Abstract
We here report a novel phage display selection strategy enabling fast and easy selection of thermostabilized proteins. The approach is illustrated with stabilization of an aggregation-prone soluble single chain T cell receptor (scTCR) characteristic of the murine MOPC315 myeloma model. Random mutation scTCR phage libraries were prepared in E. coli over-expressing the periplasmic chaperone FkpA, and such over-expression during library preparation proved crucial for successful downstream selection. The thermostabilized scTCRmut variants selected were produced in high yields and isolated as monomers. Thus, the purified scTCRs could be studied with regard to specificity and equilibrium binding kinetics to pMHC using surface plasmon resonance (SPR). The results demonstrate a difference in affinity for pMHCs that display germ line or tumor-specific peptides which explains the tumor-specific reactivity of the TCR. This FkpA-assisted thermostabilization strategy extends the utility of recombinant TCRs and furthermore, may be of general use for efficient evolution of proteins.
Evolution of improved recombinant proteins by random mutagenesis and subsequent in vitro selection has been successfully applied to a wide range of protein classes1 and in particular antibodies (Abs)2. The T cell receptor (TCR), which is highly homologous to the Ab, exhibits a delicate specificity for major histocompatibility (MHC) molecules in complex with endogenous and exogenous peptides (pMHC)3, and e.g. molecular delineation of the TCR-pMHC interaction is fundamental for the understanding of adaptive immunity in health and diseases such as cancer and autoimmunity4,5,6. Similar to Abs, molecular evolution of TCRs has also been carried out using both yeast and phage display7,8. Such engineered TCRs may be used to characterize pMHC fine-specificity as well as pave the way for improved immunotherapy9,10. In particular, the ability to raise the affinity of the pMHC-TCR interaction has revealed the potential of modified TCRs as a pMHC-specific tracker and immunomodulator11,12.
There are however still challenges regarding the evolution and expression of recombinant TCRs, as both display levels and soluble expression properties vary extensively between individual clones13,14. Thus, general use of recombinant TCRs is limited, and low throughput bacterial inclusion body expression and refolding is clone-dependent15,16. This limits the utility of modified TCRs17. Contrary, a substantial body of knowledge about the biophysical properties of the diverse Ab repertoire has been gained through the two past decades, which e.g. serves as guide for developing highly efficient engineering and expression systems2,18,19. As such, library selection after stress-induced denaturation of Ab variants has proven useful20,21,22. Combinatorial evolution has also been reported for four TCRs using yeast display, which yielded mutants with increased stability and hence increased solubility and yield9,17,23. These mutations appeared to be partially clone-specific, hence case-dependent optimization still appears necessary. Thus, recombinant TCR technology would strongly benefit from fast and easy methods for engineering and expression.
Here, we take advantage of the recent findings that over-expression of the periplasmic chaperone FkpA led to markedly improved TCR display levels on phage, as well as efficient rescue of functional, soluble TCRs in the E. coli periplasm24,25. A systematic approach was chosen to dissect the effect of FkpA over-expression on the success rate during phage display selection and screening for improved thermal stability of a soluble TCR. The TCR chosen was that of the murine T cell clone 4B2A1, which is specific for the MOPC315 tumor-specific peptide λ2315 amino acids 91–101 that is presented on the MHC class II molecule I-Ed 26,27. Functional periplasmic expression in E. coli of this particular TCR is strictly dependent on the over-expression of FkpA, but the yield is too low and heterogeneous for e.g. structural biology studies and kinetic analysis of the pMHC interaction25. By exploiting a very simple selection regime adopted from Jespers et al21, libraries of randomly mutated single chain TCR (scTCR) variants were subjected to rapid heating and cooling followed by capture of denaturation resistant scTCRs. This led to the retrieval of mutant scTCRs exhibiting markedly increased thermostability. Importantly, successful selection was completely dependent on FkpA over-expression. This chaperone effect is likely to be generic to phage display selection and hence of general importance. The current report also expands the use of the versatile phage display platform to now include TCR stability engineering, complementing the already reported use for TCR affinity maturation8.
The subsequent analysis of the individual mutations identified a single mutation, Leu to Ser in the TRBV13-2*01 germ line segment, as being mainly responsible for the biophysical improvement observed, which translated into reduced aggregation and increased soluble expression of the scTCR. In addition, a Leu to Pro substitution in the synthetic linker connecting the two variable (V) region domains of the scTCR, acted synergistically to improve stability, and was necessary to obtain sufficient expression yields and homogeneity upon purification to allow for equilibrium Kd determination and specificity analysis. Interestingly, the scTCR exhibited reduced binding to I-Ed in complex with a peptide in which the tumor antigen specific Asn96 residue had been exchanged with the germ line encoded Thr96, elucidating the T cell mediated cancer-specific immunosurveilance and reactivity to altered self28.
Results
scTCR mutagenesis and phage library preparation
An efficient mutagenesis scheme was adopted for phage library preparation. Briefly, the scTCR encoding gene cassette was subjected to random mutagenesis as described29, using Taq polymerase in the presence of the nucleotide analogues dPTP and 8-oxodGTP in addition to the four dNTPs. The mutagenized scTCR cassettes were then used as megaprimers and inserted into two pIII phagemid display vectors by a whole plasmid PCR as described30. The two phagemids used, namely pSEX and pFKPDN, differ in that pFKPDN, but not pSEX, contains a fkpA expression cassette24. The libraries were rescued with either VCSM13 or HyperPhage helper phages, resulting in low or high valence scTCR display31. Based on their transformation frequencies, the individual libraries consisted of 2.3 × 105 and 2.5 × 105 unique clones, respectively. Ten clones from each library were sequenced to investigate the mutagenic load, and the mutation frequency found to range from 0.1–2.3% (average 1.2%) corresponding to 1–18 randomly distributed point mutations per scTCR cassette.
