Abstract
Formation of plant virus membrane-associated replication factories requires the association of viral replication proteins and viral RNA with intracellular membranes, the recruitment of host factors and the modification of membranes to form novel structures that house the replication complex. Many viruses encode integral membrane proteins that act as anchors for the replication complex. These hydrophobic proteins contain transmembrane domains and/or amphipathic helices that associate with the membrane and modify its structure. The comovirus Co-Pro and NTP-binding (NTB, putative helicase) proteins and the cognate nepovirus X2 and NTB proteins are among the best characterized plant virus integral membrane replication proteins and are functionally related to the picornavirus 2B, 2C, and 3A membrane proteins. The identification of membrane association domains and analysis of the membrane topology of these proteins is discussed. The evidence suggesting that these proteins have the ability to induce membrane proliferation, alter the structure and integrity of intracellular membranes, and modulate the induction of symptoms in infected plants is also reviewed. Finally, areas of research that need further investigation are highlighted.
Keywords: integral membrane proteins, viral replication complexes, intracellular membranes, protein–membrane interactions, secoviridae, picornavirales, plant–virus interactions, membrane remodeling
CHARACTERIZATION OF COMOVIRUS AND NEPOVIRUS REPLICATION COMPLEXES AND IDENTIFICATION OF PUTATIVE MEMBRANE ANCHORS
Positive-strand RNA viruses replicate in large complexes that are associated with host intracellular membranes (Salonen et al., 2005; Sanfacon, 2005; Miller and Krijnse-Locker, 2008; den Boon and Ahlquist, 2010; Laliberte and Sanfacon, 2010; Nagy and Pogany, 2012 ). Some viruses require host membrane proteins to target their replication proteins to the membranes (Yamanaka et al., 2000). However, many viruses encode proteins that interact with membranes directly and modify their intrinsic structure. These proteins have membrane association domains and contain protein–protein and/or protein–RNA interaction domains that allow them to recruit the viral RNA, other viral replication proteins, or host factors to the membranes. Well-characterized plant virus membrane proteins include the tombusvirus 33–36 kDa proteins, bromovirus 1a protein, potyvirus 6K protein, and tymovirus 140 kDa protein (Schaad et al., 1997; den Boon et al., 2001; Weber-Lotfi et al., 2002; Prod’Homme et al., 2003; Turner et al., 2004).
The family Secoviridae (order Picornavirales) includes the genera Comovirus, Fabavirus, Nepovirus, Sequivirus, Waikavirus, Cheravirus, Sadwavirus, and Torradovirus (Sanfacon et al., 2011). The best characterized members of the family are Cowpea mosaic virus (CPMV, comovirus), Grapevine fanleaf virus (GFLV, nepovirus), and Tomato ringspot virus (ToRSV, nepovirus; Pouwels et al., 2002a; Sanfacon et al., 2006). These viruses use a polyprotein strategy to express their proteins and have a replication block consisting of a nucleotide-binding protein (NTB), a genome-linked protein (VPg), a proteinase (Pro), and an RNA-dependent RNA polymerase (Pol; Figure 1C). Although they share these properties with picornaviruses (including the well-characterized poliovirus), nepo- and comoviruses differ in that they have bipartite genomes. The RNA1-encoded polyprotein contains all protein domains necessary for replication and RNA1 can replicate independently of RNA2 (Vos et al., 1988; Viry et al., 1993 ).
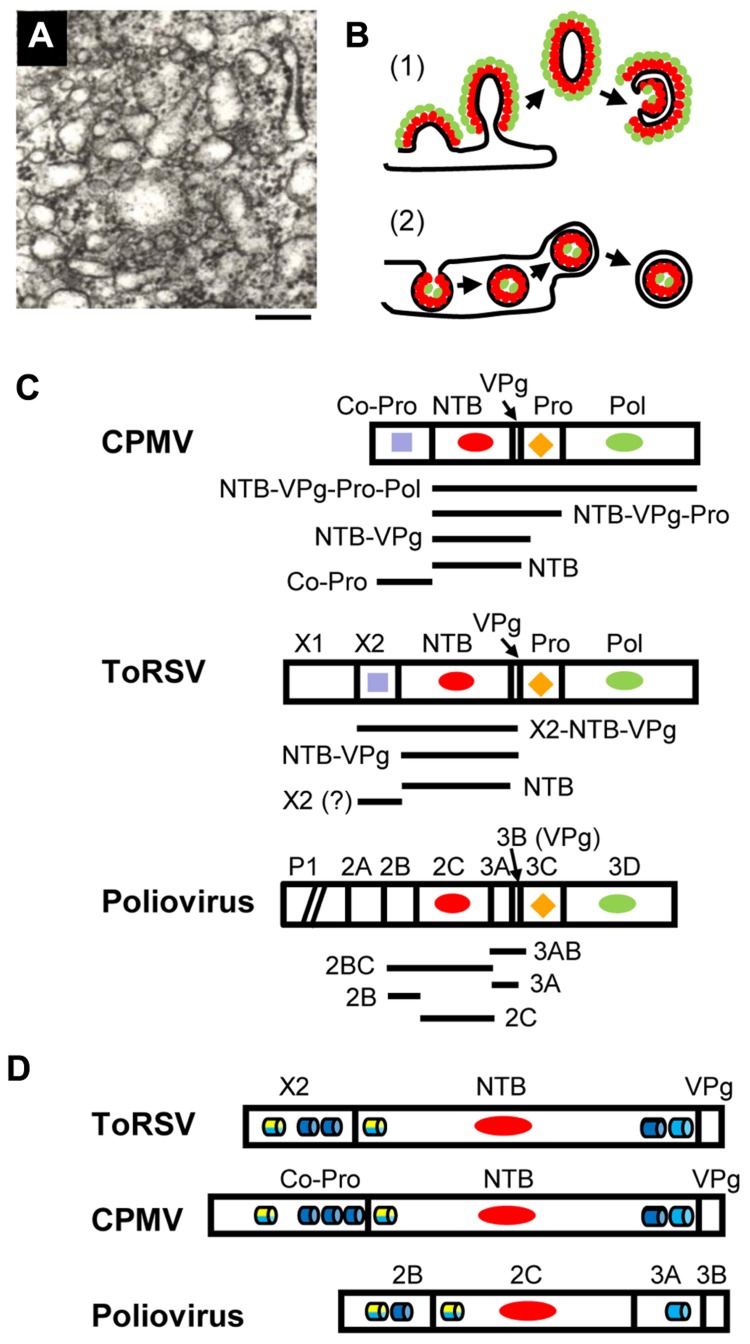
FIGURE 1.
