SUMMARY
Genetic switches are critical components of developmental circuits. Because temperate bacteriophages are vastly abundant and greatly diverse, they are rich resources for understanding the mechanisms and evolution of switches and the molecular control of genetic circuitry. Here, we describe a new class of small, compact, and simple switches that use site-specific recombination as the key decision point. The phage attachment site attP is located within the phage repressor gene such that chromosomal integration results in removal of a C-terminal tag that destabilizes the virally-encoded form of the repressor. Integration thus not only confers prophage stability, but also is a requirement for lysogenic establishment. The variety of these self-contained integration-dependent immunity systems in different genomic contexts suggests that these represent ancestral states in switch evolution from which more complex switches have evolved. They also provide a powerful toolkit for building synthetic biological circuits.
INTRODUCTION
Genetic circuits typically use input-responsive bi-stable switches to determine phenotypic readouts (Chatterjee et al., 2008). Perhaps the best-studied switch is that determining the outcome of infection by the temperate bacteriophage lambda: either lytic growth involving phage replication and host lysis, or lysogeny, a stable state in which lytic functions are switched off, the phage genome replicates as a component of the host bacterium, and the resulting lysogens are immune to superinfection (Oppenheim et al., 2005). Two central components provide commitment to the outcome of this decision, repressor (cI) for lysogeny and Cro for lytic growth (Hochschild et al., 1986). However, the decision itself is determined by the status of the transcriptional activator cII, which is subject to proteolytic processing by host proteases (Kobiler et al., 2002). Under conditions in which protease levels are low, cII accumulates, activates transcription of both cI and integrase, and lysogeny is established. When protease levels are high, cII is degraded, activation does not occur, and lytic growth proceeds. Various factors influence the decision, including the phage-encoded cIII protein (Halder et al., 2007) and Oop RNA (Krinke et al., 1991), as well as the multiplicity of infection and physiological state of the cell. Further details of the lambda genetic circuitry are reviewed elsewhere (Dodd et al., 2005; Little, 2010; Oppenheim et al., 2005).
Bacteriophages are the most abundant life forms in the biosphere (Suttle, 2007). The phage population is both old and dynamic, and not surprisingly there is huge genetic diversity (Hatfull and Hendrix, 2011). The temperate life style is prevalent, and phages thus likely harbor the greatest variety of genetic switches in biology. Although these may share common elements conferring commitment to lytic or lysogenic growth, it is likely that they have evolved a variety of evolutionary solutions for wiring the circuitry to place the switch in each of the two stable positions, and for flipping the switch from one position to the other.
A large collection of sequenced mycobacteriophages presents an opportunity to examine the generality of lytic-lysogenic circuitry, and how these complex systems have evolved. Over 220 sequenced mycobacteriophage genomes are available and can be grouped into at least 25 types that are distinct at the nucleotide sequence level [Clusters A-Q and 8 singleton phages; (Hatfull, 2012), unpublished observations]. More than half of these encode a recognizable int gene and are presumed to be temperate, although the immunity systems have only been studied in the Cluster A phages L1 (Sau et al., 2004), L5 (Brown et al., 1997; Donnelly-Wu et al., 1993) and Bxb1 (Jain and Hatfull, 2000). Although there are notable departures from the canonical lambda system [for example, there are large numbers of asymmetric repressor binding sites (Brown et al., 1997; Pope et al., 2011)], other aspects of prophage integration and excision are similar (Lewis and Hatfull, 2003; Pena et al., 2000). In the mycobacteriophages described below, bioinformatic analyses suggest more dramatic departures from the lambda system.
We describe here a type of genetic circuitry in which phage immunity is dependent on integration, and in which core components of the lambda switch are absent. A relatively small (~2 kbp) self-contained immunity unit containing all of the functions needed to set and then maintain the switch in one of two states – leading to either lysogeny or lytic growth – is present in diverse mycobacteriophage genomes and examples can be predicted in other prophages. In these systems, integrase plays a decision-making central role by recombination within the repressor gene, and both integrase and the viral form of the repressor are subject to targeted degradation by host proteases. This illuminates the origin and evolution of genetic switches and provides new components for the construction of complex synthetic genetic circuitries (Bonnet et al., 2012; Friedland et al., 2009; Lu et al., 2009).
RESULTS AND DISCUSSION
Non-canonical immunity organizations in mycobacteriophage genomes
Many sequenced mycobacteriophages encode a tyrosine-integrase (Hatfull, 2010, 2012). In some, the immunity functions are organized such that putative repressor genes are located immediately upstream of int (Fig. 1A). This is observed in a group of six closely related phages within Cluster G (Hatfull, 2012; Sampson et al., 2009), three phages in Subcluster I1, two phages within Cluster N (Hatfull, 2012) and the Cluster P genome BigNuz (Hatfull, 2012). In general, there is nucleotide sequence similarity between genomes within a cluster, but not between clusters (Hatfull et al., 2006). We will focus here on three representative types, BPs, Brujita, and Charlie. Each of these is temperate, but forms lightly turbid plaques reflecting relatively low frequencies of lysogeny (~5%), and is heteroimmune with the others (see Fig. 2).
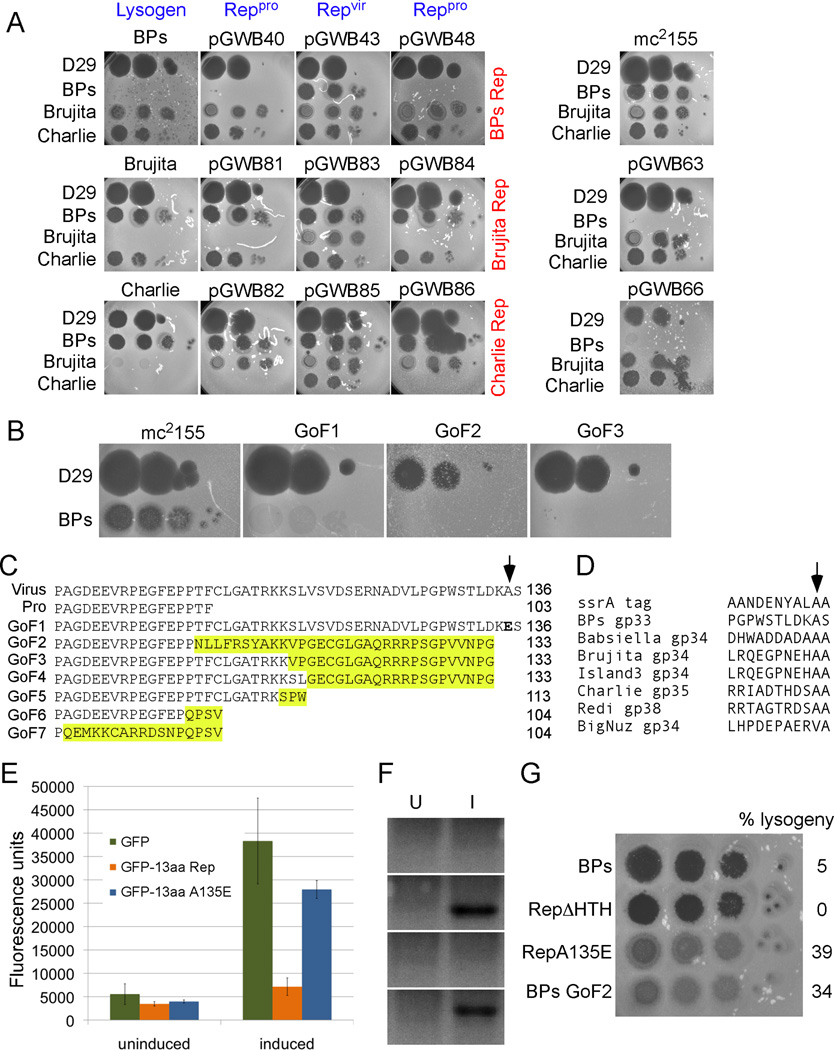
Figure 1. Mycobacteriophages with integration-dependent immunity systemst.
