Abstract
The bacterial Type VI Secretion System (T6SS) functions as a virulence factor capable of attacking both eukaryotic and prokaryotic target cells by a process that involves protein transport through a contractile bacteriophage tail-like structure. The T6SS apparatus is composed, in part, of an exterior sheath wrapped around an interior tube. Here we report that in living cells the cytoplasmic ATPase called ClpV specifically recognizes the contracted T6SS sheath structure causing its disassembly within seconds. ClpV imaging allowed spatial and temporal documentation of cell-cell interactions (termed "T6SS dueling") that likely mark the location of repeated T6SS-mediated protein translocation events between bacterial cells.
The bacterial Type 6 Secretion System (T6SS) is a dynamic apparatus that translocates proteins from predator to prey cells by a mechanism analogous to phage tail contraction (1–3). In Vibrio cholerae, two proteins (VipA and VipB) build a phage tail sheath-like tubular structure in the cytosol of predator cells that exists in two conformations, extended and contracted (3). Contraction of the extended VipA/VipB sheath is thought to drive the T6SS spike and inner tube complex out of the effector or 'predator' cell and into an adjacent target or 'prey' cell (3). Disassembly of the cytoplasmic contracted sheath requires ClpV in vivo (3), a AAA+ ATPase that binds VipA/VipB tubules in vitro and can remodel these structures in the presence of ATP (4, 5). In Pseudomonas aeruginosa, ClpV1-GFP localizes to discrete foci that depend on T6SS function (6). Although ClpV binds VipA/VipB tubules in vitro (4, 5), the ability of this protein to interact with other T6SS components has not been demonstrated in vivo. Accordingly, we imaged ClpV localization in intact cells to examine its possible association with dynamic T6SS structures in vivo such as extended and contracted T6SS sheaths and base plates.
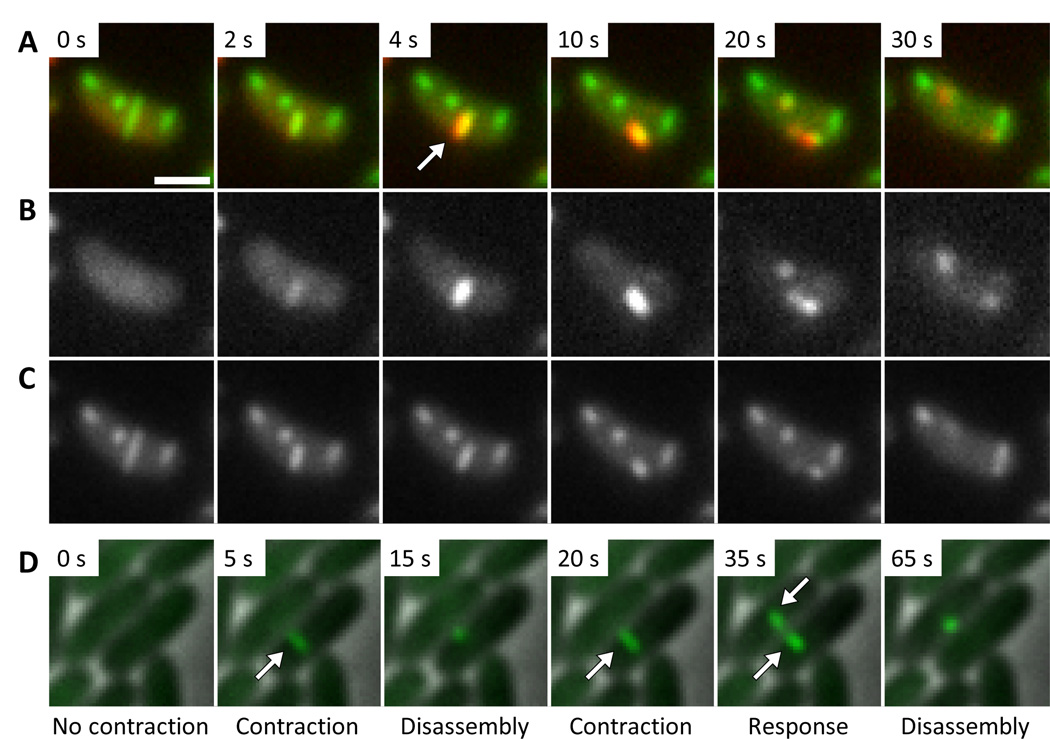
We used time-lapse fluorescence microscopy to follow ClpV localization in live V. cholerae 2740-80 cells. Functional ClpV-super folder GFP (sfGFP) and mCherry2 fusion proteins assembled at random times into short structures that disappeared in 10’s of seconds. In the ΔVipA background, ClpV was evenly distributed in cytosol suggesting that the short ClpV structures were dependent on T6SS sheaths (Fig. S1, Video S1). We used functional VipA-sfGFP (3) and ClpV-mCherry2 fusions to image ClpV and T6SS sheaths simultaneously. Extended VipA-sfGFP containing sheaths were not co-localized with ClpV-mCherry2, while contraction of a sheath led to immediate co-localization of ClpV-mCherry2 with the whole contracted sheath (Fig. 1, Video S2, S3). Half of the ClpV associated with the contracted sheath between 683 ms and 1273 ms (average 952 ms, standard deviation 164 ms, n = 10, Fig. S2, Video S4). The disassembly of the contracted sheath required between 22 and 46 seconds (average 32.5 s, standard deviation 6.1 s, n = 40), measured from the moment of contraction to the moment when both ClpV and VipA signal were no longer co-localized to one spot (Video S2 and S3).