Selection for increased thermostability
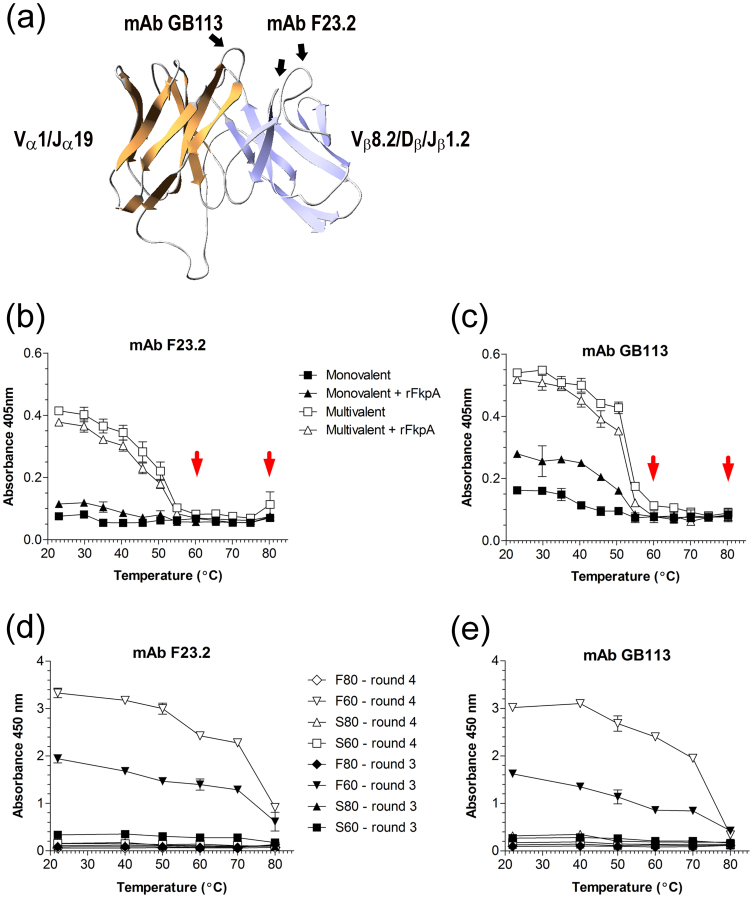
To determine the temperature at which the wt scTCR denatured, phages displaying wt scTCR at low and high valence were heated in a temperature gradient from room temperature to 80°C and tested in ELISA for binding to the mAbs GB113 and F23.2, respectively (Fig. 1a). We found the phages to gradually loose binding to the mAbs as the temperature increased (Fig. 1b and c). There was a distinct transition interval between 50°C and 60°C however, where the mAb reactivity sharply dropped. Therefore, the phages were heated to either 60°C (low stringency) or 80°C (high stringency) before panning, which resulted in 4 different groups: S60 (phage produced without FkpA over-expression and heated to 60°C), S80 (phage produced without FkpA over-expression and heated to 80°C), F60 (phage produced with FkpA over-expression and heated to 60°C), and F80 (phage produced with FkpA over-expression and heated to 80°C). The four groups (S60, F60, S80 and F80) were subjected to a total of four selection rounds, where the mAb bait and block conditions were altered in each round (Table 1). The mAb concentration was kept constant at 6.7 μM throughout, corresponding to 1.2 × 1013 capturing molecules, thus greatly exceeding the phage input. The F60 sample showed the strongest, yet modest total enrichment of about 30-fold from round 1 to 4 (Table 1). Furthermore, phage denaturation followed by mAb capture in ELISA using the polyclonal selection outputs from round 3 and 4 confirmed an increasing population of phages with mAb binding ability at elevated temperature in the F60 library only (Fig. 1d and e).
Figure 1.
(a) A homology model presented in ribbons of the scTCR 4B2A1 was generated using Swiss-Model (swissmodel.expasy.org/) and shows the suggested structure of the scTCR.The postulated mAb bindings sites onto the CDRs is indicated (b and c) 1010 cfuampR/ml phages were heated in a temperature gradient ranging from room temperature to 80°C. After heating and cooling, the samples were tested for binding to mAb F23.2 (b) and GB113 (c) in a phage capture ELISA. The red arrows indicate the temperatures (60°C and 80°C), which were used for library denaturation prior to selection. The ELISA experiment was performed in triplicate and the mean ± SD is shown. (d and e) Polyclonal phage capture ELISA after three and four rounds of selection. 1010 cfuampR/ml phages were heated in a temperature gradient ranging from room temperature to 80°C. After heating in different temperature intervals and cooling, parallel aliquots with normalized phage content were tested for binding to mAbs F23.2 (d) and GB113 (e) in a phage capture ELISA. The ELISA experiment was performed in duplicate and the mean ± SD is shown. Open and closed symbols are display without (prefixed S followed by the temperature to which the given library was exposed for denaturation) and with (prefixed F followed by the temperature to which the given library was exposed for denaturation) FkpA over-expression during phagemid rescue, respectively.
Table 1. Summary of selection.
| Library | Selection round | Bait | Block | Input (I) | Output (O) | Ratio (O/I) | Enrichmenta | Total enrichmentb |
|---|---|---|---|---|---|---|---|---|
| S60 | 1 | F23.2 | Skim milk | 1.2 × 1010 | 6.5 × 105 | 5.4 × 10−5 | ||
| 2 | GB113 | BSA | 1.1 × 1011 | 2.7 × 107 | 2.5 × 10−4 | 4.6 (I) | ||
| 3 | F23.2 | BSA | 2.1 × 1011 | 1.7 × 105 | 8.1 × 10−7 | 0.003 (II) | ||
| 4 | GB113 | Skim milk | 2.9 × 1011 | 4.6 × 105 | 1.6 × 10−6 | 2.0 (III) | 0.03 | |
| S80 | 1 | F23.2 | Skim milk | 1.2 × 1010 | 1.6 × 104 | 1.3 × 10−6 | ||
| 2 | GB113 | BSA | 1.3 × 1011 | 9.5 × 104 | 7.3 × 10−7 | 0.6 (I) | ||
| 3 | F23.2 | BSA | 3.5 × 1011 | 2.5 × 106 | 7.1 × 10−6 | 9.7 (II) | ||
| 4 | GB113 | Skim milk | 3.8 × 1011 | 5.0 × 103 | 1.3 × 10−8 | 1.8 (III) | 10.5 | |
| F60 | 1 | F23.2 | Skim milk | 1.9 × 1010 | 2.0 × 103 | 1.1 × 10−7 | ||
| 2 | GB113 | BSA | 1.1 × 1011 | 1.8 × 105 | 1.6 × 10−5 | 145.5 (I) | ||
| 3 | F23.2 | BSA | 2.4 × 1011 | 5.1 × 106 | 2.1 × 10−5 | 1.3 (II) | ||
| 4 | GB113 | Skim milk | 3.1 × 1011 | 1.0 × 106 | 3.2 × 10−6 | 0.2 (III) | 28.4 | |
| F80 | 1 | F23.2 | Skim milk | 1.9 × 1010 | NAc | NAc | ||
| 2 | GB113 | BSA | 1.9 × 1011 | 6.2 × 105 | 3.3 × 10−6 | NA | ||
| 3 | F23.2 | BSA | 2.3 × 1011 | 1.6 × 105 | 7.0 × 10−8 | 0.02 (II) | ||
| 4 | GB113 | Skim milk | 2.4 × 1011 | 1.5 × 104 | 6.3 × 10−8 | 0.9 (III) | 0.018 |
aEnrichment was defined as the ratio (O/I) from the selection round in question divided with the ratio from the previous round, e.g. I = round 2/round 1.
bTotal enrichment: I × II × III.
cThe output was below the threshold of the titration protocol (≥6.67 × 103).
Screening for thermostabilized soluble scTCR mutants
The output of all four groups (S60, F60, S80 and F80) after selection round 4 were batch cloned into vectors for soluble expression25. To further study the influence of FkpA over-expression, all were cloned into expression vectors either with (pFKPEN) or without (pHOG) the gene encoding fkpA, resulting in a total of eight independent transformations. Then, 46 single colonies from each were expanded in 400 μl expression cultures, separated into the medium and periplasmic fractions, and tested for binding to mAbs GB113 and F23.2 in ELISA (Supplementary Table 1). Binding was verified in an up-scaled protocol. While we found no binders in the libraries produced without FkpA over-expression or heated to 80°C, the library produced with FkpA over-expression and heated to 60°C yielded 22% positive clones. The number further increased to 89% positive clones when FkpA also was over-expressed during soluble expression.