Membrane replication proteins encoded by CPMV, ToRSV and poliovirus. (A) Electron micrograph showing the proliferation of single-membrane vesicles in ToRSV-infected Nicotiana clevelandii plants. The bar indicates 200 nm. (B) Models for the formation of viral replication complexes. (1) In cells infected with poliovirus or coxsackie B3 virus, viral integral membrane proteins (red ovals) induce positive curvature of the membrane allowing the budding of tubular structures. Other viral replication proteins (e.g., polymerase, green ovals) interact with the viral membrane proteins. Host factors and viral RNA (not shown) associate with the replication complex by protein–protein and protein–RNA interactions. Single-membrane vesicles may bud out and form double-membrane vesicles after the internal collapse of the single-membrane vesicle and subsequent membrane fusion to allow its circularization. Late in infection, double-membrane vesicles are predominant in picornavirus-infected cells. This model is based on electron tomography observations from Belov et al. (2012) and Limpens et al. (2011). (2) In cells infected with many plant and animal viruses, induction of negative membrane curvature results in membrane invagination and formation of spherules in the lumen of the membrane. In plant, spherules have been observed in association with membranes from the ER (brome mosaic virus), chloroplast (turnip yellow mosaic virus), peroxisome (tomato bushy stunt virus) and mitochondria (carnation Italian ringspot virus). The spherules are connected to the cytoplasm by a neck. Viral integral membrane proteins (red ovals) line the interior of the spherule. The viral polymerase (green ovals) as well as other viral proteins, host factors and the viral RNA (not shown) are enclosed in the spherule. Release of the vesicle in the lumen of the membrane may be followed by budding of a double-membrane vesicle into the cytoplasm. This model has been discussed in recent reviews (den Boon and Ahlquist, 2010; Laliberte and Sanfacon, 2010; Nagy and Pogany, 2012). (C) Organization of replication protein domains in the polyproteins of CPMV, ToRSV, and poliovirus. The RNA1-encoded polyproteins of CPMV and ToRSV are shown. For poliovirus, the polyprotein encoded by the single genomic RNA is shown, although the P1 region (containing the structural proteins) is truncated as indicated by the diagonal bars. Vertical lines represent the protease cleavage sites. Conserved motifs are: RNA-dependent RNA polymerase (Pol, green ovals), protease (Pro, orange diamond), nucleotide-binding protein (NTB, red oval), Co-Pro and X2 (purple square). Horizontal bars under each polyprotein represent integral membrane proteins that have been detected in virus-infected cells. The mature ToRSV X2 protein is shown with a question mark. Although likely, its presence in infected cells could not be confirmed due to the lack of antibodies. (D) The regions of the polyprotein containing the putative membrane anchors are shown for each virus. Predicted membrane-association domains are indicated with blue barrels (hydrophobic helices) or with yellow/blue barrels (amphipathic helices, with the yellow half representing the polar/charged hydrophilic side of the helix and the blue half representing the hydrophobic side of the helix).
Plant cells infected by como- and nepoviruses are characterized by the presence of numerous membraneous vesicles, which are derived from the endoplasmic reticulum (ER; Carette et al., 2000; Ritzenthaler et al., 2002; Han and Sanfacon, 2003). In CPMV-infected cells, vesicles first appear throughout the cytoplasm, but later coalesce in a large perinuclear structure (Carette et al., 2002a). Actin microfilaments are probably involved in this process (Carette et al., 2002a). Perinuclear membrane aggregates are also observed in ToRSV- and GFLV-infected cells (Ritzenthaler et al., 2002; Han and Sanfacon, 2003). Viral replication proteins, de novo RNA synthesis and dsRNA intermediates co-localize with these structures, indicating that they are the site of viral replication (de Zoeten et al., 1974; Carette et al., 2000,2002a; Ritzenthaler et al., 2002; Han and Sanfacon, 2003).
Vesicles induced in como- and nepovirus-infected cells are irregularly shaped, vary in size and are usually surrounded by a single-membrane (Carette et al., 2000; Ritzenthaler et al., 2002; Figure 1A). These vesicles are similar to those observed in early stages of infection by poliovirus and coxsackie B3 virus (both picornaviruses). Three-dimensional reconstruction of these early picornavirus-induced structures revealed that they are branching tubular structures rather than closed vesicles (Limpens et al., 2011; Belov et al., 2012). Positive membrane curvature induced by viral membrane proteins allows budding of tubular structures from the surface of the membrane. Replication proteins accumulate on the outside of the single-membrane structures (Figure 1B, model 1). That GFLV- and poliovirus-induced vesicles are immunoprecipitated by antibodies against viral replication proteins, is consistent with this model (Bienz et al., 1994; Carette et al., 2000). In contrast, membrane structures induced by many plant viruses (including bromo-, tombus-, and tymoviruses) are formed by membrane invagination and require negative membrane curvature. These replication complexes are sheltered inside spherules that are connected to the cytoplasm by a neck (Figure 1B, model 2).