(A) Genome segments are aligned by regions encoding Int, Rep and Cro. Genes are shown as boxes above (transcribed rightwards) or below (transcribed leftwards) each genome, and are colored according to Phamily assignments using Phamerator (Cresawn et al., 2011); Phamily number is shown above or below each gene with the number of phamily members (i.e. homologues) in parentheses. Pairwise nucleotide sequence similarity is represented by spectrum-coloring between adjacent genomes, with violet being the most similar and red the least. The genomes are sorted into the clusters according to overall genome similarity: BPs and Halo, Cluster G; Island 3, Babsiella, and Brujita, Subcluster I1; BigNuz, Cluster P; Charlie and Redi, Cluster N (Hatfull, 2012).
(B) Integration results in reconfiguration of phage repressor genes. The attP common core (green box) lies within the repressor (rep) genes such that strand exchange results in a truncated version of the repressor adjacent to attL, and the 3’ end of the gene (arrowhead) next to attR. (See Figures S1, S2, Tables S1, S2)
Figure 2. Viral and prophage repressor activities.
(A) Serial dilutions of phages D29 (control), BPs, Brujita, and Charlie were spotted onto BPs, Brujita, and Charlie lysogens (first column) and onto strains carrying integrated plasmids (columns 2–4) expressing different forms of the BPs, Brujita, and Charlie repressors in rows 1 – 3 respectively. Plasmids in columns 2 express prophage repressors (Reppro) from their normal integration site, and in columns 3 and 4 express viral repressors (Repvir) or Reppro integrated at attB -L5. Plasmid pGWB63 has a stop codon such that the viral gene expresses the prophage repressor; pGWB66 expresses the viral repressor form from the hsp60 promoter and replicates extrachromosomally. M. smegmatis mc2155 is the control strain. Note that the Charlie lysogen confers partial but non-reciprocal immunity to Brujita, and is not repressor-mediated.
(B) Gain of function mutants (GoF) of the BPs viral repressor. Mutant derivatives of pGWB43 (see panel A) that have gained immunity were isolated, plasmids recovered, and retransformed into M. smegmatis. The full gain of immunity by mutants GoF2 and GoF3, and the partial recovery of immunity in GoF1 is shown.
(C) GoF2-7 contain frameshift mutations near the 3’ end of the viral repressor and generate altered C-termini (yellow). GoF1 has a single amino acid substitution at the penultimate alanine residue (arrow). Protein lengths (in amino acids) are shown.
(D) Alignment of the ssrA proteolytic tag and the last 11 amino acids of the repressor proteins of phages from Clusters G (BPs), I1 (Brujita, Babsiella and Island3), N (Charlie and Redi), and P (Bignuz) phages. Arrow indicates the highly conserved penultimate alanine residue.
(E) The C-terminal 13 resides of BPs Rep tagged to the C-terminus of GFP lead to a loss of fluorescence upon induction. An A135E substitution at the penultimate residue restores GFP activity. Data are represented as mean +/− SD.
(F) Western blot of uninduced (U) and induced (I) M. smegmatis expressing no GFP (control, top panel), untagged GFP (second panel), GFP-13aa Rep (third panel), GFP-13aa A135E (bottom panel) with anti-GFP.
(G) Serial dilutions of BPs, mutants with a deletion of the helix-turn-helix motif in the repressor (RepΔHTH), with the stabilizing A135E substitution, or with a frameshift mutation in the repressor (GoF2) were spotted onto lawns of M. smegmatis mc2155. Lysogeny frequencies were determined as described in Experimental Procedures.
The immunity regions of these phages each encode a putative repressor (rep ; genes 33, 34, and 35, in BPs, Brujita, and Charlie respectively; Fig. 1A) transcribed leftwards, which is relatively small (<140 aa) and contains a predicted helix-turn-helix DNA-binding motif (Table S1). Downstream of the repressor is int, separated from rep by a 120–200 bp intergenic non-coding region. To the right of rep and divergently transcribed from it is a Cro-like protein that is small and also contains a predicted helix-turn-helix DNA binding motif (Fig. 1A, Table S1). The Int proteins are non-canonical (see below) and the common core of the phage attachment site (attP) – readily identified as sharing 35–40 bp of homology to a chromosomal attB site overlapping a tRNA gene – lies wholly within the rep gene (Fig. 1B). BPs, Brujita, and Charlie utilize different attB sites, overlapping host tRNA genes tRNA-Arg (Msmeg_6349), tRNA-Thr (Msmeg_6152) and tRNA-Lys (Msmeg_5758) respectively (Fig. 1B; Fig. S1), as confirmed by sequencing of the attL or attR junctions; strand exchange occurs at the 5’ side of the common cores, as in phage L5 (Peña et al., 1996) (Fig. S1). These features are central to the genetic circuitry governing the lyticlysogenic decision in these phages as described below.
Consequences of integration on repressor function
The location of attP within rep is particularly intriguing (Fig. 1B). As a consequence, integration results in interruption of rep such that the prophage-encoded products are truncated versions of the viral repressor. In BPs, the prophage form (gp33103 or Rep103) is a precise 33-residue truncation of the viral form (c136 or Rep136), the Charlie prophage-encoded protein (gp35103 or Rep103) is 20 residues shorter than the virally-encoded form (gp35123 or Rep123), and the Brujita prophage form (gp34100 or Rep100) is truncated by 28 residues relative to the virally-encoded form (gp34128 or Rep128) but the extreme C-terminal residue is a glutamic acid rather than the virally-encoded glutamine residue (Fig. S2).