A total of 15 candidate clones, all originating from the F60 library and showing the highest expression levels were sequenced. They were found to group into four unique mutants, A to D, of which three occurred repetitively (A, #8; B, #1; C, #3; D, #3), and upon closer scrutiny, the A and C mutants had the overall highest expression levels (Supplementary Table 2). Clone A had 8 point mutations resulting in 1 amber stop (Q31STOPα), 3 silent mutations and 4 amino acid substitutions. Of these, two were found in CDR loops, namely F25SVα (S*) in CDR1α and I50SVα (S¤) in CDR2α, one was found in the linker connecting the two V domains, namely L115P (P), and one was found in Vβ, namely L214SVβ (S). Clone C had 3 point mutations resulting in 1 amber stop (Q5STOPα) and 2 amino acid substitutions in Vβ, namely K199RVβ (R) and L214SVβ (S), the latter shared with the A clone (Fig. 2). The three silent mutations found in the A clone were analyzed against the GenBank E. coli K12 codon usage and all three represent changes from frequently used to less frequently used codons.
Figure 2. Sequence alignment of the wt and selected A and C clones.
The amino acid sequence of the 4B2A1 scTCR is shown with the corresponding coding sequence aligned with the two mutant clones. Mutations and their phenotype are indicated below the coding sequences. CDR and frame work (FR) annotations are according to Hare et al54. The 4B2A1 TCR V domain composition is [Vα1, Jα19/Vβ8.2, Dβ, Jβ1.2]55 with the corresponding genotype [TRAV7D-3*01, TRAJ40*01/TRBV13-2*01, TRBD1*01, TRBJ1-2*01]. The linker between the Vα and Vβ domains in the scTCR is indicated in white font43.
FkpA over-expression results in elevated and even display levels
To delineate the contribution of the individual amino acid substitutions identified, we made single and combination mutants by in vitro mutagenesis of the mother clone. These new clones were then inserted into the original phagemids with or without the gene encoding fkpA. The selected A and C clones harbored mutations denoted S*S¤PS and RS, respectively. A combination mutant of the A and C clones harboring all five mutations S*S¤PRS, was named A+C. The single mutants were named according to the amino acid targeted, as S*, S¤, S, R and P. The combination mutants were named PR, PS, PRS, and S*S¤P.
We then analyzed and compared the display levels of wt and mutant scTCRs in the presence and absence of FkpA over-expression at low valence display by SDS PAGE/western blotting detected against pIII (Fig. 3a). Indeed, varying display levels were seen when phagemid rescue was done in the absence of FkpA over-expression (Fig. 3a, upper panel). Importantly, both A and C exhibited a higher display level than the wt mother clone. The highest display level was however seen with the PRS clone. When the clones were rescued with FkpA over-expression, all exhibited a high and fairly even display level (Fig. 3a, lower panel), the one exception being the artificially reconstructed P mutant.
Figure 3. Analysis of scTCR-pIII display levels, thermostability profile and soluble expression.
The A and C clones harbor the S*S¤PS and RS mutations, respectively. A combination mutant of the A and C clone harboring all five mutations S*S¤PRS, is named AC. The single mutants are named according to the amino acid targeted, as S*, S¤, S, R and P. The combination mutants are named PR, PS, PRS, and S*S¤P. (a) Phagemids encoding the wt and mutant scTCRs were rescued with VCSM13 helper phage yielding low valence display. Samples corresponding to 1 × 1010 cfuampR/lane were separated by non-reducing SDS-PAGE (4–12%) followed by western blotting and pIII detection. Samples rescued without and with over-expression of FkpA are shown in the upper and lower panel, respectively. (b) scTCR thermostability profile when displayed on phage. To generate the temperature denaturing slopes of the multivalently displayed scTCR versions, aliquots of 109 cfuampR/ml of each sample were heat denatured in a temperature gradient and assessed for mAb GB113 reactivity by ELISA as described in Methods. For each sample, 50% residual mAb binding was determined and denoted the apparent melting temperature, Tm. Secondly, the maximum mAb GB113 response (the A450 nm value) of the wt scTCR Vαβ was set to 1 and the corresponding values of the remaining samples normalized accordingly. Hence, the relative maximum mAb reactivity as a function of Tm for each sample is shown. (c) Small scale expression profile of soluble scTCR. 15 μl of normalized periplasma fractions were separated on non-reducing SDS-PAGE (4–12%) followed by western blotting and anti-His6 detection. The upper panel is developed with supersignal west femto, and the lower panel is developed with supersignal west pico.
scTCR stability assessed by phage denaturation
To map the actual stabilizing effect of the mutations, we measured mAb GB113 binding of the panel of phages described above with multivalently displayed scTCRs, rescued in the presence of FkpA after increasing temperature challenge. An apparent Tm was defined for each phage as the temperature at which the phage retained 50% of maximum GB113 binding (Supplementary Fig. S1). We then calculated relative GB113 binding as the ratio between maximum GB113 binding of the mutant and that of the wt scTCR. This was plotted as a function of apparent Tm (Fig. 3b). As we have previously shown that the scTCR with the Vα domain N-terminally (denoted Vαβ) exhibits superior display compared to Vβα24, we included both variants in the assay. Here, the apparent ΔTm was 9°C in favor of Vαβ, strongly pointing towards increased intrinsic stability compared to the Vβα (Fig. 3b and Table 2).
Table 2. scTCR stability analysis by phage heat denaturation and DSF.
| Clone | Mutation | Apparent Tma | ΔTmc | Maximum responsed | Tmb |
|---|---|---|---|---|---|
| wt Vβα | NA | 29°C | −9°C | 0.130 | ND |
| wt Vαβ | NA | 38°C | NA | 0.650 | 52.1°C |
| S*S¤PS | F25SVα/I50SVα/L115Plinker/L214SVβ | 45°C | 7°C | 2.021 | ND |
| RS | K199RVβ/L214SVβ | 49.2°C | 11.2°C | 2.484 | ND |
| S | L214SVβ | 48.3°C | 10.3°C | 2.788 | 56.2°C |
| P | L115Plinker | 44°C | 6°C | 0.992 | ND |
| PS | L115Plinker/L214SVβ | 54°C | 16°C | 2.784 | 57.4°C |
| PRS | L115Plinker/K199RVβ/L214SVβ | 53.8°C | 15.8°C | 2.851 | 57.1°C |
aTm = temperature of which 50% mAb GB113 binding was observed. The values were computed from the mean of two independent experiments and are hence given as single values.
bTm = determined by DSF.
cDifference in Tm compared to wt scTCR Vαβ.
dMaximum A450 nm signal observed in mAb GB113 binding. The values were computed from the mean of two independent experiments and are hence given as single values.