Of the replication proteins encoded by como- or nepovirus RNA1, two contain obvious hydrophobic regions: the comovirus Co-Pro and NTB proteins and the cognate nepovirus X2 and NTB proteins (Figure 1D). In infected cells, mature proteins co-exist with stable intermediate polyproteins (Figure 1C). The CPMV Co-Pro is only detected as a mature protein due to efficient cleavage between Co-Pro and NTB. However, NTB is found either as a mature protein or within various intermediates (NTB–VPg, NTB–VPg–Pro, and NTB–VPg–Pro–Pol; Wellink et al., 1986). In contrast, processing at the nepovirus X2–NTB cleavage site is inefficient in vitro leading to the accumulation of X2–NTB and X2–NTB–VPg in addition to X2 and NTB (Wang and Sanfacon, 2000; Wetzel et al., 2008). In ToRSV-infected cells, NTB, NTB–VPg, and X2–NTB–VPg are tightly associated with ER membranes active in viral replication (Han and Sanfacon, 2003). In contrast, only a sub-population of a polyprotein containing the VPg, Pro, and Pol domains (VPg–Pro–Pol’) is associated with replication-competent membranes and this association is peripheral, suggesting that it requires an interaction between VPg–Pro–Pol’ and a membrane protein (Chisholm et al., 2007). Similarly, only a fraction of VPg–Pro–Pol is membrane-bound in CPMV-infected cells (Dorssers et al., 1984). When expressed individually, the ToRSV X2, NTB and NTB–VPg and the CPMV Co-Pro and NTB–VPg associate with ER membranes, while proteins containing the ToRSV or CPMV VPg, Pro, and Pol domains remain in the soluble cytoplasmic fraction (Carette et al., 2002b; Zhang et al., 2005; Zhang and Sanfacon, 2006;Chisholm et al., 2007 ). Thus, the CPMV Co-Pro and NTB and ToRSV X2 and NTB and/or intermediate polyproteins containing these protein domains are likely to act as membrane anchors for the replication complex.
The nucleotide-binding motif of the nepo- and comovirus NTB is related to that of the poliovirus 2C protein (Figure 1C). The nepo- and comovirus NTB also contain a hydrophobic C-terminal domain, which is absent in 2C (Figure 1D). The poliovirus 3A protein (immediately downstream of 2C in the polyprotein) has a hydrophobic domain that corresponds to the C-terminal region of the nepo- and comovirus NTB, although polyproteins containing both 2C and 3A are not detected in infected cells (Figure 1C; Cameron et al., 2010). The ToRSV X2, and CPMV Co-Pro are highly hydrophobic and share a signature sequence (F-x27-W-x11-L-x23-E; Rott et al., 1995), which is also found in the cognate proteins of nepo-, como-, faba-, and cheraviruses (Sanfacon et al., 2011). Co-Pro is a protease co-factor that slows the processing of the CPMV RNA1 polyprotein (Peters et al., 1992). However, there is no experimental evidence that X2 regulates the nepovirus protease activity (Wang and Sanfacon, 2000; Wetzel et al., 2008). Thus, the conserved motif may be important for another common activity of Co-Pro and X2. The poliovirus 2B protein is located immediately upstream of 2C (Figure 1D) but does not share sequence motifs with X2 and Co-Pro, other than a general hydrophobicity.
MEMBRANE MODIFICATIONS AND SYMPTOMS INDUCED BY THE COMOVIRUS Co-Pro AND NTB–VPg
When overexpressed from a viral vector, the CPMV NTB–VPg or Co-Pro induces the formation of small ER-derived perinuclear bodies (Carette et al., 2002b). Proliferation of the cortical ER is also observed after overexpression of Co-Pro. These structures resemble the ER modifications observed in early stages of natural CPMV infections but differ from the large perinuclear structures present later in infection. Thus, both proteins may act together to induce the larger structures in natural infections. NTB and NTB-containing intermediate polyproteins co-immunoprecipitate with Co-Pro, suggesting that Co-Pro and NTB interact with each other (Wellink et al., 1986). This situation is reminiscent of that observed with poliovirus membrane proteins. While 3A, 2C, and 2BC each induce ER modifications, co-expression of 3A and 2BC together is required to induce vesicles that are similar to those observed in natural poliovirus infections (Suhy et al., 2000). Protein–protein interactions among these proteins are well-documented (Teterina et al., 2006; Yin et al., 2007).