Are there functional consequences to the rearrangement of the rep genes by site-specific integration? To address this we constructed strains expressing either the viral or prophage forms of the repressors and tested them for immunity (Fig. 2A). All the prophage forms – regardless of whether they are integrated into their natural attB site or elsewhere – confer immunity to superinfection (Fig. 2A). In contrast, the longer viral repressors fail to confer immunity. To eliminate the possibility that inactivity of the full-length gene results from negative regulatory cis-acting elements in the extreme 3’ end of the gene, we introduced a termination codon within full-length BPs 33 to generate a predicted 103-residue product, while retaining the full-length coding sequence; full immunity is restored (pGWB63, Fig. 2A). Overexpression of the virally-encoded repressors also restores near complete immunity (pGWB66, Fig. 2A).
Because the viral BPs repressor does not confer immunity, gain of function (GoF) mutants can be readily isolated as recombinants that can grow on solid media seeded with a repressor-defective mutant of BPs. Seven independent mutants were recovered and characterized (Fig. 2B); six have frameshift mutations between codons 94 and 119 in rep, resulting in loss of the C-terminal tail and substitution with out-of-frame residues (Fig. 2B, C). One of the mutants has a single amino acid substitution in the penultimate residue (A135E), implicating the extreme C-terminus in loss of function of the viral form.
Alignment of the extreme C-terminal sequences of the repressors (Fig. 2D) shows limited similarity to the M. smegmatis ssrA tag that is added to incompletely translated proteins targeting them for degradation by ClpXP (Raju et al., 2012). Few residues are well conserved, although the two terminal alanine residues important for recognition by E. coli ClpXP (Flynn et al., 2001) are present in most of the repressors (Fig. 2D). This alignment, the GoF mutant in the penultimate residue of BPs gp33136, and the partial restoration of immunity by overexpression, suggests that the viral repressors are non-functional due to degradation by ClpXP. This is supported by the observation that fusion of the C-terminal 13 residues of BPs gp33136 to GFP results in strong reduction in activity, and that the A135E substitution largely restores GFP activity (Fig. 2E). Western analysis shows reduction of GFP with the Rep tag and restoration with the A135E substitution (Fig. 2F).
To determine the functional consequences resulting from targeted degradation of the virally-encoded form of the repressor, we engineered both the A135E and the GoF2 substitution into the BPs genome. Both mutants have normal lytic growth but form plaques that are visibly more turbid than wild-type BPs due to substantial increases in lysogeny (Fig. 2G). These immunity organizations thus have important consequences for the lytic-lysogenic decision.
Repressor control of lytic gene expression
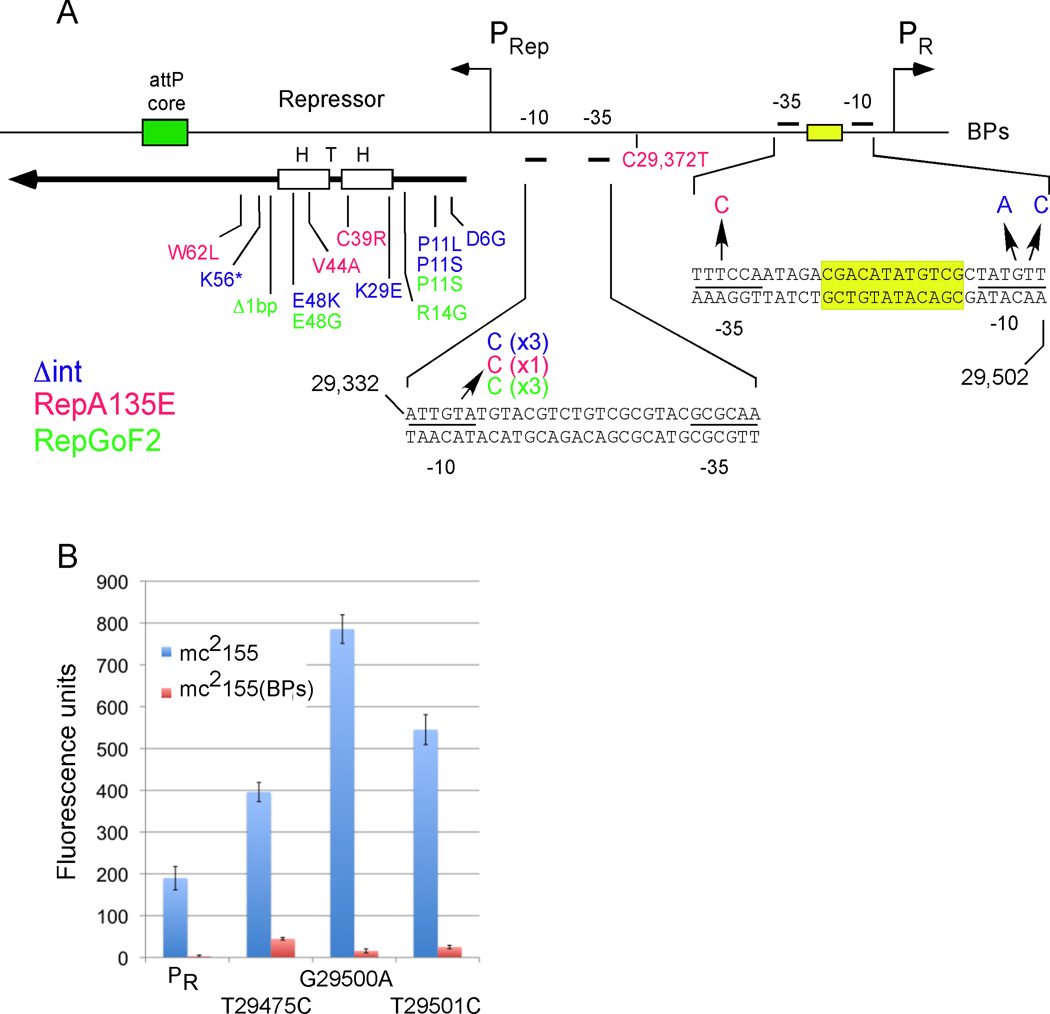
Divergent transcription of the repressors and the presumed lytic genes to their right suggests that expression signals are located in the intergenic region (Fig. 3A). BPs, Brujita, and Charlie all have predicted sigA-like rightwards-facing promoters located immediately upstream of the first rightwards-facing gene (Fig. 3A). When the BPs 33-34 intergenic region is fused to a reporter gene (oriented to measure rightwards transcription, Fig. 3A, B) moderate expression is observed, and mutations within predicted −10 and −35 promoter motifs reduce activity (data not shown); the transcription start site of this PR promoter was confirmed (Fig. 3C). PR is tightly down-regulated in a BPs lysogen, and BPs gp33103 alone is sufficient for full repression (Fig. 3B). The viral gp33136 form of the repressor fails to repress under the same conditions in which it fails to confer immunity, unless the stabilizing A135E substitution is present (Fig. 3B).