Figure 1. ClpV co-localizes with contracted sheath.
3×3 µm field of cells is shown. Bar in A is 1 µm and applies to A–D. (A–C) V. cholerae ClpV-mCherry2 + pBAD24-VipA-sfGFP. (A) merge of ClpV-mCherry2 and VipA-sfGFP signals. (B) ClpV-mCherry2 signal. (C) VipA-sfGFP signal. Additional frames and cells are shown in Videos S2 and S3. (D) P. aeruginosa ΔretS/ClpV1-GFP, additional frames and cells are shown in Videos S6 and S7.
The Y664A mutation in the pore of ClpV blocks VipA/VipB disassembly but still allows binding of ClpV to VipB in vitro (4) while the F87R mutation of N-terminal domain of ClpV blocks VipB recognition in vitro (5). We found that in vivo ClpV-Y664A-mCherry2 were co-localized with VipA-sfGFP to short non-dynamic structures that were likely contracted T6SS sheaths (Fig. S3, Video S5). In contrast, ClpV-F87R-mCherry2 was distributed uniformly in the cytosol with only VipA-sfGFP localized into contracted non-dynamic sheaths (Fig S3, Video S5). Localization of these ClpV mutants and the change in the dynamics of VipA-containing structures is consistent with published in vitro biochemical data (4, 5) and suggest that in vivo, the N-terminus of VipB is exposed on the surface of the contracted sheath just prior to its disassembly.
In P. aeruginosa, mutation of the regulatory gene retS allows expression of one of its T6SS loci (6). To assess the dynamics of T6SS in P. aeruginosa we imaged a ClpV1-GFP fusion protein in a retS mutant (6) by time-lapse fluorescence microscopy. In contrast to V. cholerae, only a subset of P. aeruginosa cells actively formed and disassembled ClpV1-GFP containing complexes during the observation period; the formation and dynamics of these structures required the VipA homolog PA0083 (Fig. S1, Video S6 and S7). ClpV1-GFP structures often assembled and disassembled repeatedly in apparently the same subcellular location (Fig 1D, Video S7, segments 1–5) indicating that, in contrast to V. cholerae (3), multiple T6SS apparatuses assemble in close proximity, or more likely, T6SS 'base plate components' (3) are recycled by P. aeruginosa.
Interestingly, P. aeruginosa cells apparently responded to T6SS activity occurring in a neighboring sister cell with an increase in their own T6SS dynamics (Fig. 1D, Video S6 and S7). Over time the coincidence of T6SS activity between pairs of sister cells (termed "T6SS dueling") became the dominant category of T6SS activity observable in the P. aeruginosa population (Table S1, Fig. S4). Spatially concurrent T6SS activity could not be documented between V. cholerae sister cells because this species exhibited much higher levels of T6SS activity in nearly all cells (Fig. S4). The spatial and temporal coincidence of T6SS activity in adjacent P. aeruginosa cells strongly suggests that a signal was being transferred between cells precisely at the position of the initial T6SS activity. The P. aeruginosa T6SS is thought to transfer peptidoglycan-hydrolyzing T6SS substrates into sister cells that express immunity proteins to their action (7). We hypothesize that cellular attack by a T6SS apparatus mediated by translocation of T6SS components (e.g., the spike/inner tube complex, or effector proteins) into nearby adjacent sister cells induces local cell envelope alterations (e.g., membrane perturbation, mild peptidoglycan hydrolysis, or protein phosphorylation (7, 8)) that trigger the formation of a T6SS apparatus in the vicinity of such alterations (see Fig. S5).
In conclusion, ClpV imaging provides evidence that P. aeruginosa likely recycles T6SS membrane base plate components and can sense T6SS activity in nearby cells. Because T6SS dueling events were spatially and temporally linked, we hypothesize that they likely mark the exact location of T6SS translocation of protein components (e.g., VgrG and/or effector proteins) between cells. T6SS dueling may reflect social interactions between heterologous T6SS+ species that coexist in the same niche.
Supplementary Material
Acknowledgments
We thank T. G. Bernhardt and N. T. Peters for suggestions on the use of fluorescence microscopy resources and B. Ho and K. Roberts for helpful discussions. This work was supported by NIAID grants AI-018045 and AI-26289 to J.J.M.
Abbreviations
- T6SS
Type 6 secretion system
- GFP
green fluorescent protein
- sfGFP
super folding green fluorescent protein
- ATP
Adenosine triphosphate
References
- 1.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. Embo J. 2009;28:315. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pietrosiuk A, et al. Molecular Basis for the Unique Role of the AAA+ Chaperone ClpV in Type VI Protein Secretion. J Biol Chem. 2011;286:30010. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell AB, et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silverman JM, et al. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun. 2001;69:4681. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalf WW, et al. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 11.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.