When comparing the wt scTCR with the mutants, there was an increase in apparent ΔTm of 7°C and 11.2°C for the A and C clone, respectively (Table 2). Interestingly, there was an even larger increase in apparent ΔTm of 16°C when removing the two CDR mutations in the Vα domain of the A clone (PS mutant). From the single mutation analysis, it was evident that both the P and the S mutant contributed to the increased thermostability. Furthermore, all mutants exhibited increased mAb GB113 reactivity as compared to the wt, and the largest effect was seen with the S mutant. Combining this with the linker mutation (PS), and in addition the R mutation (PRS), yielded two very similar mutants both exhibiting more than 4-fold increase in mAb GB113 reactivity combined with an increased ΔTm of approximately 16°C as compared to the wt.
Expression and characterization of soluble scTCR mutants
To test how the increase in thermostability translated into improved soluble scTCR expression, the panel of mutants were re-cloned into the fkpA-containing expression vector (pFKPEN) for soluble expression. After small-scale expression, the periplasmic fractions were tested for presence of scTCR by SDS-PAGE and western blotting (Fig. 3c). Of the wt variants, only the Vβα was found in the periplasm, which is in line with previous observations25. Furthermore, a dramatic increase in soluble scTCR expression between this wt and the mutants was seen, and all the mutants, except S*S¤P, were found in the periplasm. As in the phage stability analysis (Fig. 3b), the largest single effect was seen with the S mutation. Similarly, the P and R mutations added to the effect in a synergistic manner as seen for the PS and PRS mutants. The originally selected A clone appeared very similar to the PS mutant.
Based on the small-scale expression analysis, the S, PS and PRS mutants were chosen for further analysis in an up-scaled 400 ml expression followed by affinity purification (IMAC) of the periplasmic fractions. For each sample, the eluted material was concentrated and equal amount (normalized on total protein by A280 nm) analyzed by SDS PAGE/western blotting detected against the His6-tag (Fig. 4a). Again, the hierarchy of expression was as follows: PRS ≥ PS > S > wt.
Figure 4. Analysis of soluble scTCR yield, aggregation profile and secondary structure topology.
(a) Large scale expression profile of soluble IMAC purified scTCR. 0.5 μg of IMAC purified soluble wt and mutant scTCR were separated by non-reducing SDS-PAGE (12%) followed by western blotting and His6 detection. An empty lane was included as a control. The western blot was developed with supersignal west pico. (b) Superdex 200 size exclusion chromatography of 0.250 ml IMAC purified wt and mutant scTCRs with a concentration of 2 mg/ml. Monomer fractions are indicated with an arrow. (c) Analytical size exclusion of concentrated monomer fractions from (b), equal volumes (0.2 ml) of all samples were applied to the column and monomer fractions indicated with an arrow. (d) Circular dichroism (CD) spectra of SEC purified monomeric soluble wt and mutant scTCRs as well as a control scFv Ab fragment collected at 20°C. MRE, mean residual ellipicity.
The IMAC eluates were further purified by size-exclusion chromatography (SEC) and the monomeric fractions isolated. To allow for semi-quantitative comparison of the monomeric yields, the SEC analysis was run with equal sample concentration and in-put volume. We found the mutations to profoundly increase the monomeric yields (Fig. 4b), and no evidence of higher aggregates was seen for the PS and PRS mutants (Fig. 4c). They gave close to identical monomeric production yields of 4.4 and 4.1 mg/L, respectively, which corresponds to ~3.5 fold increase in production yield compared with the wt (Supplementary Table 3).
Structural analysis by circular dichroism (CD) spectroscopy on monomeric SEC purified samples normalized to equal total protein concentration showed an improvement in ellipticity spectra between the wt and the S, PS and PRS mutants on par with that of a well expressed functional scFv control (Fig. 4d). The subsequent secondary topology analysis confirmed an ordered α-helix and β-sheet content and a corresponding reduction in the random coil content for all three mutant scTCRs (Supplementary Table 4). The protein stability of the monomeric SEC purified samples was measured by differential scanning fluorimetry (DSF)32, and a clear difference was seen when comparing the wt and the S, PS and PRS mutants (Supplementary Fig. S2). The wt started to unfold at 40°C and reached a 50% unfolding (Tm value) at 52.1°C, whereas the S, PS and PRS started to unfold at 45°C, yielding Tms of 56.2°C, 57.4°C and 57.1°C, respectively (Table 2). Although the absolute Tm values differ, the same stability hierarchy was seen as with the phage displayed scTCRs (Table 2).
Functional analysis of scTCR mutants
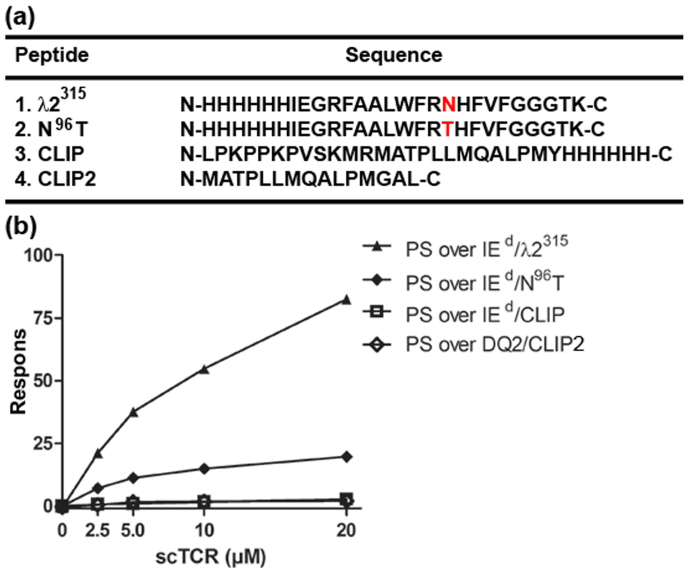
The stability engineered scTCRs were then analyzed for functionality and specificity. We used surface plasmon resonance (SPR) with soluble recombinant pMHC II, (I-Ed/λ2315, aa89–103) immobilized on the chip (Supplementary Fig. S3). The soluble wt, S, PS and PRS scTCR variants were injected and compared for binding to the ligand. All mutants as well as wt scTCR bound I-Ed/λ2315. However, both the wt and S mutant exhibited biphasic binding profiles typical for functional affinity effects due to aggregation and/or oligomerization. The scTCR variants PS and PRS, on the other hand, exhibited binding profiles typical of a monomeric TCR-pMHC interaction. This result was in good agreement with the biochemical analysis in which increased stability conferred by the mutations translated into stable Ig folds and true monomeric forms of the purified scTCR (Fig. 4c and d). The equilibrium dissociation constants (Kd) of both the PS and PRS mutant could therefore be determined as described in Methods and was estimated to be 5.8 ± 2.6 and 7.0 ± 3.3 μM (Kd ± SEM) (Table 3), respectively, which is identical within the experimental variation and in the range typical for agonistic TCR-pMHC interactions3. The PS mutant was then chosen for further analysis and injected over immobilized I-Ed/λ2315, I-Ed/CLIP, DQ2.5/CLIP2 and I-Ed/λ2315 N96T. In the latter pMHC complex, the peptide has the tumor specific amino acid Asp96 residue exchanged with the germ line encoded Thr96. The PS mutant showed high degree of specificity, as it did not bind the irrelevant I-Ed/CLIP or DQ2.5/CLIP2. Moreover, a significantly reduced binding to the germ-line reconstituted I-Ed/N96T complex was observed (Fig. 5).