Ectopic overexpression of CPMV Co-Pro or NTB–VPg induces local necrosis in plant (Carette et al., 2002b). Interestingly, CPMV does not cause necrosis in natural infection, even though Co-Pro and NTB–VPg accumulate in infected cells. Accumulation of these proteins in electron-dense bodies, which are probable sites of protein aggregation, may help reduce their toxicity (Carette et al., 2002b). Comparison of the symptomatology induced by chimeric constructs of two isolates of bean pod mosaic virus (another comovirus) also points to Co-Pro and NTB as symptom severity determinants (Gu and Ghabrial, 2005). Chimeric constructs containing Co-Pro or NTB from the severe isolate induce increased symptomatology and accumulate to higher level than the mild isolate. Co-Pro and NTB may regulate the rate of virus replication, in agreement with their proposed role in replication complex assembly. Although the severe symptoms may be due to increased accumulation of viral products, possibly triggering plant defense responses, it may be a direct consequence of the membrane alterations induced by NTB and Co-Pro. Poliovirus 2B and 3A induce apoptosis when overexpressed (Madan et al., 2008). At least for 2B, the induction of apoptosis was correlated with its viroporin activity, which affects the integrity of various membranes, including mitochondrial membranes (Madan et al., 2008,2010). Although a sub-population of the CPMV NTB–VPg targets chloroplast membranes (Carette et al., 2002b), there is no experimental evidence that mitochondria are targeted. Further studies will be necessary to investigate possible correlations between membrane alterations and symptomatology induced by the comovirus NTB and Co-Pro proteins and to determine whether the nepovirus X2 and NTB proteins can alter membrane structures and induce symptoms.
MEMBRANE TOPOLOGY OF THE ToRSV X2 AND NTB: EVIDENCE FOR OLIGOMERIZATION AND VIROPORIN ACTIVITY
Membrane association of integral membrane proteins can be directed by transmembrane α-helices, which are highly hydrophobic, or by amphipathic α-helices. Amphipathic helices initially insert parallel to the membranes with their hydrophobic face inserted in the lipid bilayer (Figures 2A,B). Oligomerization of amphipathic helices can lead to the formation of aqueous pores whereby the hydrophilic faces of the helices orient toward the pore and the hydrophobic faces interact within the membrane environment (Gonzalez and Carrasco, 2003; Figure 2B). Hydrophobic intra- and intermolecular interactions among amphipathic and adjacent hydrophobic helices can stabilize the oligomers (Figure 2B), as suggested for the poliovirus 2B protein (Agirre et al., 2002; Martinez-Gil et al., 2011).
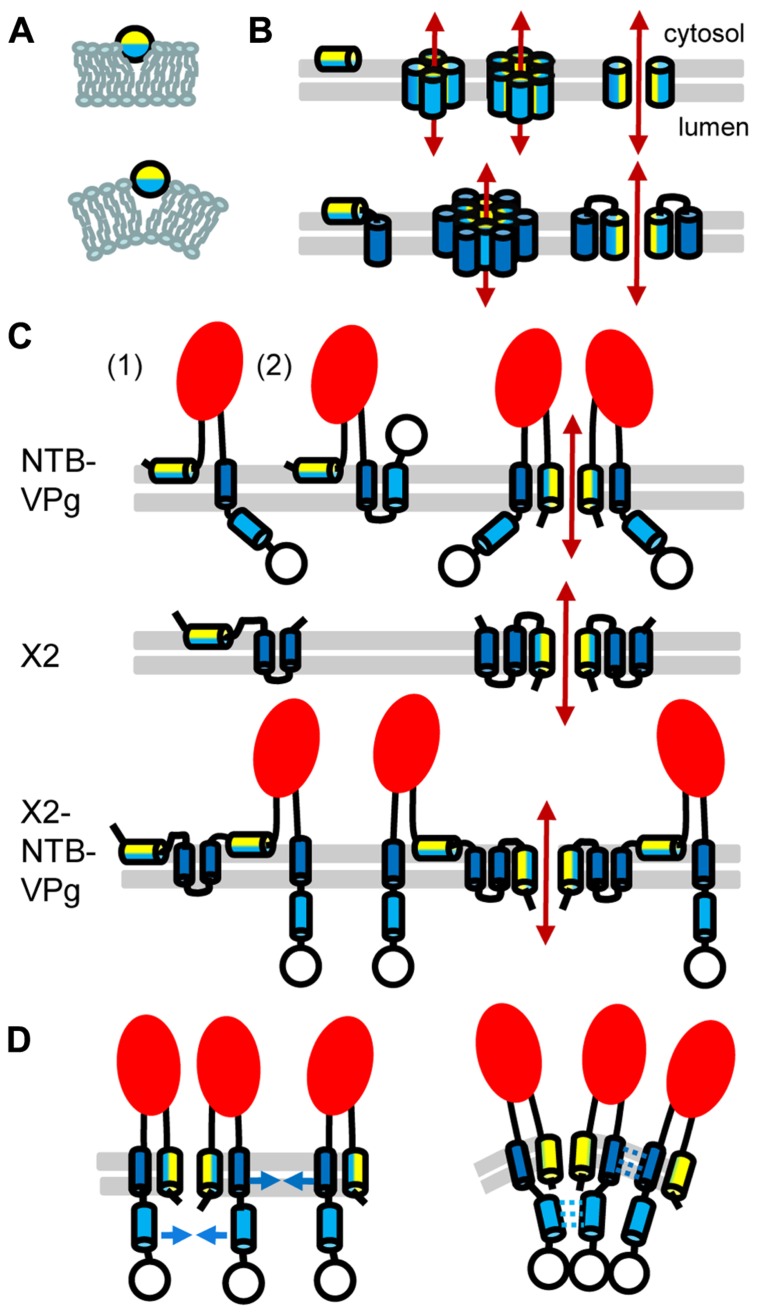
FIGURE 2.