Figure 3. Divergent transcription in BPs.
(A) Organization of the rep-cro intergenic region showing the PR and PRep promoters, the OR operator, and the locations of repressor-insensitive mutations within OR (see Figure S3).
(B) The promoters PR and P6 (located between genes 5 and 6) were fused to mCherry and fluorescence units determined in M. smegmatis mc2155, a BPs lysogen [mc2155(BPs)], or strains expressing Rep136 , Rep103 or RepA135E. The PR promoter activities of two repressor insensitive mutants (A29486C, T29489C) are also shown. Data are represented as mean +/− SD.
(C) The trace derived by sequencing PCR products of ligated BPs gp34 (cro) mRNA shows that transcription starts at the first codon; cro coding sequence is shown in green. Cro is expressed from a leaderless mRNA.
(D) Dilutions of either BPs or a D29 control phage were spotted onto lawns of strains carrying plasmids pGB109 or pGB110. The sequence below shows the mutations and substitutions (red) at each of the two rep plausible start sites in each plasmid. Loss of the upstream start codon (in pGB109) destroys immunity.
(E) BPs Rep103-DNA complexes were formed and separated by native gel electrophoresis. DNA substrates are: 366 bp with the entire 33-34 intergenic region, 139 bp containing OR, similar fragments containing either mut 1 or mut 2 in OR (A29486C and T29489C respectively), or a 110 bp DNA containing the 26-27 intergenic region (O27). BPs Rep103 concentrations are: none, 0.16 µM, 0.54 µM, 1.6 µM, 5.4 µM, 16 µM, and 54 µM. Migration of DNA and complexes (cmplx) are indicated, and the multiple complexes formed with the 33-34 substrate are labeled C1 – C4. Controls with non-specific DNA and competition with OR-specific oligonucleotides are shown in Figure S4.
(F) Promoter activity of PRep and a PRep T29336C mutation (see Fig. 6) is shown, as in panel B. Data are represented as mean +/− SD.
(G) Determination of the PRep transcription start site, as in panel C. The coding sequence of gene 33 (rep) is shown in green. Rep also is expressed from a leaderless mRNA.
To determine the site of action of BPs Rep, we took advantage of the observation that although a strain expressing BPs gp33103 confers immunity to BPs superinfection (e.g. pGWB40 & pGWB48, Fig. 2A), mutants that are insensitive to the repressor and which form plaques on this strain can be readily isolated. Thirteen independent mutants (corresponding to 10 different mutations) were isolated and characterized, all of which were found to contain changes in the 33–34 intergenic region (Fig. S3). Eight of these contain inversions, deletions, or small duplications, and two contain single nucleotide substitutions (Fig. 3A, Fig. S3). The latter both lie within a 12bp sequence with dyad symmetry located between the PR −10 and −35 motifs, and which we propose is the operator OR (Fig. 3A). In the promoter reporter plasmid, both mutants were found to have lower levels of repression in the presence of gp33103 relative to the wild-type operator, but were not completely de-repressed; both also have modestly higher promoter activity (Fig. 3B).
The translation start site for the BPs repressor gene was confirmed (Fig. 3D) and BPs gp33103 was shown to bind to 33-34 intergenic DNA (366 bp) as well as to a smaller fragment (139 bp) containing OR (Fig. 3E). The binding pattern to the 139 bp fragment is simple and specific (Fig. 3E, S4), and repressor-insensitive mutations reduce the binding affinity ~10-fold, consistent with modest derepression of PR in vivo (Fig. 3B). Interestingly, the Cluster G phage Angel naturally contains the equivalent substitution as BPs OR-T29489C (Fig. 3A) and is also repressor-insensitive (data not shown). Although Rep may have additional functions, these data support a critical role in binding to OR to repress PR in the integrated prophage. Preliminary data suggest that the viral form of the repressor binds similarly compared to the prophage form.
Although there is only a single copy of the 12 bp OR sequence in the 33-34 intergenic region, there are three additional identical 12 bp copies elsewhere in the BPs genome, located between 5 and 6, 26 and 27, and 54 and 55 (defined as O6, O27 and O55 respectively); each of these is a binding site for BPs Rep (Fig. 3E; Fig. S4). Because gene 27 encodes the endolysin, O27 may simply play a role in silencing the otherwise lethal lysis system during lysogeny, and bioinformatic, reporter gene fusions, and RNAseq analysis all fail to identify a promoter in this region (data not shown). However, both the intergenic regions between 5 and 6 and between 54 and 55 have promoter activity and are BPs gp33103–regulated (Fig. 3B, data not shown). The control of virion structural genes by Rep is somewhat surprising. We note that a phage mutant in which O6 has been mutationally altered, maintains the ability to lysogenize (R. King, GWB and GFH, unpublished observations).
It is plausible that one or more of these other operators regulates repressor action at OR through long-range DNA looping, as observed for OL and OR of phage lambda (Lewis et al., 2011). However, we note that the inter-site distance is greater (the closest site, O27 is 5.6 kbp away; lambda OL and OR are separated by 2.1 kbp), that we failed to recover any repressor-insensitive mutations outside of the BPs 33–34 intergenic region, and that although cognate operators can be predicted in Brujita and Charlie, there are no additional copies outside of the regulatory intergenic region. We also note that the repressor-insensitive mutants are virulent and capable of infecting a BPs lysogen, although individual plaques are extremely small (Fig. S3).
Repressor synthesis
In phage lambda, repressor synthesis is central to the genetic switch and is different for the establishment and the maintenance of lysogeny (Oppenheim et al., 2005). This is critical to the circuitry, because without this separation the dependency on cII would be lost. The question thus arises as to whether separate promoters are responsible for repressor expression in lysogenic establishment and maintenance in these mycobacteriophages. To address this, we placed the BPs 33–34 region upstream of a reporter (oriented such that mCherry is in an equivalent position to rep) and showed there is robust promoter activity ~2-fold higher than PR and is active in the absence of other phage-encoded functions, including Rep (Fig. 3F). A putative sig-A promoter was identified upstream of rep (Fig. 3A), which we designate as PRep, a base substitution within the putative −10 motif eliminates promoter activity, and the transcription start site was mapped (Fig. 3F, G). This promoter is not responsive to the prophage or viral repressor forms, but is modestly up-regulated by the stabilized viral repressor mutant, RepA135E (Fig. 3F). We propose that PRep is responsible for both lysogenic establishment and maintenance.
Simple integrases using simple attachment sites
The integration systems used by these phages are non-canonical and differ from the well-studied lambda and related systems in several important ways. First, the Int proteins lack the N-terminal arm-type binding domain present in lambda and in most other Integrases (Fig. 4A) (Landy, 1989; Lee and Hatfull, 1993), confirmed through mutational analysis of potential translation start sites (Fig. S5). Second, they contain an extended C-terminal tail, 36–79 residues C-terminal to the conserved catalytic tyrosine (Fig. 4A), in contrast to 14 residues in lambda Int. With the possible exception of BPs Int and its close relatives, these extensions are insufficiently long to include a DNA-binding motif and thus are unlikely to provide arm-type binding activity. These integrases are thus members of the subset of tyrosine recombinases – like Cre, FLP and XerC/D – that do not share the bivalent DNA binding property of λ and other phage Integrases, which enables formation of higher-order protein-DNA architectures central to control of recombination directionality. Third, the attP sites are non-canonical and the Brujita attP requirements are within a 74 bp segment containing the common core (Fig. 4B); Charlie attP also requires <100 bp (data not shown). The apparent absence of arm-type Int binding sites is consistent with the atypical Int organization. These observations raise the question as to how directionality of recombination is regulated in the absence of bivalent Int binding (Landy, 1989).
Figure 4. The role of integrase in the establishment of lysogeny.
(A) Organization of non-canonical mycobacteriophage integrases. Each contains a proposed catalytic domain (red) and a core-type DNA binding domain (blue), but lack the arm-type DNA binding domain of λ Int (green). However, they contain C-terminal extensions (yellow) (see Figure S5).
(B)attP requirements for integration were determined by co-transformation of attP plasmids with int-expressing plasmid pLB03 (Table S4) and selection for the attP plasmid. Transformation frequencies for plasmids pBL06, pBL08, pBL10, pBL14, pBL26, pBL15, pBL23, pBL24, pBL39 (top to bottom with BPs coordinates shown) are expressed as percentages of pBL06 (6.7 × 103 cfu/µg DNA). Deletion beyond the right side of the attP core (at 29,211) is predicted to interfere with tRNA function. Competency of cells using plasmid pMH94 was 2 × 105 transformants/µg DNA.
(C) Serial dilutions of BPs and BPs mutants were spotted on a lawn of M. smegmatis mc2155. Lysogeny frequencies were determined as described in Experimental Procedures and reported as % recovery. The mutant (Δint) that removes sequences downstream of the repressor yields 100% recovery of colonies – reflecting the plaque turbidity – but does not form recoverable stable lysogens (Figure S6).
(D) Integration-proficient plasmids carrying a selectable marker (KanR) and phage cassette including attP and int stably transform by chromosomal integration. The transformation frequencies (transn/µg DNA) of various plasmids are shown. Substitutions stabilizing either Brujita or Charlie Int (in pGWB87 and pES14 respectively) substantially increase transformation efficiency.
(E) Int C-termini are aligned with the M. smegmatis ssrA tag, showing their spacing from the catalytic tyrosine. Arrow indicates the penultimate residue where substitutions stabilize Int. The BPs C-terminus is atypically longer and does not have a canonical ssrA tag. Charlie Int has a valine residue at the penultimate position which likely accounts for the 50-fold higher transformation efficiency of pGWB82 relative to pGWB81 (see panel D).
(F) Addition of 5 C-terminal residues of Brujita Int destablizes GFP and the A296E substitution restores activity. Western blots show that activities reflect GFP protein levels (data not shown), and addition of the C-terminal 60 aa of BPs Int to GFP has the same effect (data not shown). Data are represented as mean +/− SD.
(G) Brujita mutants with deletions of either int(Δint) or rep (Δrep) make clear plaques and do not form lysogens. Both a stabilized form of Int and a catalytically defective Int fail to form lysogens (see Figure S7).
(H) Transformants of pGWB81 (wt Int) or pGWB87 (IntA296E; see panel D) were grown either with or without selection for the specified number of generations and the proportion of colonies retaining the plasmid determined.
Integration is required for lysogenic establishment
Because prophage establishment is required for active repressor synthesis, site-specific integration must be a determining component of the lytic-lysogenic switch. Furthermore, if lysogenic establishment is not dependent on activation of repressor synthesis (as with lambda cII) how is the outcome of infection decided? To address this we constructed a BPs mutant with an alanine substitution in the catalytic tyrosine of Int (Y256A); this mutant forms completely clear plaques, and is completely defective in lysogeny (Fig. 4C). Integration is thus required for immunity and lysogeny; two additional mutants with termination codons in Int have the same phenotype (Fig. 4C). However, a mutant in which the entire region downstream of the rep gene (including int and int-rep intergenic region; see Fig. 1B) is deleted forms plaques with increased turbidity (Fig. 4C), and although healthy stable lysogens cannot be recovered, 100% of cells survive infection to form small colonies (Fig. S6). This phenotype is repressor-dependent (Fig. 4C) and perhaps results from loss of an undefined regulatory element downstream of rep that normally prevents overexpression of Rep from replicating viral genomes (see pGWB66, Fig. 2A).
If establishment of lysogeny is dependent on integration and Int is expressed from the constitutive promoter PRep (Fig. 1B), what prevents lysogeny being the outcome of all infections? A clue is provided by the behavior of non-replicating integration-proficient plasmids that contain the rep-int segments of BPs, Brujita, or Charlie (Fig. 4D). These plasmids transform M. smegmatis through chromosomal integration, but do so at greatly reduced frequencies relative to canonical integration systems (Fig. 4D) (Lee et al., 1991). We note that the extended C-termini of these Int proteins (Fig. 4A) – with the exception of BPs – contain ssrA-like tags (Fig. 4E) similar to those on the viral repressors, suggesting that proteolytic degradation, presumably by the host ClpXP system, contributes to the low transformation frequencies. Consistent with this, addition of five C-terminal Brujita residues destabilizes GFP and activity is restored by a glutamic acid substitution at the penultimate residue (A296E) (Fig. 4F). A stabilizing glutamic acid substitution in Brujita Int results in an 800-fold increase in transformation efficiency, and substitutions at the Charlie Int C-terminus also increase transformation efficiency (Fig. 4D). BPs Int has a longer C-terminus with no obvious ssrA-like tag, and although its 64 C-terminal residues destabilize GFP, the location of the signal is not clear (data not shown).
Control of integration and excision
Targeted proteolysis of Int suggests that it is the key determinant of the lytic-lysogenic switch – analogous to the function of lambda cII – in addition to conferring prophage stability. To explore this further we examined the phenotype of a phage Brujita mutant with the stabilizing IntA296E substitution. Although we anticipated that this substitution would increase lysogeny, we observed the opposite effect; clear plaques and no stable lysogens recovered (Fig. 4G; Fig. S7). Although puzzling, a model in which the directionality of recombination is controlled solely by the amount of integrase present, such that the stabilizing Int mutant also mediates efficient excision, could explain this. To test this, we examined the stability of plasmid transformants generated with the wild-type Brujita integrating plasmid (pGWB81) or the stabilized Int derivative (pGWB87) (Fig. 4H). After 26 generations of unselected growth, the IntA296E integrated plasmid remained in only 27% of cells, whereas the wild-type plasmid was fully maintained. These data suggest that excision does not require an RDF (an RDF gene could be embedded within int, but exhaustive BLAST searches failed to support this) and that wild-type prophage maintenance simply results from insufficient Int for excision, the consequence of separation of int from PRep by recombination (Fig. 1B) and targeted degradation of Int. How int is expressed during prophage induction is not clear, but could result from an unidentified (and perhaps activated) promoter between attR and int (Fig. 1B), in response to expression of the adjacent bacterial genes (Msmeg_6348, Msmeg_6151, and Msmeg_5759 in BPs, Brujita and Charlie respectively; Fig. S1), or from stress-related regulation of the host proteases.
Commitment to lytic growth
Modulation of integrase activity through proteolysis must play a central role in the lyticlysogenic decision, although there is expected to be a counterbalancing function that provides commitment to lytic growth. In phage lambda this is provided by the Cro protein, which acts antagonistically to the cI repressor, shutting down repressor synthesis though binding to lambda OR. We propose that the first genes of the early lytic operons of the mycobacteriophages discussed here play similar roles, and all are small with predicted helix-turn-helix DNA binding motifs (Table S1). We note, however, that a Cro-like phenotype could be observed either through direct influence on repressor levels, or by acting to promote excisive recombination. To test this we constructed strains containing either BPs gene 34 (cro) and the 33-34 intergenic region, or the same region but containing the OR mutation T29489C (to derepress cro expression; Fig. 3A). We then examined plaque morphologies and measured lysogenization frequencies on these strains. We observed a marked reduction in lysogeny of BPs, with the greatest effect generated by the derepressed mutant (Fig. 5A). Similar phenotypes were observed with BPs mutants, including Δint, demonstrating that this phenotype does not arise from the Cro-mediated stimulation of excision. The mechanism of this Cro-mediated antagonism of lysogeny is not clear, although in promoter reporter assays BPs Cro does not modulate PRep activity (Fig. 5B).
Figure 5. BPs Cro antagonizes lysogeny.
(A) Serial dilutions of BPs and BPs mutants were spotted onto M. smegmatis mc2155 and strains expressing BPs cro from the wild type promoter PR (pGWB76) or the repressor insensitive mutant PR T29489C (pGWB78). Lysogenization frequencies of phages on M. smegmatis pGWB78 were determined and can be compared with the parent strain in Figures 2G and 4C.
(B) The activities of PR and PRep promoter reporter plasmids were measured in a strain expressing Cro from PR (red bars) or from the depressed PR (green bars), both on a lysogen and non-lysogen. Data are represented as mean +/− SD.
The integrase-repressor cassette contains all the components for the lytic-lysogenic decision
To determine whether there are additional requirements for immunity outside of the BPs intrep-cro cassette, we isolated a collection of clear plaque mutants defective in lysogeny. Because wild-type BPs plaques are insufficiently turbid to easily identify clear plaque variants, we initiated the screen with three mutant parents that give more turbid plaques and have higher lysogenization frequencies, BPsRepA135E, BPsGoF2, and BPsΔint (see Figs. 2 and 4). A total of 25 independent clear plaque mutants were isolated and mapped. All of these have mutations within the int-rep-cro cassette suggesting that this contains all the information required for lysogeny and that there are no counterparts of lambda cII or cIII (Fig. 6A). Moreover, the phenotypes of all of the mutants (with one exception, see below) can be readily accounted for.
Figure 6. All clear-plaque mutations map within the BPs 32–34 locus.
(A) Independent clear plaque mutants were isolated using three mutant parent phages that have increased turbidity, RepA135E (red), RepGoF2 (green) and Δint (blue), and the mutations mapped. The locations of mutations or amino acid substitutions in repressor are shown.
(B) Effects of clear plaque mutations on PR activity. Data are represented as mean +/− SD.
Not surprisingly, almost one half of the mutations are within the repressor gene itself (Fig. 6A). Two of these 13 mutants have frameshift and nonsense mutations respectively, and the other 11 have single amino acid substitutions. Only five of these are within the putative HTH DNA binding motif, and another five lie between the N-terminus and the HTH motif, suggesting an important role for this part of the protein. Seven independent clear plaque mutants recovered from all three parents have the identical single base substitution (T29336C) in the −10 element of the PRep promoter that strongly reduces PRep activity (Fig. 3F). Because lysogeny is effectively eliminated in these PRep mutants, PRep is critical for maintenance of repressor synthesis.
Three of the mutants have single base substitutions in PR, two in the −10 motif and one in the −35 (Fig. 6A). These mutants all have a modest promoter-up phenotype, but are also partially derepressed, so that expression is increased 8–10-fold in a lysogen (Fig. 6B). These mutations are thus similar to the two repressor-insensitive mutations mapping in OR (Fig. 3), although the PR −35 up-promoter mutant (which was isolated in a BPsRepA135E background) is defective in lysogenic establishment rather than maintenance, and forms lysogens at low frequency (<1%). Presumably repression is sufficient to support maintenance, but the increased expression of Cro promotes a lytic outcome of infection. The same phenotype is observed for the mutation at position 29,372 (Fig. 6A) but why this mutation leads to a clear plaque phenotype is unclear. It is plausible that when the viral repressor escapes proteolysis that it binds to this region (see Fig. 3F) and activates PRep to stimulate Int expression and lysogenic establishment. This is supported by modest stimulation of PRep by the stabilized RepA135E mutant (Fig. 3F).
A model for a simple genetic circuit
These observations support a simple but elegant model for the regulation of the lytic and lysogenic life cycles in these mycobacteriophages. The end-points of the decision are similar to those in phage lambda, where expression of repressor maintains lysogeny, confers immunity to superinfection, and shuts down expression of Cro (Fig. 7); expression of Cro provides commitment to lytic growth and antagonizes the action of the repressor. However, the decision process is distinctly different. Upon infection, both PR and PRep promoters are active, initiating synthesis of Cro (from PR) as well as Int and repressor (from PRep) (Fig. 7); the balance of these promoter activities is important, with PRep twice as active as PR (Fig. 3). The outcome of infection is determined by the stability of Int. If Int activity is relatively high – either at high moi or when cellular protease levels are low – then integration into the host genome occurs and the active form of repressor is made, shutting down PR and expression of Cro. If insufficient active Int is expressed, then integration does not occur, and accumulation of Cro results in shutting down of Int and Rep synthesis, and lytic growth proceeds.