Table 3. SPR-derived steady-state affinity for TCR variants over I-Ed/λ2315.
aThe steady-state affinity constant was obtained using an equilibrium (Req) binding model supplied by the BIAevaluation software.
bND, not determined.
Figure 5. SPR specificity analysis of the scTCR PS mutant.
Soluble scTCR PS (0, 2.5, 5, 10 and 20 μM) were tested for binding to immobilized I-Ed/λ2315 molecules (370 RU), I-Ed molecules with a germline reconstituted λ2315 peptide mutant, I-Ed/N96T (452 RU), as well as irrelevant control I-Ed/CLIP (445 RU) and a DQ2.5/CLIP2 (432 RU) molecules at 25°C. The SPR response of each interaction were normalized based on amount of active I-Ed on the chip as described in “Methods” and displayed as response. (a) The peptide Ag sequence of the different I-Ed/λ2315 epitope variants as well as the CLIP and CLIP2 peptides. The mutated aa residue at position 96 (P5) is shown in red. (b) scTCR PS binding to immobilized I-Ed/λ2315, I-Ed/N96T, IEd/CLIP and DQ2.5/CLIP2.
Discussion
Compared to the structurally similar Ab, TCRs are generally unstable and difficult to produce in soluble form or as fusion to the coat proteins of yeast and phage. When TCRs are expressed as soluble molecules, and in particular as scTCRs, hydrophobic patches are displayed which promote aggregation33. Furthermore, low affinity between the α and β domains may lead to inefficient chain pairing34. In previous studies, we showed that FkpA over-expression has a major effect on display levels on phage, as well as on soluble periplasmic expression of scTCRs24,25. In this study, we report an improved phage display based thermostability engineering protocol for selection of stable scTCRs exploiting FkpA chaperone assistance35.
As starting point for the library construction, a phagemid harboring the scTCR 4B2A1 with the Vα domain N-terminally was chosen, as this domain orientation showed superior pIII display in a previous analysis24. Random mutations were then introduced in the scTCR by a PCR method that yields high mutagenic load29. The selection regime used was adopted from the work of Jespers et al21. In that study, multivalent display was found to be critical for successful selection, and this was obtained by use of a phage genome vector. In many situations, phagemid-based display is preferable for phage display selection of folded domains, such as Abs and TCRs36, and we therefore used phagemid display and obtained multivalency by use of a pIII-devoid helper phage31. Thus, the protocol may be readily implemented to a range of standard phagemid display platforms without reformatting to a phage genome vector.
Selection of scTCR 4B2A1 variants was done using two specific mAbs, GB113 and F23.2, as capture bait. Before library selection, the temperature at which displayed wt scTCR denatured was determined as loss of binding to the two specific mAbs in ELISA. In addition, the effect of FkpA over-expression on low and high valence display was investigated. FkpA over-expression was found to increase low valence display levels during virion rescue. Here, pIII fusions compete with helper phage derived pIII, and the majority of the virions are devoid of a fusion altogether37. The effect of FkpA over-expression would thus appear to increase the fraction of pIII fusions on the virions. Upon multivalent display, this FkpA effect should not be operational, which was indeed found to be the case, as no difference in mAb reactivity was seen whether or not FkpA was over-expressed during rescue. However, when the effect of FkpA over-expression was tested on selection performance using multivalent display, selection of improved TCR variants was critically dependent on such FkpA over-expression. Thus, it appears that the FkpA foldase exerts its function both by preventing premature aggregation and by improving the quality of the final folded domain fusion. The latter supports the notion that some of the folding intermediates bound to FkpA may subsequently be released to enter a productive folding pathway38.
The mAb F23.2 binds to a large conformational epitope encompassing both CDR1 and CDR2 of the Vβ8.2 segment39,40. The binding profile of GB113 suggests CDR3α to be a key element of its epitope41,42. The use of both should thus reduce the likelihood of selecting CDR loop mutants, which might well affect pMHC specificity. However, the CDR1 and CDR2 loops of the α chain, as well as the CDR3 loop of the β chain, were not protected, and the two scTCR variants selected for further analyses, A and C, indeed had amino acid changes in CDR1α and CDR2α. Βoth changes were from hydrophobic to hydrophilic amino acids. The E. coli XL1-Blue host used during selection has the supE genotype, and hence suppresses the amber stop at low frequency into Gln. Both clone A and C contained such amber mutations in Gln codons, thus exhibiting clear signs of selection for down-regulation of expression, indicative of host toxicity. New clones were generated to characterize the contribution of each amino acid change. Neither contained amber, nor silent mutations, and this eliminated differences in display levels and soluble expression caused by differences in translation efficiency.
The two scTCR variants selected, A and C, both contained two point mutations outside of CDRs that were chosen for further analyses; namely L214SVβ found in both clones, L115P found in clone A, and K199RVβ found in clone C. The L214SVβ mutation resulted in an alteration of a hydrophobic surface exposed amino acid to a hydrophilic amino acid. This particular residue has earlier been selected during yeast display stability engineering of a TCR harboring the same Vβ gene segment, namely Vβ8.223 (here denoted L80SVβ). The L115P mutation is located in the N-terminal end of the 21 amino acid long and flexible synthetic linker sequence connecting the V domains. This particular linker has been optimized for scFv expression and behaves favorably compared to several other commonly used linkers43. The L115P mutation should thus be general to our system and the Pro residue likely acts by restricting the linker flexibility, hence providing a favorable entropic contribution. The most pronounced effect on stability was seen for L214SVβ. However, further improvement was seen when the L115P mutation was added to make a double mutant. No additional effect was found upon adding the third mutation, making a triple mutant. Thus, K199R may well be a neutral bystander mutation. Both the double and the triple mutants had high display levels, apparent Tm and soluble periplasmic expression yields. The two CDR mutations, one in CDRα1 and one in CDRα2, did not contribute to the TCR stability and was excluded from further studies.
The TCR chosen for engineering was derived from the murine T cell clone 4B2A1, which is specific for a MOPC315 tumor-derived peptide, (amino acids 91–101 of λ2315) presented on the MHC class II molecule I-Ed 28,44,45. Using SPR, we were able to obtain cognate kinetic data from the binding of the double and triple mutants to immobilized ligand. The equilibrium dissociation constants (Kd) were estimated to be in the range typical for TCR-pMHC interactions3. Importantly, both the single L214SVβ mutant and the wt showed clear signs of biphasic binding to the pMHC ligand, typical of functional affinity effects associated with aggregation. The double mutant showed high degree of specificity, as it did not bind irrelevant pMHC or I-Ed with irrelevant peptide, and binding was reduced to a peptide where the idiotype-related and tumor specific amino acid in λ2315 was exchanged with the equivalent found in germline encoded λ2. This is in line with previous reports on the functional specificity of the 4B2A1 T cell clone26,27, and suggests a TCR-pMHC affinity threshold that differentiates between the germ line and tumor-specific peptides and explains the absence of germ line reactivity in vivo.
Thermal challenge in combination with phage display is a simple and easy way to induce domain destabilization, followed by selection of mutants that resist aggregation21. As case-specific stabilization often appears necessary to obtain improved soluble TCR variants, our FkpA-enhanced selection regime therefore offers a straight forward way to stabilize TCRs, and very likely, also other protein classes.