Topology model for ToRSV membrane replication proteins. (A) Model for the parallel insertion of an amphipathic helix. The hydrophobic side of the helix (blue) inserts in one leaflet of the lipid bilayer while the polar/charged hydrophilic side of the helix (yellow) is exposed to the cytosolic face of the membrane. This insertion displaces the lipid headgroup, causing the acyl chain to reorient and inducing positive membrane curvature. (B) Model for the oligomerization of amphipathic helices and formation of an aqueous pore. In the top panel, an amphipathic helix is inserted parallel to the lipid bilayer (horizontal gray lines) of the membrane (left). Formation of an aqueous pore (double-ended red arrow) requires oligomerization of four or six amphipathic helices (middle). In the aqueous pore, the hydrophilic side of the helix (yellow) is exposed toward the pore, while its hydrophobic side (blue) is oriented toward the membrane lipid bilayer. A simplified representation of the pore shows only two molecules to better visualize each side of the amphipathic helix relative to the pore (right). In the bottom panel, a membrane protein consisting of an amphipathic helix and a hydrophobic helix (blue) is shown. After initial membrane insertion of the monomer with the amphipathic helix parallel to the membrane (left), an aqueous pore is formed by oligomerization of the amphipathic helix (middle). The hydrophobic helix of each molecule is located on the outside of the pore alongside the amphipathic helix (model shown for a hexamer). Hydrophobic interactions between the hydrophobic side of the amphipathic helix and the hydrophobic helix stabilize pore formation. A simplified representation of the pore shows only two molecules (right). (C) Predicted topologies for NTB–VPg, X2, and X2–NTB–VPg shown for monomers (left) or oligomers (right). Two possible topologies are shown for NTB–VPg monomers (1 and 2, see text). To simplify the figure, only two molecules are shown in the oligomer models. However, at least four molecules would be necessary to form an aqueous pore (as shown in B). The open circle represents the VPg domain and the red oval indicates the conserved NTB motif. (D) Model for the induction of positive membrane curvature by hydrophobic interactions of membrane proteins oligomers, shown for NTB–VPg. On the left, blue arrows represent possible hydrophobic interactions. These interactions (shown by broken blue lines on the right) would induce positive membrane curvature. Similar hydrophobic interactions are predicted to occur in X2 or X2–NTB–VPg oligomers (not shown).
The hydrophobic C-terminal domain and a predicted N-terminal amphipathic helix of the ToRSV NTB protein (Figure 1D) are each sufficient to target GFP fusion proteins to ER membranes in plant cells or to direct the insertion of NTB or NTB–VPg into canine microsomal membranes in vitro (Wang et al., 2004; Zhang et al., 2005). These domains are conserved in the sequence of NTB from other nepo- and comoviruses (Figure 1D). The C-terminal hydrophobic region of the ToRSV NTB contains a highly hydrophobic α-helix, which traverses the membrane. The VPg domain of NTB–VPg is translocated in the membrane lumen (topology 1, Figure 2C), allowing the recognition of a naturally occurring N-glycosylation site (Wang et al., 2004; Zhang et al., 2005). The luminal orientation of the C-terminal region of NTB–VPg was confirmed by proteinase K protection assays using membrane-fractions of ToRSV-infected cells (Han and Sanfacon, 2003). However, these results do not exclude the possibility that a sub-population of the protein adopt an alternate topology. In vitro, a second weakly predicted transmembrane α-helix traverses the membranes when the first transmembrane helix is deleted (Wang et al., 2004). In an alternate topology (topology 2, Figure 2C), the NTB C-terminal hydrophobic region traverses the membrane twice allowing a cytosolic orientation of the VPg. Experiments are required to determine whether this alternate topology exists in infected cells. Alternative topologies for NTB–VPg could regulate the presentation of the VPg to the cytoplasmic face of the membrane where protein–protein interactions and viral replication take place.
The N-terminus of NTB is translocated in the membrane lumen, suggesting oligomerization of the amphipathic helix and pore formation (Zhang et al., 2005; Figure 2C). Pore formation may be enhanced by hydrophobic interactions between the N-terminal amphipathic helix and the C-terminal transmembrane helix. Denaturing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of NTB–VPg or of fragments containing the amphipathic or transmembrane helices revealed the presence of additional bands that correspond in size to oligomers. Membrane proteins can conserve their oligomeric structure in the presence of denaturing agents, due to strong hydrophobic interactions (DeGrado et al., 2003). The potential NTB–VPg oligomers were glycosylated, suggesting that oligomerization occurred within the membranes (Wang et al., 2004; Zhang et al., 2005). Although the topology model of NTB–VPg oligomers suggests the formation of an aqueous pore, further experimentation is required to test whether the protein affects the membrane integrity in vivo.
In plant cells, ER-targeting of ToRSV X2 is directed by two strongly predicted transmembrane helices and a putative amphipathic helix (Zhang and Sanfacon, 2006; Figure 1D). These features are conserved in the X2 from other nepoviruses. Similarly, three transmembrane helices and one amphipathic helix are predicted in the CPMV Co-Pro (Carette et al., 2002b; Zhang and Sanfacon, 2006; Figure 1D). The topology of ToRSV X2 was examined in vitro (Zhang and Sanfacon, 2006). The two predicted transmembrane helices were found to traverse the membrane, forming a hairpin and resulting in a cytosolic orientation of the C-terminus of X2 (Figure 2C). The N-terminus of X2 was translocated to the membrane lumen. Analysis by SDS-PAGE of full-length or truncated X2 suggests that, as for NTB–VPg, protein oligomerization occurs through hydrophobic interactions (Zhang and Sanfacon, 2006). A topology model of X2 oligomers implies the formation of an aqueous pore by oligomerization of the amphipathic helix (Figure 2C). However, in vivo evidence in support of this model is still lacking. Due to its highly hydrophobic nature, it has not been possible to produce antibodies against X2. Thus, although the presence of mature X2 in ToRSV-infected cells is likely, it could not be confirmed. However, polyproteins corresponding to the expected molecular mass for X2–NTB–VPg were detected with anti-NTB and anti-VPg antibodies (Han and Sanfacon, 2003). Efforts are underway to develop ToRSV infectious clones, which may allow the insertion of epitope tags in X2 to confirm its presence in ToRSV-infected cells and examine its topology in vivo (Chisholm and Sanfacon, unpublished). Although insertion of hydrophilic epitope tags into hydrophobic membrane proteins can hinder their function, a recent study described tolerated insertion sites in poliovirus membrane proteins (Teterina et al., 2011a).
The topology models for X2 and NTB–VPg pose some problems when applied to the X2–NTB–VPg polyprotein. The cytosolic orientation of the C-terminus of X2 is in apparent conflict with the luminal orientation of the N-terminus of NTB. However, the presence of two strong transmembrane domains in the X2 domain of X2–NTB–VPg may prevent the membrane translocation of the NTB amphipathic helix, forcing it to insert parallel to the membranes (Figure 2C). Thus, processing at the X2–NTB cleavage site may influence the orientation of the NTB amphipathic helix and alter the ability of NTB and/or X2 to modify intracellular membranes. The impact of proteolytic cleavage on membrane topology was demonstrated for the poliovirus 3A and 3AB (Fujita et al., 2007). Using a fluorescence quenching method, 3AB was shown to adopt a single topology, in which the hydrophobic domain is parallel to the membrane. In contrast, 3A adopts two possible orientations, one of which traverses the membrane. It was suggested that the hydrophilic VPg domain prevents the membrane translocation of the 3A hydrophobic domain in 3AB. Regulated cleavage of the poliovirus 2BC also impacts its membrane-modification activities. Although 2B, 2C, and 2BC can target to membranes, only 2BC induces a proliferation of membraneous vesicles (Suhy et al., 2000). On the other hand, poliovirus mutants with decreased processing efficiency at the 2BC cleavage site have reduced membrane permeabilization activity, suggesting that the release of mature 2B from 2BC is essential for its viroporin function (van Kuppeveld et al., 1996).
INTERACTION OF VIRAL MEMBRANE PROTEINS WITH HOST FACTORS: TOWARD A MECHANISM FOR MEMBRANE MODIFICATION
The experimental evidence points to a role for como- and nepovirus membrane replication proteins in altering host membranes and assembling the replication complexes. Positive membrane curvature can be induced by parallel insertion of amphipathic helices (Figure 2A) or by intra- and intermolecular hydrophobic interactions among membrane protein oligomers (as shown for NTB–VPg, Figure 2D; McMahon and Gallop, 2005).
Host factors are also likely to play an important role. The secretory pathway is hijacked by poliovirus to help the formation of membraneous vesicles, resulting in an inhibition of host protein transport (Hsu et al., 2010). The 2B and 3A proteins inhibit the secretory pathway (Doedens and Kirkegaard, 1995). 3A interacts with several components of the secretory pathway, including ACBD3, a Golgi adaptor protein (Greninger et al., 2012; Sasaki et al., 2012) and GBF1, a guanine nucleotide exchange factor that activates Arf1, a cellular GTPase and regulator of the secretory pathway (Wessels et al., 2006; Belov et al., 2008; Teterina et al., 2011b). Arf1 is also the known target of brefeldin A, an inhibitor of the secretory pathway that blocks poliovirus infection (Irurzun et al., 1992; Maynell et al., 1992). The 3A–GBF1 and 3A–ACBD3 interactions may assist in the recruitment of P1KIIIβ, an enzyme involved in phospholipid synthesis, to the replication complex (Hsu et al., 2010; Greninger et al., 2012; Sasaki et al., 2012). P1KIIIβ would alter the membrane lipid composition, possibly affecting the membrane curvature and facilitating the formation of virus factories. However, the sensitivity of picornaviruses to brefeldin A varies greatly and the GBF1–3A interaction is not conserved for all picornaviruses, suggesting that the interaction between viruses and the host secretory pathway varies.
How do these findings apply to como- and nepoviruses? Replication of CPMV and GFLV is hindered by cerulenin (Carette et al., 2000; Ritzenthaler et al., 2002), an inhibitor of type II fatty acid synthase, suggesting that de novo phospholipid synthesis is required for membrane proliferation, possibly involving changes in membrane lipid composition. GFLV and CPMV replication is inhibited by brefeldin A (Pouwels et al., 2002b; Ritzenthaler et al., 2002). However, the interaction of nepo- and comoviruses with the secretory pathway is not well understood and their ability to block protein secretion has not been investigated. Two SNARE-like proteins from Arabidopsis thaliana were shown to interact with the CPMV NTB–VPg (Carette et al., 2002c). Although their function is not known, they may regulate membrane fusion and vesicle formation. Identification of additional interaction partners of the nepo- and comovirus membrane proteins will be essential to better understand membrane remodeling directed by these proteins.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by an NSERC discovery grant. I wish to thank Joan Chisholm for critical reading of the manuscript and Andrew Wieczorek for taking the EM picture (shown in Figure 2A), while he was in my lab.