Figure 7. A model for integration-dependent bacteriophage immunity.
During infection by phages using integration-dependent immunity systems, the promoters PR and PRep are both active, leading to synthesis of Cro, Repressor (Rep) and Integrase (Int). The outcome of infection is decided by the stability of Int determined through its C-terminal ssrAlike tag. If Int fails to accumulate due to proteolysis, then only the viral form of Rep is made which in turn is degraded through its C-terminal ssrA-like tag; Cro accumulates, antagonizes repressor action, and lytic growth ensues. A putative function located between the rep and int genes (inhib?) may also down-regulate repressor synthesis from the viral genome. If Int escapes proteolysis (either at high moi or when cellular proteases are at low levels) then integration occurs, the active truncated form of the repressor is made which binds to OR and downregulates PR and cro-expression ceases. Lysogeny is thus established. Repressor is made constitutively from PRep, which is not autoregulated, and binds with relatively low affinity. Spontaneous induction may occur in conditions where Rep falls below critical levels, or by expression of Int, perhaps from the M. smegmatis genes adjacent to attR, Msmeg_6348, Msmeg_6151, or Msmeg_5759 for the three attB sites described here (See Figure S1).
A notable feature of this model is the central role played by Int. Not only does Int report the status of the infection through proteolysis, but it is a key determinant of prophage maintenance (Fig. 7). Because the site-specific recombination system is simple and there is no obvious role for an RDF, the stability of Int also determines the directionality of recombination. Once sufficient Int is made to promote integration, Int synthesis is interrupted by uncoupling of int from PRep; any residual Int is presumably degraded and the prophage is maintained simply through lack of sufficient Int to promote excision. The question then arises as to how Int is made when the phage is induced into lytic growth. None of the phages described here are inducible by DNA damaging agents, although spontaneous induction is observed and phage titers in the culture of a lysogen approach 1010 pfu/ml. Although spontaneous induction could occur when repressor levels fall below a critical level, an intriguing possibility is that induction occurs solely through expression of Int. This could result from a cryptic promoter between the rep and int genes, or perhaps from induced expression of the adjacent chromosomal genes (Msmeg_6348, Msmeg_6151, and Msmeg_5759 for the three attB sites described here; Fig. S1).
Evolution of the genetic circuitry
These integration-dependent phage immunity systems provide important clues as to how the more complex lambda-like systems could have evolved (Little, 2010), and we propose that these are excellent candidates for ancestral forms of genetic switches. First, they have relatively few components and are contained within small compact genetic units ~2 kbp long. Moreover, they are self-contained and can thus move laterally throughout the phage population to introduce new immune specificities. Second, there is no requirement for additional factors such as cII, cIII and Xis, all of which could have been added as subsequent elaborations. It is less simple to imagine how integration-dependent immunity systems could have been derived with multiple simultaneous accommodating mutations. Finally, we note that not only are multiple examples of these integration-dependent immunity systems found among diverse mycobacteriophages, but bioinformatic analyses suggest that it is quite widespread. For example, we have identified at least five other examples of prophages that we predict have integration-dependent immunity systems, with all or most of the five characteristic features: 1) a putative repressor gene encoding a product of ~100 residues, 2) an adjacent tRNA gene transcribed towards the repressor gene, 3) direct repeats of 30–40 bp in the 3’ end of the adjacent tRNA gene and 40–70 kbp away (defining the size of the prophage), 4) an int gene near to the second repeat which both lacks an N-terminal domain, and contains a putative degradation tag at its C-terminus, and 5) a predicted viral form of the repressor that also contains a degradation tag (Table S2).
Concluding remarks
The vast, dynamic, and diverse population of bacteriophages is a rich resource for discovering genetic novelty, and the genetic circuits described here provide an elegant solution for how biological switches can be regulated with a small number of genetic components. Perhaps not surprisingly, the key component to establishing the bistable switch is recombination – perhaps the simplest of all possible means of generating alternative states by exchanging segments of DNA. This is thus consistent with a view in which recombinases such as phage integrases have played key roles in evolution, because of their capability of generating different regulatory states. We also note that the availability of multiple small self-contained genetic switches as described here will provide a powerful collection of components for the construction of complex synthetic genetic circuits, with considerable promise for biological counting and data storage systems (Bonnet et al., 2012; Friedland et al., 2009; Lu et al., 2009). Moreover, introduction of recombination modules into other repressors could increase the numbers and types of regulatable systems.
EXPERIMENTAL PROCEDURES
Growth of bacteria and phages
M. smegmatis and recombinants were grown in 7H9 broth or 7H10 agar (Difco) with appropriate antibiotics; 1mM CaCl2 was included for phage infections. Phages were propagated as described previously (Sarkis and Hatfull, 1998). Lysogenization frequencies were determined by plating serial dilutions of M. smegmatis onto plates seeded with 2 × 109 pfu of phage compared to non-seeded plates. Immunity to phage infection was tested by spotting serial dilutions of phage lysates onto lawns of M. smegmatis. Phages and plasmids used in this study are described in Tables S3 and S4 respectively.
Isolation of gain-of-function BPs repressor mutants
Plasmid pGWB43 was isolated from E. coli XL1-Red mutator cells (Stratagene) and independent plasmid preparations were transformed into M. smegmatis mc2155. Colonies were recovered on plates seeded with 2 × 109 pfu of BPs 33ΔHTH, plasmids recovered by electroduction in E. coli, and the int-rep region sequenced.
Isolation of BPs mutants
Repressor-insensitive BPs mutants were isolated by plating independently grown lysates with M. smegmatis mc2155 carrying plasmids pGWB40 or pGWB48; plaques were recovered and purified. The complete genome was sequenced for three mutants (BPs102a, BPs102e, BPs102g) and a smaller region (either BPs coordinates 29,176–29,914, or 29,042–29,914) for the other mutants. Spontaneous independent clear plaque mutants were isolated from lysates of BPsΔint, BPsRepA135E, BPsRepGoF2 plated onto M. smegmatis mc2155 to a density of about 104 pfu/plate; clear plaque mutants were identified at frequencies of 10−4 – 10−5. The int-cro regions were sequenced following PCR amplification (BPs coordinates 27,586–29,914).
Integration and stability
attP requirements for integration were determined by co-transformation of M. smegmatis mc2155 with attP-containing plasmids and int-expressing plasmid pLB03. Plasmid stability was determined by growing M. smegmatis containing pBL06, pGWB81, and pGWB87 to saturation with selection, diluting 1:10,000 into drug-free medium and growing to saturation twice. Cultures were plated for individual colonies and tested for drug resistance.
Fluorescence of C-terminally tagged GFP constructs and promoter-mCherry reporter fusions
C-terminally tagged GFP plasmids were constructed in nitrile inducible vectors (Pandey et al., 2009), transformed into M. smegmatis mc2155 and GFP expression induced. Promoter fusions to mCherry were constructed in vector pLO86, transformed into mc2155 or mc2155(BPs), grown for 48hrs at 37°C and fluorescence measured. Activity is reported as fluorescence units (LAU/mm2)/OD of culture.
Transcription start site mapping
Total RNA was isolated from M. smegmatis cultures containing plasmids with either PR- or PRep-mCherry transcriptional fusions. Total RNA was treated with tobacco acid pyrophosphatase, circularized with T4 RNA ligase, and reverse transcribed. The ligated junction was PCR amplified and sequenced.
DNA binding of BPs Rep103
BPs Rep103 was cloned in vector pLC3 and expressed as a maltose binding protein fusion (MBP). Following purification, cleavage, and removal of MBP, Rep103 was concentrated to 1.9 mg/ml and stored in 50% glycerol at −20°C. Binding reactions contained Rep103 dilutions, 20 ng of radiolabeled DNA substrate, and 1µg calf thymus DNA. Protein-DNA complexes were separated on a 5% native polyacrylamide gel.
Supplementary Material
Acknowledgements
We are grateful to Christina Ferreira and Carlos Guerrero for superb technical assistance, to Mariana Piuri for constructing pLO86, and Eric Rubin for providing mCherry. We also thank Scarlett Shell and Sarah Fortune for assistance with transcription start site mapping. This work was supported by National Institutes of Health grants GM093901 and AI059114.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KL, Sarkis GJ, Wadsworth C, Hatfull GF. Transcriptional silencing by the mycobacteriophage L5 repressor. Embo J. 1997;16:5914–5921. doi: 10.1093/emboj/16.19.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Kaznessis YN, Hu WS. Tweaking biological switches through a better understanding of bistability behavior. Curr Opin Biotechnol. 2008;19:475–481. doi: 10.1016/j.copbio.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Shearwin KE, Egan JB. Revisited gene regulation in bacteriophage lambda. Curr Opin Genet Dev. 2005;15:145–152. doi: 10.1016/j.gde.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Donnelly-Wu MK, Jacobs WR, Jr, Hatfull GF. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Levchenko I, Seidel M, Wickner SH, Sauer RT, Baker TA. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc Natl Acad Sci U S A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder S, Datta AB, Parrack P. Probing the antiprotease activity of lambdaCIII, an inhibitor of the Escherichia coli metalloprotease HflB (FtsH) J Bacteriol. 2007;189:8130–8138. doi: 10.1128/JB.00820-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF. Mycobacteriophages: genes and genomes. Annu Rev Microbiol. 2010;64:331–356. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Complete Genome Sequences of 138 Mycobacteriophages. J Virol. 2012;86:2382–2384. doi: 10.1128/JVI.06870-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Hendrix RW. Bacteriophages and their Genomes. Current Opinions in Virology. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A, Douhan Jd, Ptashne M. How lambda repressor and lambda Cro distinguish between OR1 and OR3. Cell. 1986;47:807–816. doi: 10.1016/0092-8674(86)90523-4. [DOI] [PubMed] [Google Scholar]
- Jain S, Hatfull GF. Transcriptional regulation and immunity in mycobacteriophage Bxb1. Mol Microbiol. 2000;38:971–985. doi: 10.1046/j.1365-2958.2000.02184.x. [DOI] [PubMed] [Google Scholar]
- Kobiler O, Koby S, Teff D, Court D, Oppenheim AB. The phage lambda CII transcriptional activator carries a C-terminal domain signaling for rapid proteolysis. Proc Natl Acad Sci U S A. 2002;99:14964–14969. doi: 10.1073/pnas.222172499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke L, Mahoney M, Wulff DL. The role of the OOP antisense RNA in coliphage lambda development. Mol Microbiol. 1991;5:1265–1272. doi: 10.1111/j.1365-2958.1991.tb01900.x. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Lee MH, Hatfull GF. Mycobacteriophage L5 integrase-mediated site-specific integration in vitro. J Bacteriol. 1993;175:6836–6841. doi: 10.1128/jb.175.21.6836-6841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D, Le P, Zurla C, Finzi L, Adhya S. Multilevel autoregulation of lambda repressor protein CI by DNA looping in vitro. Proc Natl Acad Sci U S A. 2011;108:14807–14812. doi: 10.1073/pnas.1111221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Hatfull GF. Control of Directionality in L5 Integrase-mediated Sitespecific Recombination. J Mol Biol. 2003;326:805–821. doi: 10.1016/s0022-2836(02)01475-4. [DOI] [PubMed] [Google Scholar]
- Little JW. Evolution of complex gene regulatory circuits by addition of refinements. Curr Biol. 2010;20:R724–R734. doi: 10.1016/j.cub.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Raman S, Proff R, Joshi S, Kang CM, Rubin EJ, Husson RN, Sassetti CM. Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb) 2009;89:12–16. doi: 10.1016/j.tube.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena CE, Kahlenberg JM, Hatfull GF. Assembly and activation of site-specific recombination complexes. Proc Natl Acad Sci U S A. 2000;97:7760–7765. doi: 10.1073/pnas.140014297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña CE, Stoner JE, Hatfull GF. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J Bacteriol. 1996;178:5533–5536. doi: 10.1128/jb.178.18.5533-5536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, et al. Expanding the Diversity of Mycobacteriophages: Insights into Genome Architecture and Evolution. PLoS ONE. 2011;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012;8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T, Broussard GW, Marinelli LJ, Jacobs-Sera D, Ray M, Ko CC, Russell D, Hendrix RW, Hatfull GF. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology. 2009;155:2962–2977. doi: 10.1099/mic.0.030486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis GJ, Hatfull GF. Mycobacteriophages. Methods Mol Biol. 1998;101:145–173. doi: 10.1385/0-89603-471-2:145. [DOI] [PubMed] [Google Scholar]
- Sau S, Chattoraj P, Ganguly T, Lee CY, Mandal NC. Cloning and sequencing analysis of the repressor gene of temperate mycobacteriophage L1. J Biochem Mol Biol. 2004;37:254–259. doi: 10.5483/bmbrep.2004.37.2.254. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.