Methods
Library construction
The pFKPDN-scTCR Vαβ4B2A1 plasmid24 was used as a template to introduce random mutations into the scTCR Vαβ4B2A1 cassette by the method of Zaccolo et al46 using the JBS dNTP-Mutagenesis Kit (Jena Bioscience, Jena, Germany). The scTCR Vαβ4B2A1 cassette was amplified using the primer pair scTCR_frwd (5′-CTCAGCCGGCCATGGCC-3′) and scTCR_rev (5′-TTTGGATCCAGCGGCCGC-3′) flanking the NcoI/NotI cassette (RE sites underlined). The manufacturers procedure were followed using 15 fmol template DNA, 0.4 μM of each primer and 30 cycles of amplification. Template DNA was eliminated by DpnI (New England Biolabs Ipswich, MA, USA) digestion and purified with the QIAquickTM PCR purification Kit (Qiagen GmbH, Hilden, Germany). Elimination of mutagenic nucleotides were performed according to manufacturers procedure using 5 μl template, and Phusion DNA polymerase (Sigma-Aldrich,Oslo, Norway) instead of Taq. The final PCR products were run on a 2% agarose gel, purified using a QIAquickTM Gel Extraction Kit (Qiagen), 5′-phosphorylated with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) and used as megaprimers by the method of Miyazaki et al30 in a third “whole plasmid” PCR reaction to introduce the mutated scTCRs into the phagemids (pFKPDN or pSEX)24. A control reaction mix without megaprimers was prepared to monitor the background of template DNA in the library. The PCR products were ligated using T4 DNA ligase (New England Biolabs, Ipswich, MA, USA), digested with DpnI (New England Biolabs, Ipswich, MA, USA), seeDNATM precipitation (GE Healthcare, Uppsala, Sweden) and transformed into electrocompetent XL1-Blue cells (Stratagen, La Jolla, CA, USA). Libraries were amplified followed by phagemid rescue using Hyperphage (Progen Biotechnik GmbH, Heidelberg, Germany), or VCSM13 helper phages (GE Healthcare, Uppsala, Sweeden) (at MOI 4) as described43. The virions were precipitated twice with PEG/NaCl and titrated as described47. To determine the sizes of the resulting libraries, aliquots (10 μl and 0.1 μl in a total volume of 100 μl 2× YT-medium) of transformed XL1-Blue cells containing the scTCR Vαβ4B2A1 libraries were spread on LB agar plates supplemented with 100 μg/ml ampicillin and the number of colony forming units (cfu) was counted. The background of template DNA in the libraries was determined by counting cfu from the control reaction mix. To establish the frequency of mutations introduced into the scTCR Vαβ4B2A1 cassette, 10 random clones from each of the libraries were sequenced by the in-house ABI lab core facility (Dept. Molecular Biosciences, University of Oslo, Norway).
SDS-PAGE and western blotting
The display level of the scTCR Vαβ4B2A1-pIII fusions was investigated by SDS-PAGE and western blotting basically as described24. Investigation of soluble scTCRs were conducted essentially as described25 using either normalized pepriplasmic or medium inputs or 0.5 μg of IMAC purified samples.
Heat denaturation of phages and ELISA
Phages rescued with M13K07 or HyperPhage were heated in a temperature gradient from RT to 80°C using a thermal cycler, cooled to 4°C and tested for binding to both mAbs GB113 and F23.239,40,41 in ELISA basically as described48. mAbs GB113 and F23.2 were adsorbed at 250 ng/well and a 1:4 dilution of phage preparations in PBST were used.
Phage selection
Four rounds of panning were performed with alternations between target and block as follows. mAbs GB113 and F23.2 (3 ml at 1 μg/ml in PBS) were immobilized on MaxisorpTM Immunotubes followed by blocking with 3 ml 4% w/v skim milk (Acumedia, Lansing, MI, USA) or 2% w/v BSA (Sigma Aldrich) in PBS. 100 μl PEG purified phages (~1010 cfuampR) of the mutated scTCR Vαβ4B2A1 library in two different phagemids (pSEX and pFKPDN) were heated to 60°C or 80°C for 10 min, blocked in 3 ml block solution followed by incubation with pre-blocked mAb coated tubes. All incubation steps were performed with rotation. Tubes were washed sequentially with PBST (10× in rounds 1 and 2; 20× in rounds 3 and 4) and PBS (10× in rounds 1 and 2; 20× in rounds 3 and 4). The mAb bound phages were eluted by 10 min incubation with 500 μl Trypsin/EDTA mix (BioWhittaker, Lonza Group Ltd., Visp, Switzerland) and recovered by infection of E. coli XL1-Blue cultures at A600 nm 0.4. To determine the phage output, volumes of 50 μl infected cultures were titrated as described above. Phage input for next round of panning were prepared as described above.
Screening and soluble protein expression
pSEX and pFKPDN phagemids encoding potentially thermostablized scTCRs were isolated after fourth round of panning. The scTCR cassettes were moved from the phagemids to the soluble expression vectors pHOG and pFKPEN25 by NcoI/NotI cassette exchange, and transformed into electrocompetent XL1-Blue cells. Expression and isolation of medium and periplasmic fractions were performed on randomly selected single colonies basically as described25, using 96 deep well plates (Nunc, Roskilde, Denmark), 400 μl incubation volume, and 0.1 mM isopropyl-β-D-thio-galactopyranoside (IPTG) induction. Four controls were included on each plate; wt pHOG-scTCR Vαβ4B2A1, wt pFKPEN Vαβ4B2A1, pHOG21-scFv anti-phOx49 and negative control (only media and additives). Clones exhibiting increased protein yield in initial 96 deep well screen were expanded as 10 ml cultures.
Large scale expression and purification of soluble scTCR variants
Protein expression and isolation of soluble periplasmic fractions from 400 ml cultures were performed as described25. IMAC purification, concentration, dialysis against 1 × PBS buffer supplemented with 0.5 M NaCl, and 0.01% azid (pH 6.5) and protein concentration determination were performed as described50. Size exclusion chromatography (SEC) on a Superdex 200 10/300 GL column was performed on an automated ÄKTA 900 chromatography system, (GE Healthcare, Uppsala, Sweden) using a 2 mg/ml dilution in a total volume of 250 μl of each IMAC purified fractions. Pooled monomeric fractions were concentrated to equal volumes, protein concentrations determined as above and an analytical size exclusion chromatography were performed.
CD spectroscopy
CD spectra were recorded using a Jasco J-810 spectropolarimeter (Jasco International Co., Ltd., Tokyo Japan) basically as described25, using a protein concentration of 0.150 mg/ml for the scTCRs and control scFv Ab fragment (kind gifts from Affitech Research AS, Oslo, Norway). The secondary structure elements of the proteins were calculated from ellipticity data, using the spectral fitting method CONTIN/LL in the CDpro package51.