REFERENCES
- Agirre A., Barco A., Carrasco L., Nieva J. L. (2002). Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277 40434–40441 [DOI] [PubMed] [Google Scholar]
- Belov G. A., Feng Q., Nikovics K., Jackson C. L., Ehrenfeld E. (2008). A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. 10.1371/journal.ppat.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov G. A., Nair V., Hansen B. T., Hoyt F. H., Fischer E. R, Ehrenfeld E. (2012). Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 86 302–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz K., Egger D., Pfister T. (1994). Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9 147–157 [DOI] [PubMed] [Google Scholar]
- Cameron C. E., Oh H. S., Moustafa I. M. (2010). Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. 5 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Stuiver M., Van Lent J., Wellink J, Van Kammen A. (2000). Cowpea mosaic virus infection induces a massive proliferation of endoplasmic reticulum but not Golgi membranes and is dependent on de novo membrane synthesis. J. Virol. 74 6556–6563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Guhl K., Wellink J, Van Kammen A. (2002a). Coalescence of the sites of cowpea mosaic virus RNA replication into a cytopathic structure. J. Virol. 76 6235–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., van Lent J., MacFarlane S. A., Wellink J, van Kammen A. (2002b). Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 76 6293–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Verver J., Martens J., van Kampen T., Wellink J, van Kammen A. (2002c). Characterization of plant proteins that interact with cowpea mosaic virus ‘60K’ protein in the yeast two-hybrid system. J. Gen. Virol. 83 885–893 [DOI] [PubMed] [Google Scholar]
- Chisholm J., Zhang G., Wang A., Sanfacon H. (2007). Peripheral association of a polyprotein precursor form of the RNA-dependent RNA polymerase of Tomato ringspot virus with the membrane-bound viral replication complex. Virology 368 133–144 [DOI] [PubMed] [Google Scholar]
- DeGrado W. F., Gratkowski H., Lear J. D. (2003). How do helix–helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 12 647–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J. A., Chen J., Ahlquist P. (2001). Identification of sequences in Brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75 12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J. A., Ahlquist P. (2010). Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64 241–256 [DOI] [PubMed] [Google Scholar]
- de Zoeten G. A., Assink A. M, van Kammen A. (1974). Association of cowpea mosaic virus-induced double-stranded RNA with a cytopathological structure in infected cells. Virology 59 341–355 [DOI] [PubMed] [Google Scholar]
- Doedens J. R., Kirkegaard K. (1995). Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorssers L., Van der Kroll S., Van der Meer J., Van Kammen A., Zabel P. (1984). Purification of cowpea mosaic virus RNA replication complex: identification of a virus-encoded 110,000-dalton polypeptide responsible for RNA chain elongation. Proc. Natl. Acad. Sci. U.S. A. 81 1951–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Krishnakumar S. S., Franco D., Paul A. V., London E., Wimmer E. (2007). Membrane topography of the hydrophobic anchor sequence of poliovirus 3A and 3AB proteins and the functional effect of 3A/3AB membrane association upon RNA replication. Biochemistry 46 5185–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M. E., Carrasco L. (2003). Viroporins. FEBS Lett. 552 28–34 [DOI] [PubMed] [Google Scholar]
- Greninger A. L., Knudsen G. M., Betegon M., Burlingame A. L., Derisi J. L. (2012). The 3A protein from multiple picornaviruses utilizes the golgi adaptor protein ACBD3 to recruit PI4KIIIbeta. J. Virol. 86 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Ghabrial S. A. (2005). The Bean pod mottle virus proteinase cofactor and putative helicase are symptom severity determinants. Virology 333 271–283 [DOI] [PubMed] [Google Scholar]
- Han S., Sanfacon H. (2003). Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J. Virol. 77 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu N. Y., Ilnytska O., Belov G., Santiana M., Chen Y. H., Takvorian P. M., et al. (2010). Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irurzun A., Perez L., Carrasco L. (1992). Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191 166–175 [DOI] [PubMed] [Google Scholar]
- Laliberte J. F., Sanfacon H. (2010). Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48 69–91 [DOI] [PubMed] [Google Scholar]
- Limpens R. W., van der Schaar H. M., Kumar D., Koster A. J., Snijder E. J., van Kuppeveld F. J., et al. (2011). The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. MBio 2 :e00166-11. 10.1128/mBio.00166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Castello A., Carrasco L. (2008). Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 10 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V., Sanchez-Martinez S., Carrasco L., Nieva J. L. (2010). A peptide based on the pore-forming domain of pro-apoptotic poliovirus 2B viroporin targets mitochondria. Biochim. Biophys. Acta 1798 52–58 [DOI] [PubMed] [Google Scholar]
- Martinez-Gil L., Bano-Polo M., Redondo N., Sanchez-Martinez S., Nieva J. L., Carrasco L., et al. (2011). Membrane integration of poliovirus 2B viroporin. J. Virol. 85 11315–11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynell L. A., Kirkegaard K., Klymkowsky M. W. (1992). Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66 1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T., Gallop J. L. (2005). Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438 590–596 [DOI] [PubMed] [Google Scholar]
- Miller S., Krijnse-Locker J. (2008). Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P. D., Pogany J. (2012). The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 10 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. A., Voorhorst W. G., Wery J., Wellink J, van Kammen A. (1992). A regulatory role for the 32K protein in proteolytic processing of cowpea mosaic virus polyproteins. Virology 191 81–89 [DOI] [PubMed] [Google Scholar]
- Pouwels J., Carette J. E., Van Lent J., Wellink J. (2002a). Cowpea mosaic virus: effects on host cell processes. Mol. Plant Pathol. 3 411–418 [DOI] [PubMed] [Google Scholar]
- Pouwels J., Van Der Krogt G. N., Van Lent J., Bisseling T., Wellink J. (2002b). The cytoskeleton and the secretory pathway are not involved in targeting the cowpea mosaic virus movement protein to the cell periphery. Virology 297 48–56 [DOI] [PubMed] [Google Scholar]
- Prod’Homme D., Jakubiec A., Tournier V., Drugeon G., Jupin I. (2003). Targeting of the turnip yellow mosaic virus 66K replication protein to the chloroplast envelope is mediated by the 140K protein. J. Virol. 77 9124–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C., Laporte C., Gaire F., Dunoyer P., Schmitt C., Duval S., et al. (2002). Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J. Virol. 76 8808–8819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott M. E., Gilchrist A., Lee L., Rochon D. (1995). Nucleotide sequence of tomato ringspot virus RNA1. J. Gen. Virol. 76 465–473 [DOI] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kaariainen L. (2005). Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285 139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfacon H. (2005). Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can. J. Bot. 83 1529–1549 [Google Scholar]
- Sanfacon H., Zhang G., Chisholm J., Jafarpour B., Jovel J. (2006). “Molecular biology of Tomato ringspot nepovirus, a pathogen of ornamentals, small fruits and fruit trees,” in Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues (1st Edn) ed.Teixeira da Silva J. (London: Global Science Books; ) 540–546 [Google Scholar]
- Sanfacon H., Iwanami T., Karasev A., Van der Vlugt R., Wellink J., Wetzel T., et al. (2011). “Family Secoviridae”. in Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on the Taxonomy of Viruses, eds King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. (San Diego: Elsevier; ) 881–899 [Google Scholar]
- Sasaki J., Ishikawa K., Arita M., Taniguchi K. (2012). ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites. EMBO J. 31 754–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad M. C., Jensen P. E., Carrington J. C. (1997). Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16 4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhy D. A., Giddings T. H., Jr., Kirkegaard K. (2000). Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74 8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N. L., Levenson E., Rinaudo M. S., Egger D., Bienz K., Gorbalenya A. E., et al. (2006). Evidence for functional protein interactions required for poliovirus RNA replication. J. Virol. 80 5327–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N. L., Lauber C., Jensen K. S., Levenson E. A., Gorbalenya A. E., Ehrenfeld E. (2011a). Identification of tolerated insertion sites in poliovirus non-structural proteins. Virology 409 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teterina N. L., Pinto Y., Weaver J. D., Jensen K. S., Ehrenfeld E. (2011b). Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J. Virol. 85 4284–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. A., Sit T. L., Callaway A. S., Allen N. S., Lommel S. A. (2004). Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320 276–290 [DOI] [PubMed] [Google Scholar]
- van Kuppeveld F. J., van den Hurk P. J., Zoll J., Galama J. M., Melchers W. J. (1996). Mutagenesis of the coxsackie B3 virus 2B/2C cleavage site: determinants of processing efficiency and effects on viral replication. J. Virol. 70 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viry M., Serghini M. A., Hans F., Ritzenthaler C., Pinck M., Pinck L. (1993). Biologically active transcripts from cloned cDNA of genomic grapevine fanleaf nepovirus RNAs. J. Gen. Virol. 74 169–174 [DOI] [PubMed] [Google Scholar]
- Vos P., Jaegle M., Wellink J., Verver J., Eggen R., Van Kammen A., et al. (1988). Infectious RNA transcripts derived from full-length DNA copies of the genomic RNAs of cowpea mosaic virus. Virology 165 33–41 [DOI] [PubMed] [Google Scholar]
- Wang A., Sanfacon H. (2000). Proteolytic processing at a novel cleavage site in the N-terminal region of the tomato ringspot nepovirus RNA-1-encoded polyprotein in vitro. J. Gen. Virol. 81 2771–2781 [DOI] [PubMed] [Google Scholar]
- Wang A., Han S., Sanfacon H. (2004). Topogenesis in membranes of the NTB–VPg protein of tomato ringspot nepovirus: definition of the C-terminal transmembrane domain. J. Gen. Virol. 85 535–545 [DOI] [PubMed] [Google Scholar]
- Weber-Lotfi F., Dietrich A., Russo M., Rubino L. (2002). Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J. Virol. 76 10485–10496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellink J., Rezelman G., Goldbach R., Beyreuther K. (1986). Determination of the proteolytic processing sites in the polyprotein encoded by the bottom-component RNA of cowpea mosaic virus. J. Virol. 59 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E., Duijsings D., Lanke K. H., van Dooren S. H., Jackson C. L., Melchers W. J., et al. (2006). Effects of picornavirus 3A Proteins on Protein Transport and GBF1-dependent COP-I recruitment. J. Virol. 80 11852–11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel T., Chisholm J., Bassler A., Sanfacon H. (2008). Characterization of proteinase cleavage sites in the N-terminal region of the RNA1-encoded polyprotein from Arabis mosaic virus (subgroup A nepovirus). Virology 375 159–169 [DOI] [PubMed] [Google Scholar]
- Yamanaka T., Ohta T., Takahashi M., Meshi T., Schmidt R., Dean C., et al. (2000). TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. U.S.A. 97 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Liu Y., Wimmer E., Paul A. V. (2007). Complete protein linkage map between the P2 and P3 non-structural proteins of poliovirus. J. Gen. Virol. 88 2259–2267 [DOI] [PubMed] [Google Scholar]
- Zhang G., Sanfacon H. (2006). Characterization of membrane-association domains within the tomato ringspot nepovirus X2 protein, an endoplasmic reticulum-targeted polytopic membrane protein. J. Virol. 80 10847–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. C., Zhang G., Yang L., Chisholm J., Sanfacon H. (2005). Evidence that insertion of tomato ringspot nepovirus NTB–VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79 11752–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]