. doi: 10.1038/sj.bjc.6602052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee M.V., Fong E.M., Singer F.R., Guenette R.S. Bisphosphonate treatment inhibits the growth of prostate cancer cells. Cancer Res. 2001;61:2602–2608. [PubMed] [Google Scholar]
- 72.Lin Y.S., Park J., De Schutter J.W., Huang X.F., Berghuis A.M., Sebag M. Design and synthesis of active site inhibitors of the human farnesyl pyrophosphate synthase: apoptosis and inhibition of ERK phosphorylation in multiple myeloma cells. J Med Chem. 2012;55:3201–3215. doi: 10.1021/jm201657x. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Cao R., Yin F., Hudock M.P., Guo R.T., Krysiak K. Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation. J Am Chem Soc. 2009;131:5153–5162. doi: 10.1021/ja808285e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Armstrong S.A., Hannah V.C., Goldstein J.L., Brown M.S. CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J Biol Chem. 1995;270:7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- 75.Basso A.D., Kirschmeier P., Bishop W.R. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res. 2006;47:15–31. doi: 10.1194/jlr.R500012-JLR200. [DOI] [PubMed] [Google Scholar]
- 76.Epling-Burnette P.K., Loughran T.P., Jr. Suppression of farnesyltransferase activity in acute myeloid leukemia and myelodysplastic syndrome: current understanding and recommended use of tipifarnib. Expert Opin Investig Drugs. 2010;19:689–698. doi: 10.1517/13543781003801076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Law B.K., Norgaard P., Gnudi L., Kahn B.B., Poulson H.S., Moses H.L. Inhibition of DNA synthesis by a farnesyltransferase inhibitor involves inhibition of the p70(s6k) pathway. J Biol Chem. 1999;274:4743–4748. doi: 10.1074/jbc.274.8.4743. [DOI] [PubMed] [Google Scholar]
- 78.Moasser M.M., Rosen N. The use of molecular markers in farnesyltransferase inhibitor (FTI) therapy of breast cancer. Breast Cancer Res Treat. 2002;73:135–144. doi: 10.1023/a:1015209123900. [DOI] [PubMed] [Google Scholar]
- 79.Whyte D.B., Kirschmeier P., Hockenberry T.N., Nunez-Oliva I., James L., Catino J.J. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 80.Zhang F.L., Kirschmeier P., Carr D., James L., Bond R.W., Wang L. Characterization of Ha-ras, N-ras, Ki-Ras4A, and Ki-Ras4B as in vitro substrates for farnesyl protein transferase and geranylgeranyl protein transferase type I. J Biol Chem. 1997;272:10232–10239. doi: 10.1074/jbc.272.15.10232. [DOI] [PubMed] [Google Scholar]
- 81.Ohtsuka Y., Manabe A., Kawasaki H., Hasegawa D., Zaike Y., Watanabe S. RAS-blocking bisphosphonate zoledronic acid inhibits the abnormal proliferation and differentiation of juvenile myelomonocytic leukemia cells in vitro. Blood. 2005;106:3134–3141. doi: 10.1182/blood-2005-03-0972. [DOI] [PubMed] [Google Scholar]
- 82.Kaneda A., Wakazono K., Tsukamoto T., Watanabe N., Yagi Y., Tatematsu M. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410–6415. doi: 10.1158/0008-5472.CAN-04-1543. [DOI] [PubMed] [Google Scholar]
- 83.Carnero A. The PKB/AKT pathway in cancer. Curr Pharm Des. 2010;16:34–44. doi: 10.2174/138161210789941865. [DOI] [PubMed] [Google Scholar]
- 84.Omerovic J., Laude A.J., Prior I.A. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nojima D., Li L.C., Dharia A., Perinchery G., Ribeiro-Filho L., Yen T.S. CpG hypermethylation of the promoter region inactivates the estrogen receptor-beta gene in patients with prostate carcinoma. Cancer. 2001;92:2076–2083. doi: 10.1002/1097-0142(20011015)92:8<2076::aid-cncr1548>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 86.Kinoshita H., Shi Y., Sandefur C., Meisner L.F., Chang C., Choon A. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60:3623–3630. [PubMed] [Google Scholar]
- 87.Coleman R.E., Marshall H., Cameron D., Dodwell D., Burkinshaw R., Keane M. Breast-cancer adjuvant therapy with zoledronic acid. N Engl J Med. 2011;365:1396–1405. doi: 10.1056/NEJMoa1105195. [DOI] [PubMed] [Google Scholar]