Protein stability measurement by differential scanning fluorimetry (DSF)
Protein stability of the SEC purified monomeric scTCRs was measured by DSF using a Lightcycler RT-PCR machine (Roch). SYPO Orange (Sigma) was used at a 1:1000 dilution and protein concentration at 0.1 mg/ml in a volume of 25 μl. Samples were run in triplicates in 96 well Lightcycler 480 Multiwell Plate. The RT-PCR machine was programmed to ramp the temperature from 25°C to 80°C after a stabilization period for 10 min at 25°C. Data were collected every 0.5°C using the 450 nm excitation and 568 nm emission filters. Data transformation and analysis were performed using the DSF Analysis protocol essentially as described32.
Expression and purification of soluble pMHC
The soluble recombinant I-Ed/λ2315, I-Ed/N96T, I-Ed/CLIP and DQ2.5/CLIP2 molecules were generated as previously described52,53. Briefly, α- and β-chains of the I-Ed were separately expressed and purified as inclusion bodies from E. coli BL21 (DE3), followed by refolding with either the MOPC315 tumor-derived peptide λ2315 amino acids 85–105, a germline reconstituted λ2315 peptide mutant, N96T, or the CLIP peptide. The DQ2.5/CLIP2 were expressed in insect cells and purified from culture supernatant53.
Kinetic evaluation of the TCR-pMHC interaction
A Biacore T200 instrument was used together with a CM3 sensor chip (GE Healthcare, Uppsala, Sweden). All experiments were performed in HBS-EP+ buffer (GE Healthcare, Uppsala, Sweden). Soluble I-Ed/λ2315 was immobilized by amine coupling chemistry to ~400 RU, as described by the manufacturer (GE Healthcare, Uppsala, Sweden). Samples ranging from 0–15 μM scTCR were then injected over I-Ed/λ2315 as well as a negative inactivated reference cell, at a flow rate of 25 μl/min at 25°C. In all experiments, data were reference cell value subtracted. The single cycle kinetics protocol was employed for Kd determination and data evaluation done with the T200 kinetic evaluation software as described by the manufacturer (GE Healthcare, Uppsala, Sweden).
Specificity evaluation of the TCR-pMHC interaction
Soluble I-Ed/λ2315, I-Ed/N96T, I-Ed/CLIP and DQ2.5/CLIP2 were immobilized to ~400 RU, using a Biacore T200 instrument, CM3 chip and HBS-EP+ buffer as above. scTCR PS (0 μM, 2.5 μM, 5 μM, 10 μM and 20 μM) were injected over over I-Ed/λ2315, a germline reconstituted λ2315 peptide mutant, I-Ed/N96T, and a irrelevant MHC molecule I-Ed/CLIP as well as a negative inactivated reference cell, at a flow rate of 25 μl/min at 25°C. As above, data were reference cell value subtracted. To account for differences in immobilization level as well as level of functional I-Ed molecules on the chip a conformational dependet mAb, scFv-anti I-Ed (2.5 μM), were injected over each flow cell (except DQ2.5/CLIP2) and the absolute RU response were used as a measure of relative amount of active I-Ed on the chip. The subsequent SPR measurements of the TCR-pMHC interactions were normalized according to the relative amount of active I-Ed on the chip surfaces, and displayed as response. For the DQ2.5/CLIP2 the absolute response is shown.
Author Contributions
G.Å.L. and I.S. conceived the study. K.S.G., S.G.K., T.F., S.J. and G.Å.L. performed the experiments. B.B. and S.B. both provided crucial reagents. K.S.G., I.S. and G.Å.L. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary info
Acknowledgments
The authors would like to thank Sathiaruby Sivaganesh for excellent technical assistance, Elin Bergseng for reagents, Gabriel Cordara for access to the Lightcycler RT-PCR instrument and Prof. Kjetil Taskén for access to the Biacore T200 instrument. K.S.G., T.F. and G.Å.L. were financially supported by the Norwegian Research Council (Grants no. 179573, 175358 and 174796). S.J. and S.B. were financially supported by the Danish Council for Independent Research, Medical Sciences (Grant no. FSS, 10-083458).
References
- Hosse R. J., Rothe A. & Power B. E. A new generation of protein display scaffolds for molecular recognition. Protein Sci 15, 14–27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom H. R. Selecting and screening recombinant antibody libraries. Nat Biotechnol 23, 1105–1116 (2005). [DOI] [PubMed] [Google Scholar]
- Rudolph M. G., Stanfield R. L. & Wilson I. A. How TCRs bind MHCs, peptides, and co-receptors. Annual review of immunology 24, 419–466 (2006). [DOI] [PubMed] [Google Scholar]
- Garcia K. C., Adams J. J., Feng D. & Ely L. K. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol 10, 143–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa J. B. et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig K. W., Call M. J., Deng L. & Mariuzza R. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Current opinion in immunology 21, 590–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieke M. C. et al. Selection of functional T cell receptor mutants from a yeast surface- display library. Proc Natl Acad Sci U S A 96, 5651–5656 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nature biotechnology 23, 349–354 (2005). [DOI] [PubMed] [Google Scholar]
- Weber K. S., Donermeyer D. L., Allen P. M. & Kranz D. M. Class II-restricted T cell receptor engineered in vitro for higher affinity retains peptide specificity and function. Proceedings of the National Academy of Sciences of the United States of America 102, 19033–19038 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. High-affinity TCRs generated by phage display provide CD4+ T cells with the ability to recognize and kill tumor cell lines. J Immunol 179, 5845–5854 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Rohena A. et al. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nature medicine 14, 1390–1395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddy N. et al. Monoclonal TCR-redirected tumor cell killing. Nature medicine, 10.1038/nm.2764 (2012). [DOI] [PubMed] [Google Scholar]
- Richman S. A. et al. Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments. Molecular immunology (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxel G. I. et al. Some lessons from the systematic production and structural analysis of soluble (alpha)(beta) T-cell receptors. J Immunol Methods 350, 14–21 (2009). [DOI] [PubMed] [Google Scholar]
- Clements C. S. et al. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr D Biol Crystallogr 58, 2131–2134 (2002). [DOI] [PubMed] [Google Scholar]
- Boulter J. M. et al. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng 16, 707–711 (2003). [DOI] [PubMed] [Google Scholar]
- Aggen D. H. et al. Identification and engineering of human variable regions that allow expression of stable single-chain T cell receptors. Protein Eng Des Sel 24, 361–372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewert S., Honegger A. & Pluckthun A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 34, 184–199 (2004). [DOI] [PubMed] [Google Scholar]
- Rothlisberger D., Honegger A. & Pluckthun A. Domain Interactions in the Fab Fragment: A Comparative Evaluation of the Single-chain Fv and Fab Format Engineered with Variable Domains of Different Stability. Journal of molecular biology 347, 773–789 (2005). [DOI] [PubMed] [Google Scholar]
- Jung S., Honegger A. & Pluckthun A. Selection for improved protein stability by phage display. Journal of molecular biology 294, 163–180 (1999). [DOI] [PubMed] [Google Scholar]
- Jespers L., Schon O., Famm K. & Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nature biotechnology 22, 1161–1165 (2004). [DOI] [PubMed] [Google Scholar]
- Famm K., Hansen L., Christ D. & Winter G. Thermodynamically stable aggregation-resistant antibody domains through directed evolution. Journal of molecular biology 376, 926–931 (2008). [DOI] [PubMed] [Google Scholar]
- Shusta E. V., Holler P. D., Kieke M. C., Kranz D. M. & Wittrup K. D. Directed evolution of a stable scaffold for T-cell receptor engineering. Nature biotechnology 18, 754–759 (2000). [DOI] [PubMed] [Google Scholar]
- Løset G. Å., Lunde E., Bogen B., Brekke O. H. & Sandlie I. Functional phage display of two murine α/β T-cell receptors is strongly dependent on fusion format, mode and periplasmic folding assistance. Protein Eng Des Sel. 20, 461–472 (2007). [DOI] [PubMed] [Google Scholar]
- Gunnarsen K. S. et al. Periplasmic expression of soluble single chain T cell receptors is rescued by the chaperone FkpA. BMC Biotechnol 10, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogen B., Malissen B. & Haas W. Idiotope-specific T cell clones that recognize syngeneic immunoglobulin fragments in the context of class II molecules. Eur J Immunol 16, 1373–1378 (1986). [DOI] [PubMed] [Google Scholar]
- Bogen B. & Lambris J. D. Minimum length of an idiotypic peptide and a model for its binding to a major histocompatibility complex class II molecule. Embo J 8, 1947–1952 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthay A. et al. Primary antitumor immune response mediated by CD4+ T cells. Immunity 22, 371–383 (2005). [DOI] [PubMed] [Google Scholar]
- Zaccolo M., Williams D. M., Brown D. M. & Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. Journal of molecular biology 255, 589–603 (1996). [DOI] [PubMed] [Google Scholar]
- Miyazaki K. & Takenouchi M. Creating random mutagenesis libraries using megaprimer PCR of whole plasmid. BioTechniques 33, 1033–1034, 1036–1038 (2002). [DOI] [PubMed] [Google Scholar]
- Rondot S., Koch J., Breitling F. & Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol 19, 75–78 (2001). [DOI] [PubMed] [Google Scholar]
- Niesen F. H., Berglund H. & Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols 2, 2212–2221, 10.1038/nprot.2007.321 (2007). [DOI] [PubMed] [Google Scholar]
- Nieba L., Honegger A., Krebber C. & Pluckthun A. Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Eng 10, 435–444 (1997). [DOI] [PubMed] [Google Scholar]
- Pecorari F., Tissot A. C. & Pluckthun A. Folding, heterodimeric association and specific peptide recognition of a murine alphabeta T-cell receptor expressed in Escherichia coli. Journal of molecular biology 285, 1831–1843 (1999). [DOI] [PubMed] [Google Scholar]
- Bothmann H. & Pluckthun A. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J Biol Chem 275, 17100–17105 (2000). [DOI] [PubMed] [Google Scholar]
- Bradbury A. R. & Marks J. D. Antibodies from phage antibody libraries. J Immunol Methods 290, 29–49 (2004). [DOI] [PubMed] [Google Scholar]
- Clackson T. & Wells J. A. In vitro selection from protein and peptide libraries. Trends Biotechnol 12, 173–184 (1994). [DOI] [PubMed] [Google Scholar]
- Ramm K. & Pluckthun A. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J Biol Chem 275, 17106–17113 (2000). [DOI] [PubMed] [Google Scholar]
- Staerz U. D., Pasternack M. S., Klein J. R., Benedetto J. D. & Bevan M. J. Monoclonal antibodies specific for a murine cytotoxic T-lymphocyte clone. Proceedings of the National Academy of Sciences of the United States of America 81, 1799–1803 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning T. C. et al. Alanine Scanning Mutagenesis of an [alpha][beta] T Cell Receptor: Mapping the Energy of Antigen Recognition. Immunity 8, 413–425 (1998). [DOI] [PubMed] [Google Scholar]
- Bogen B., Lauritzsen G. F. & Weiss S. A stimulatory monoclonal antibody detecting T cell receptor diversity among idiotype-specific, major histocompatibility complex-restricted T cell clones. Eur J Immunol 20, 2359–2362 (1990). [DOI] [PubMed] [Google Scholar]
- Munthe L. A., Sollien A., Dembic Z. & Bogen B. Preferential positive selection of T lymphocytes which express two different TCR alpha chains, an endogenous and a transgenic. Scandinavian journal of immunology 42, 651–661 (1995). [DOI] [PubMed] [Google Scholar]
- Løset G. Å. et al. Construction, evaluation and refinement of a large human antibody phage library based on the IgD and IgM variable gene repertoire. J Immunol Methods 299, 47–62 (2005). [DOI] [PubMed] [Google Scholar]
- Munthe L. A., Corthay A., Os A., Zangani M. & Bogen B. Systemic autoimmune disease caused by autoreactive B cells that receive chronic help from Ig V region-specific T cells. J Immunol 175, 2391–2400 (2005). [DOI] [PubMed] [Google Scholar]
- Zangani M. M. et al. Lymphomas can develop from B cells chronically helped by idiotype-specific T cells. The Journal of experimental medicine 204, 1181–1191 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M. & Gherardi E. The effect of high-frequency random mutagenesis on in vitro protein evolution: a study on TEM-1 beta-lactamase. Journal of molecular biology 285, 775–783 (1999). [DOI] [PubMed] [Google Scholar]
- Løset G. Å., Kristinsson S. G. & Sandlie I. Reliable titration of filamentous bacteriophages independent of pIII fusion moiety and genome size by using trypsin to restore wild-type pIII phenotype. BioTechniques 44, 551–554 (2008). [DOI] [PubMed] [Google Scholar]
- Loset G. A., Lunde E., Bogen B., Brekke O. H. & Sandlie I. Functional phage display of two murine alpha/beta T-cell receptors is strongly dependent on fusion format, mode and periplasmic folding assistance. Protein Eng Des Sel 20, 461–472 (2007). [DOI] [PubMed] [Google Scholar]
- Kipriyanov S. M., Moldenhauer G. & Little M. High level production of soluble single chain antibodies in small-scale Escherichia coli cultures. J Immunol Methods 200, 69–77 (1997). [DOI] [PubMed] [Google Scholar]
- Qiao S. W. et al. Posttranslational modification of gluten shapes TCR usage in celiac disease. J Immunol 187, 3064–3071 (2011). [DOI] [PubMed] [Google Scholar]
- Sreerama N. & Woody R. W. Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Analytical Biochemistry 287, 252–260 (2000). [DOI] [PubMed] [Google Scholar]
- Justesen S., Harndahl M., Lamberth K., Nielsen L. L. & Buus S. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome research 5, 2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarsten H. et al. Staining of celiac disease-relevant T cells by peptide-DQ2 multimers. J Immunol 167, 4861–4868 (2001). [DOI] [PubMed] [Google Scholar]
- Hare B. J. et al. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat Struct Biol 6, 574–581 (1999). [DOI] [PubMed] [Google Scholar]
- Snodgrass H. R., Fisher A. M., Bruyns E. & Bogen B. Restricted alpha/beta receptor gene usage of idiotype-specific major histocompatibility complex-restricted T cells: selection for CDR3-related sequences. Eur J Immunol 22, 2169–2172 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary info