Abstract
Tumor cell migration is essential for invasion and dissemination from primary solid tumors and for the establishment of lethal secondary metastases at distant organs. In vivo and in vitro models enabled identification of different factors in the tumor microenvironment that regulate tumor progression and metastasis. However, the mechanisms by which tumor cells integrate these chemical and mechanical signals from multiple sources to navigate the complex microenvironment remain poorly understood. In this review, we discuss the factors that influence tumor cell migration with a focus on the migration of transformed carcinoma cells. We provide an overview of the experimental and computational methods that allow the investigation of tumor cell migration, and we highlight the benefits and shortcomings of the various assays. We emphasize that the chemical and mechanical stimulus paradigms are not independent and that crosstalk between them motivates the development of new assays capable of applying multiple, simultaneous stimuli and imaging the cellular migratory response in real-time. These next-generation assays will more closely mimic the in vivo microenvironment to provide new insights into tumor progression, inform techniques to control tumor cell migration, and render cancer more treatable.
Keywords: Cell migration, Mechanotransduction, Chemotaxis, Interstitial flow, Tumor microenvironment, Microfluidics
Introduction
The metastatic cascade is a complex, multistage process involving modulation of cell phenotype, cell migration, and dynamic homeotypic and heterotypic cell–cell interactions [1]. There are various proposed models for carcinoma progression and the formation of metastases (reviewed in [2]), and tumors likely employ multiple mechanisms in vivo. Metastatic carcinoma progression follows the general pattern of primary tumor growth, tumor cell invasion, intravasation, circulation, extravasation, and growth of the secondary tumor. However, the heterogeneous tumor microenvironment [3], plasticity of invasion [4], and hypoxia-induced genomic instability [5] are among many factors that contribute to tumor heterogeneity, and there is debate as to the source and onset of the metastatic phenotype [6, 7]. Most cancer-related deaths result from the formation of metastases [8], which are difficult to detect and can remain dormant for years after treatment of the primary tumor [9, 10]. The formation of these metastases by disseminated tumor cells is preceded by and requires tumor cell invasion at the primary tumor site, a ubiquitous step in early tumor progression that represents a viable target for therapy [11].
Disseminating carcinoma cells navigate through the tumor microenvironment, across the basement membrane and into the surrounding stroma. Migration is a highly orchestrated process in which cells are guided both by internal and external signals. Mechanical signals, sensed by integrins [12] and other adhesion receptors [13], and chemical signals, sensed by chemokine and growth factor receptors [14], influence the migration of tumor cells. Hence, understanding the mechanisms that guide cell migration in response to various stimuli in the tumor and stromal microenvironments is key to developing therapies that prevent tumor cell migration and render cancer more treatable.
Because it is difficult to isolate the effects of an individual stimulus on cell migration in vivo, in vitro models have emerged as powerful tools for investigating tumor cell migration. These reductionist in vitro assays isolate a subset of stimuli that can be examined in detail to enhance our overall understanding of the important chemical and mechanical signals that guide tumor cell migration. A key assumption in many reductionist experiments is that chemical and mechanical stimuli act in parallel. However, the migrating cell acts as a signal integrator, sensing simultaneous stimuli, activating intracellular pathways, and responding through organized processes that culminate in the extension of protrusions and subsequent migration. Furthermore, tumor cells invade stromal tissue through a variety of mechanisms, and the process of migration is dynamic and a function of tissue substrate [4]. As we look to develop the next generation of assays for tumor cell migration, it is important to consider the crosstalk between chemical and mechanical stimuli, and the role it plays in guiding the migration of tumor cells.
We start by summarizing the experimental and computational approaches that have been developed to study tumor cell migration, and we highlight their benefits and shortcomings. We then discuss the results of these studies and introduce the various stimuli that guide tumor cell migration. We focus on single migrating tumor cells, and we broadly divide the stimuli into mechanical and chemical cues. We conclude by highlighting recent data demonstrating that chemical and mechanical stimuli are not independent and the crosstalk among them strongly influences cell migration. In conjunction with computational models, assays that allow the application of several, simultaneous stimuli will provide insight into tumor cell migration in vivo and help in the development of new methods to control and limit cell migration, improving the efficacy of cancer therapy.
Experimental methods to study tumor cell migration
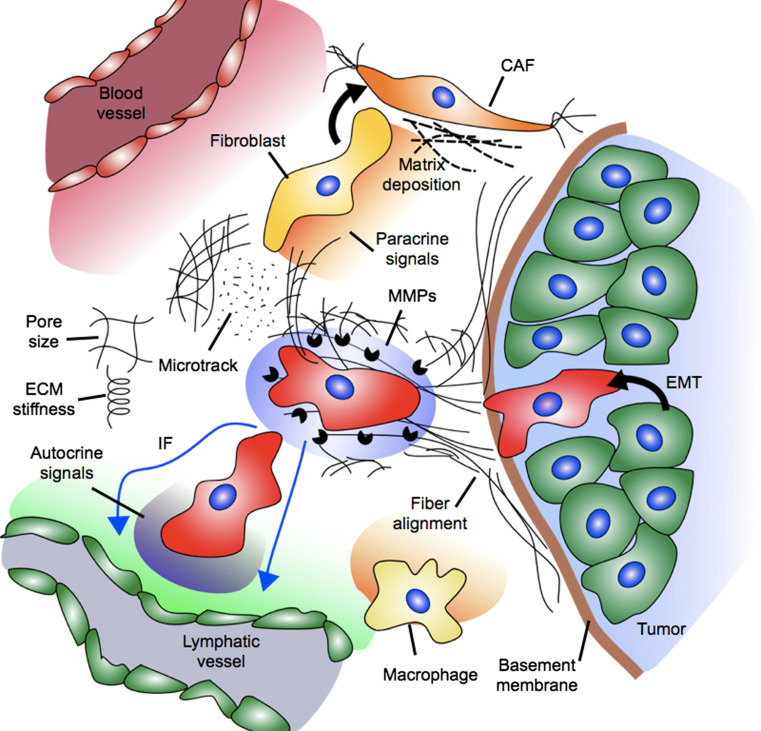
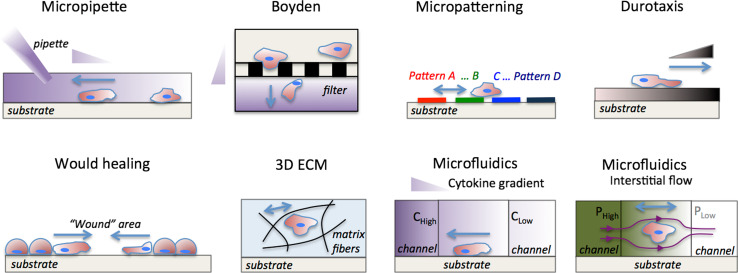
The tumor microenvironment is comprised of a dynamic network of extracellular matrix (ECM) proteins [15] bathed in interstitial fluid and a host of associated cells including fibroblasts, bone marrow-derived cells, endothelial cells, and infiltrating immune cells (reviewed in [16]). These stromal cells remodel the ECM and provide mechanical and chemical signals to the tumor cells. The many components and the dynamic nature of the tumor microenvironment (Fig. 1) contribute to its complexity, but investigation of the effect of individual stimuli on migration requires an environment in which the mechanical and chemical properties can be tuned precisely with reproducibility. The requirement for such control has led to the development of in vitro assays that mimic aspects of the in vivo tissue. In vitro studies are well suited for dissecting the signaling pathways that govern cell migration in response to a particular factor of interest, while in vivo studies can be utilized to investigate the relevance of these signaling pathways in the intrinsic tumor microenvironment during different steps of the metastatic cascade. Different experimental methods to assay tumor cell migration in vitro are presented schematically in Fig. 2 and summarized in Table 1 with their key advantages/limitations, parameters that can be manipulated, and practical information for implementation.
Fig. 1.
A host of biochemical and biophysical factors influence the migration of tumor cells. Mechanical signals include stiffness of the extracellular matrix (ECM), the pore size of the ECM, solid stress, fiber alignment, and fibroblast-generated matrix tension and microtracks. Fibroblasts are activated to assume the cancer-associated fibroblast (CAF) phenotype, and these cells secrete altered matrix components and generate tension. Chemical signals include autocrine gradients, MMPs, oxygen tension, and paracrine signals from the vasculature, lymphatics, and stromal cells (e.g., macrophages)
Fig. 2.
Experimental methods for investigating factors that influence tumor cell migration. In vitro tumor cell migration assays (reviewed in Table 1). Micropipette assay [21]: a pipette is placed in the vicinity of the cell and a chemoattractant solution is injected into the culture medium establishing a growth-factor gradient. Boyden [17] (or Transwell) chamber : cells are seeded in suspension in the top chamber and migrate through the porous filter (black rectangles) in response to a chemokine gradient, which is established by the different culture medium concentrations in the top and bottom chambers. Micropatterning [29]: cells are seeded on patterns of different geometry, size and surface coatings and their migration characteristics are monitored. Durotaxis [40]: cells are seeded on a substrate of variable stiffness and respond by changing traction forces, cell spread area, and migration direction. Wound healing [26]: a “wound” is formed on a confluent tumor monolayer, and the wound closure dynamics are monitored. 3D ECM [39]: cells are seeded inside the 3D ECM and migrate depending on the ECM architecture (stiffness, pore size, and ligand concentration); ECM fibers are outlined with black curved lines. Microfluidics [28, 32, 147, 206]: cytokine gradients can be established in a 3D matrix by flowing different chemokine concentration (C high − C low) solutions in the left and right microchannels; Interstitial flow can be established by adjusting the hydrostatic pressure (P high − P low) in the left and right microchannels; streamlines are indicated with dark magenta lines. Micropipette, Boyden chamber and microfluidics assays enable control of biochemical gradients. Durotaxis, 3D ECM and microfluidics assays enable control of biophysical forces (ECM stiffness and interstitial flow). Wound healing and micropatterning assay enable control of intercellular distances, whereas only micropatterning assays enable control of substrate topography. Growth-factor gradients are indicated by the purple triangles. ECM stiffness gradients are indicated by the dark brown triangle. Blue arrows indicate direction of tumor cell migration, and pressure gradients are indicated by the shades of green
Table 1.
Comparison of in vitro experimental approaches to study tumor cell migration
| Assay | Key advantages/disadvantages | Applications | Important parameters that can be controlled | Implementation guidelines | References |
|---|---|---|---|---|---|
| Transwell system or Boyden chamber |
+High throughput +Easy to use −Live imaging −Gradient control −Cell population |
Single and collective cell migration Co-culture of 2 cell types |
Pore filter and protein coating characteristics, Chemoattractant to stimulate chemotaxis |
No special equipment (commercially available, e.g., Corning, BD Biosciences) | [17] |
| Would healing |
+High throughput +Live imaging −2D substrate −Gradient control requires modifications |
Collective cell migration |
Wound area Substrate coating |
No special equipment (commercially available, e.g., Essen BioScience) | [26] |
| Durotaxis assay |
+Live imaging −2D substrate |
ECM control |
ECM coating ECM stiffness |
Polyacrylamide substrates (Bio-Rad) | [40] |
| Micropipette |
+Local stimulation +Live imaging −2D substrate −Low throughput −Temporal gradient decay |
Chemotaxis Axon guidance |
Injection parameters Substrate coating |
Special equipment (micromanipulator, e.g., Narishige) | [21] |
| 3D ECM |
+Cell-ECM signaling −Confocal imaging −Gradient control requires modifications |
EMT Tumor spheroid Proteolytic migration |
ECM concentration (pore size, stiffness, ligand density) ECM layer thickness |
Commercially available matrices (e.g., BD Biosciences) | [39] |
| Micropatterned |
+Cell–cell signaling +Easy to use +Live imaging −Low cell numbers −Gradient control requires modifications |
ECM topography Cell–cell interactions |
Pattern dimensions Pattern coating |
Microstencil, custom designs require wafer master (Stanford Microfluidic Foundry) and nanoimprint lithography (Scivax) | [29] |
| Microfluidic |
+Fluid flow +Gradients +Cell–cell signaling +Live imaging −Complex to use −Low cell number −Biochemical assays |
Microenvironment Temporal control of input signals |
Flow-rate Gradients |
Custom designs require wafer master (Stanford Microfluidic Foundry) Special equipment (syringe pumps: e.g., Harvard Apparatus; plasma cleaner: e.g., Harrick Plasma) Commercial microchannels (Ibidi, Millipore) |
[28, 32, 147, 206] |
Traditional in vitro assays
Single cell migration in response to soluble biochemical factors has been traditionally assayed using Boyden chambers [17], or modifications of the original design (e.g., Zigmond [18] and Dunn [19] chambers). Boyden chambers, also known as Transwell systems, incorporate a stiff, porous membrane between two cell culture chambers. Soluble factor gradients can be established across the membrane, and the number of tumor cells that migrate through the membrane, from one chamber to the other, is used as a metric for migration (Fig. 2). To improve upon the traditional Transwell assay design, a hydrogel, often Matrigel or collagen type I, can be placed on top of the membrane to study invasion through a 3D matrix [17]. An alternative method to investigate cell migration in response to localized chemokine gradients is the micropipette assay [20, 21], where a micromanipulator-controlled micropipette is used to dispense predefined chemokine solutions in close proximity to cells on a 2D substrate. However, chemokine gradients generated by micropipette are transient and difficult to quantify.
Collective cell migration can be assayed by using a cell scraper [22] or a micropipette [23] to generate a “wound,” or an area without cells in a cellular monolayer. Cell motility is assayed by monitoring the time required for the wound area to be covered by the collectively migrating cells. Alternatively, cell damage can be avoided by positioning a barrier to migration, allowing cells to migrate in the area covered by the barrier upon its removal [24]. These assays are known as “wound healing assays” and are typically limited to two-dimensional substrates with uniform stimulation conditions, although stimulus gradients can be created through modified assays involving microfabrication [25]. An important advantage of these assays is the ability to perform live cell imaging and conduct multiple parallel assays, enabling high throughput data generation [26].
Microfluidic in vitro assays
The limitations of conventional assays to precisely control microenvironmental stimuli have led to the development of novel migration assays using microfabrication technology and 3D ECM matrices. In particular, the ability to accurately position fluid streams in microfluidic channels has enabled the development of microfluidic wound-healing assays [27], and multi-channel systems that allow the seeding of multiple cell types in 2D or 3D [28]. Other approaches utilize microcontact printing methods to create patterned substrate-bound gradients [29]. These novel assays enable the creation of user-defined microenvironments, where the magnitude, direction, and temporal characteristics of biochemical and biophysical stimuli can be precisely controlled.
Recent advances in microfluidic technology have made it possible to create novel assays that allow precise control of the cellular microenvironment [30]. Microfluidic assays can also be readily adapted for live cell imaging to reveal tumor cell migration dynamics, in response to critical factors in the tumor microenvironment, such as chemokine gradients [31], interstitial flow [32], electrical fields [33], and paracrine and juxtacrine interactions with other cell types [34, 35]. Cell seeding in microfluidic platforms is performed by loading cells suspended in fluid or hydrogel solution, and cell motility is monitored after establishment of chemokine gradient or flow conditions. Microfluidic platforms enable the accurate control of these microenvironmental factors, allowing the establishment of well-defined combinatorial conditions to dissect the interplay of mechanical and chemical signals. Small sample volumes and low cell numbers in microfluidic assays pose challenges for traditional biochemical assays, and often data analysis in microfluidic migration assays is performed using live cell imaging and immunofluorescence staining for proteins of interest.
Macroscale models
Mesenchymal cells in their intrinsic environment interact with a 3D ECM, characterized by physical parameters, including pore size and stiffness [36], and chemical parameters, including adhesion site density and bound ligand concentration. The development of novel biomaterials has enabled the creation of 3D environments that provide control of both the chemical and physical parameters of the ECM to study the effects of these factors on cell migration [37]. Hydrogel matrices, such as collagen type I, Matrigel and synthetic matrices have been used to investigate how ECM physical properties (e.g., stiffness [38] and tumor cell proteolytic activity [39]) influence tumor cell invasion. In these assays, tumor cells are uniformly seeded inside a homogeneous 3D ECM and their migration characteristics are monitored in real-time or after a given time period [39]. These are termed “macroscale migration assays” because of the inability to control biochemical and biophysical factors at the single cell level compared to microfluidic- or micropatterned-based assays. Elegant methods have also been developed to investigate the effects of stiffness gradients (durotaxis) on cell migration [40].
Micropatterned models
Integration of microfabrication technology with ECM patterning has enabled the creation of 1D, 2D, or 3D micropatterned substrates to selectively define the topography for culturing single or multiple cell types under well-defined cell positioning [41]. Micropatterned substrates can be fabricated using different techniques, such as microstencil stamping [42] or optical patterning [43]. Cell seeding in micropatterned assays is achieved in a similar way as microfluidic platforms, where cells are positioned in different areas of the device by pipetting cell suspensions. Interestingly, advanced micropatterning methods have also been developed that allow for active control of the substrates and impose tunable constraints on cell migration [44]. Doyle et al. [43] developed an assay to investigate tumor cell migration along 1D paths, and Irimia et al. [45] studied the effect of physical geometrical constraints on tumor cell migration persistence. These technologies can recreate a more physiologically relevant tumor microenvironment, including multiple cell types [46], mimicking tumor cell migration along fibers in the tumor stroma and invasion in 3D [47]. Although these micropatterned assays offer scalability and the capability to culture large cell numbers, they do not typically include fluid flow, and hence the application of chemical gradients and localized mechanical (e.g., fluid shear stress) stimuli to cell subpopulations is generally not possible.
Tumor cell migration assays in vivo
Intravital imaging of live animals at sufficient resolution to identify individual tumor cells has allowed monitoring tumor cell migration in the native tumor microenvironment in vivo [48, 49]. In these studies, human tumor cells are transplanted into different model organisms such as mouse [50], zebrafish [51], and chicken embryo [52], or alternatively genetically engineered mouse tumor models incorporating constitutively expressed fluorophores [53, 54] are employed. Intravital studies using multiphoton microscopy enable deep imaging (~mm) into the tissue and also allow for second harmonic imaging to visualize ECM fibers [53]. Wyckoff et al. [55] have developed an in vivo chemotaxis migration assay to be used in combination with monitoring tumor cell motility, which allows for the collection of the tumor cells as they migrate inside a cytokine-filled needle.
Computational modeling of tumors
Computational methods have been developed to augment experiments and gain deeper insight into tumor development. Cancer progression has been modeled with a variety of approaches (Fig. 3), ranging in length scale from microscopic and discrete models that consider individual cells, to macroscopic models that treat the tumor mass as a continuum (reviewed in [56]). Such computational models provide insight into biological processes and guide experimental development. For example, Baxter and Jain [57] demonstrated that interstitial flow velocity varies significantly for constant interstitial fluid pressure when the ratio of transvascular to interstitial resistance is varied. Interstitial fluid pressure can be measured experimentally, while flow velocity is more difficult to assay. However, flow is a strong regulator of drug delivery, so these computational models provide insight into the barriers to drug delivery and inform more effective cancer treatment [58]. Although these models have provided valuable insight into solid tumor progression, the treatment of a tumor as a continuum does not allow explicit consideration of single cell behavior or the effectors that regulate the behavior of individual tumor cells.
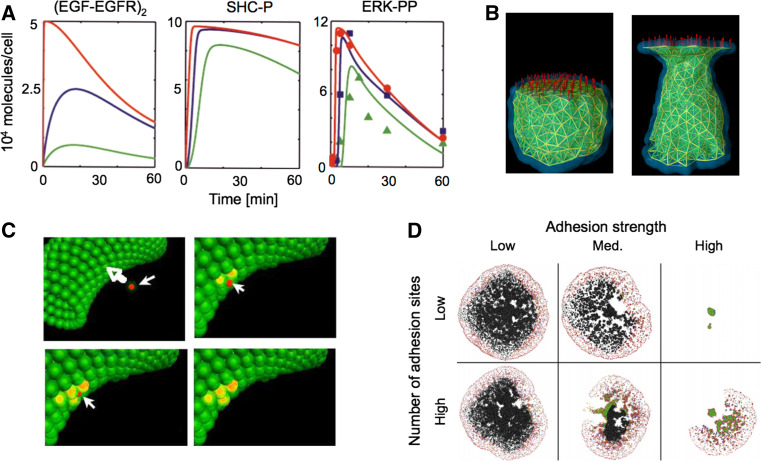
Fig. 3.
Computational methods provide insight into tumor progression. a Cell signaling models capture the dynamics of protein activation and are useful in identifying key signaling molecules and events, such as the role of EGF concentration on ERK-1/2 phosphorylation. Colored lines represent different concentrations of EGF (red, blue, and green are 50, 0.5, and 0.125 ng/ml EGF, respectively). Symbols in panel 3 represent experimental data, demonstrating that the model well-captures the dynamics of EGF-induced ERK-1/2 phosphorylation (SHC-P is phosphorylated SHC, ERK-PP is phosphorylated ERK. Adapted with permission from [64]). b The subcellular element model recapitulates cell-level phenomena by discretizing the cell into a series of nodes and defining mechanical potentials that govern the interaction of the nodes. The panels demonstrate elongation of a cell under 1 nN of tensile force; the filaments indicate interactions between nodes, which lie at the intersection of the filaments (adapted with permission from [63]). c A multi-scale agent-based model captures the dynamics of tumor intravasation. The subset of models that contributed to the overall multi-scale model was comprised of a simplified set of differential equations for intracellular adhesion signaling, a modified Hertz model for intercellular adhesion mechanics, and the Langevin equation for multi-cellular interactions. The panels demonstrate a tumor cell (red) approaching the endothelium (green), forming nascent N-cadherin bonds (yellow), disrupting endothelial cell VE-cadherin bonds, and traversing the endothelium (adapted with permission from [66]). d An off-lattice hybrid discrete-continuum approach modeled the growth of a whole tumor and invasion at the tumor periphery. The model was comprised of a continuum model for nutrient transport, and a discrete model for single cell behavior such as motility, adhesion, and proliferation. The model demonstrated that single cell adhesion plays a role in determining the fate and morphology of the tumor mass (adapted with permission from [59])
To improve on continuum models, Jeon et al. [59] developed a hybrid discrete-continuum model that implemented a continuum sub-model for chemical transport within the tumor and a discrete sub-model for individual cell behavior such as cell adhesion strength (Fig. 3d). Because the hybrid model explicitly considers cell–cell adhesion sites, the model produced realistic tumor morphologies that could not be generated by previous continuum models. Several studies have also implemented a force-based, microscale approach to effectively model the biphasic haptotactic and haptokinetic migratory response for tumor cells in 3D matrices, and these models have provided insight into the parameter space of adhesion and ECM stiffness that governs 3D migration speed [60, 61]. Although these models establish a framework for studying cell migration through 3D matrices, they are limited by certain assumptions such as fixed values for matrix density and cell adhesivity to the matrix. Furthermore, the model described by Zaman et al. lacked consideration of soluble ligand fields and thus could not account for chemotaxis. Anderson et al. [62] developed an agent-based model that predicts clonal evolution of a tumor as a function of the microenvironment properties. Such agent-based models have the advantages of simulating multiple interacting cells, but require the input of numerous parameters or assumptions, not all of which can be precisely or independently determined.
Subcellular models provide insight into mechanical and cellular processes that are difficult to measure experimentally. The subcellular element model successfully captures cell-level mechanical properties of cells by modeling the cell as a subset of nodes whose interaction is governed by mechanical potentials between the nodes (Fig. 3b) [63]; however, this model lacks chemical reaction kinetics and cannot capture signal transduction. On the other hand, chemical reaction models provide insight into signal transduction and have long been used to predict the effect of a drug on cell function (Fig. 3a) [64], but these models lack the mechanical consideration to adequately capture cell migration.
Cell migration is inherently a multi-scale process, involving both mechanical and chemical signal integration, and coarse-graining provides a technique for modeling multi-scale phenomena. Contact guidance in migrating cell populations has been effectively modeled by treating the cells as a continuum, where the continuum properties are derived from single cell migration models [65]. To further improve this multi-scale approach, Ramis-Conde et al. [66] developed a hybrid discrete-continuum model and explicitly considered cell signaling in the discretely modeled cells (Fig. 3c). The signaling model allowed dynamic modulation of cell–cell adhesion properties, which allowed simulation of intravasation. However, by considering signaling on a whole-cell level, the authors could not capture localized signaling events or simulate the effects of signaling gradients, which are critical in carcinogenesis and metastasis formation. Parameter estimation and assumption validation for computational models remains an issue for maintaining the physiologic relevance of the models (a list of parameters relevant to tumor cell migration is provided in Table 2), but advanced experimental platforms such as microfluidics provide resolution over parameters that are difficult to estimate with traditional assays. For example, Irimia et al. [67] recently coupled their computational model for neutrophil chemotaxis to experiments performed in a microfluidic device to aid in parameter estimation and model verification. Although computational models have offered insight into the complex and interactive mechanisms that regulate tumor cell migration, there remains room for new approaches that simultaneously encompass both chemical and mechanical stimuli, and are capable of bridging scales from molecular to cellular to tissue levels.
Table 2.
Approximate values for parameters governing tumor cell migration
| Parameter | Value | Notes | References |
|---|---|---|---|
| Stiffness | |||
| Normal stromal ECM stiffness | 200 Pa | Unconfined compression with electromechanical indenter | [12] |
| Mammary adenocarcinoma ECM stiffness | 4 × 103 Pa | Explants of tumor tissue from MMTV-Her2/neu, Myc, and Ras transgenic mice measured by unconfined compression | [12] |
| Colon adenocarcinoma stiffness | 4 × 103 Pa | Explants of tumor tissue from L5147T mouse xenografts measured by confined compression | [207] |
| Glioblastoma stiffness | 26 × 103 Pa | Explants of tumor tissue from U87 mouse xenografts measured by confined compression | [207] |
| Reconstituted basement membrane (Matrigel, stock concentration) | 175 Pa | Unconfined compression with electromechanical indenter | [12] |
| Tissue culture plastic | 109 Pa | Elastic modulus | [208] |
| Collagen gel elastic modulus | 8 Pa | 0.5 mg/ml collagen density, storage modulus measured by parallel plate rheology | [108] |
| 328 Pa | 2 mg/ml measured with electromechanical indenter | [12] | |
| Collagen fiber extensional modulus | 9 × 109 Pa | Force applied to fiber, strain measured by X-ray crystallography | [109] |
| Cell stiffness | 1 × 100 to 1 × 103 Pa | Highly dependent on cell type, culture substrate stiffness, and mechanical testing method. Lower stiffnesses measured by particle tracking microrheology for cells embedded within collagen gel [96]. Higher stiffnesses measured by AFM on metastatic cancer cells isolated from pleural fluids [209] | [96, 209] |
| Length scales | |||
| Basement membrane pore size in vivo | 50–100 nm | Measured by electron microscopy | [210] |
| Tumor-associated matrix pore size | 20–130 nm | Measured by electron microscopy of human melanoma implanted subcutaneously. Two distinct regions of matrix were observed. Bundles of aligned collagen fibrils with interfibrillar distances of 20–42 nm and bundles of poorly organized fibrils with interfibrillar distances of 75–130 nm | [211] |
| <150 nm | Herpes simplex virus (HSV, ~150 nm in diameter) did not penetrate into collagen-rich areas of human melanoma implanted into mouse skinfold chambers. Treatment with collagenase increased transport of HSV into the tumor | [212] | |
| Collagen gel pore size | 2 × 103 nm | 4.0 mg/ml collagen gel polymerized at 37 °C | [110] |
| 11 × 103 nm | 1.0 mg/ml collagen gel polymerized at 22 °C | [110] | |
| Tumor cell dimensions 3D | 50 × 103 nm | Major axis length of MDA-MB-231 in fibroblast derived ECM, measured from image provided by reference | [213] |
| Mesenchymal migrating cell microtrack | 10–15 × 103 nm | Diameter of defect in ECM generated by proteolytically active migration | [4] |
| Collagen fiber bundle diameter | 2–10 × 103 nm | Measured in vivo | [214] |
| Collagen fibril diameter | 35 nm | Collagen type I gel polymerized at pH7 measured by emission scanning electron microscope | [215] |
| 75 nm | Measured by electron micrograph from in vivo tissue samples | [214] | |
| Transport properties | |||
| Chemokine and growth factor diffusivity | 130–160 × 10−8 cm2/s | CCL5, CCL17, CCL21 in dilute solution | [117] |
| 10 × 10−7 cm2/s | EGF in dilute agarose gel | [216] | |
| Oxygen diffusivity | 5 × 10−5 cm2/s | In cell culture medium | [217] |
| Collagen gel permeability | 10−13 m2 | Measured in a 2 mg/ml collagen gel in a microfluidic device with a modified PIV technique | [32] |
| 10−13 to 10−14 m2 | 2.5 mg/ml | [171] | |
| Colon adenocarcinoma permeability | 3.4 × 10−16 m2 | Explants of tumor tissue from L5147T mouse xenografts measured by confined compression and assuming a poroelastic model for deformation | [207] |
| Glioblastoma permeability | 4.8 × 10−16 m2 | Explants of tumor tissue from U87 mouse xenografts measured by confined compression and assuming a poroelastic model for deformation | [207] |
| Interstitial flow speed | 0.1–4.0 × 10−6 m/s | Measured in vivo by FRAP | [167, 168] |
| Computational model for IF in solid tumors | <20 × 10−6 m/s | The magnitude of IF speed depends on ratio of transvascular to interstitial flow resistance. Larger resistance to flow in the interstitium creates high IF speed at the tumor periphery | [57, 166] |
| Forces | |||
| Myosin II motor force | 1.4–3.5 pN | Measured using an optical trap | [218, 219] |
| Force generated by actin polymerization | 5 pN/filament | Dependent on experimental method and platform | [13] |
| α5β1/fibronectin bond strength | 0.1–100 pN | Dependent on loading rate and method. Lower strengths measured with magnetic tweezers, higher strengths measured with AFM | [13] |
| Cellular contractile force | 5 × 103 pN | Epithelial cell edge measured on fibronectin-coated pillars | [220] |
| 300 × 103 pN | MDA-MB-231 total magnitude of contractile force measured on compliant substrate | [221] | |
| 2 × 106 pN | Fibroblast whole cell on collagen-coated compliant substrate | [222] | |
| Intratumoral IFP | 4–38 mmHg | Mean IFP measured for various tumors measured in vivo. Brain tumors are characterized by the lowest IFP with a mean of 4.6 mmHg, and renal cell carcinomas are characterized by the highest IFP with a mean of 38 mmHg. Individual mammary carcinomas varied from 4.0 to 53.0 mmHg, with a mean of 23.7 mmHg. IFP is a function of tumor size and of position within the tumor | [166, 223] |
| Normal IFP | 0–0.4 mmHg | Control measurement within rat thigh muscle | [223] |
Mesenchymal migration
The experimental platforms and computational methods discussed above have provided much insight into the mechanisms by which tumor cells migrate in the surrounding stroma. In epithelial tissues, cells are tightly connected to their neighbors and form polarized cell sheets with high levels of E-cadherin expression. During the formation of metastatic lesions in carcinomas, epithelial cells undergo genetic and epigenetic changes that induce a phenotypic transition, in which cells lose E-cadherin expression [68], reducing homophilic cell–cell connections, and express vimentin [69], N-cadherin [70], and other fibroblastic markers (reviewed in [71]). This phenotypic switch is known as the epithelial to mesenchymal transition (EMT), and results in cells of epithelial origin acquiring a phenotype more amenable to migration and invasion into the surrounding tissue. The migratory phenotype is varied and dynamic, with invading cells migrating individually or collectively and modulating the mechanism of migration with changes in the stromal properties (review of migration mechanisms [4]).
Mesenchymal cell migration is an orchestrated process, requiring ECM remodeling, extension of protrusions through actin polymerization, formation of new adhesions, molecular motor-mediated cell contraction, and the release of adhesions at the trailing edge (reviewed in [72, 73]). Migration through 3D matrices is termed invasion, and tumor cell invasion is mediated by invadopodia, specialized Src-kinase-dependent protrusions that are selectively observed in metastatic cells and extend into the ECM to probe the proximal chemical and mechanical microenvironment [74]. In general, cell migration is a stochastic process [75], and in the absence of an external gradient or directional cue, cells migrate randomly [76]. However, when presented with a directional cue, such as a chemokine gradient, the internal signaling machinery becomes polarized and cells migrate with directional bias [77]. Several metrics are commonly used to quantify aspects of cell migration beyond speed and direction. Detailed persistence metrics such as root-mean-squared speed, directional persistence time, and a random motility coefficient can be determined by fitting experimental mean-squared displacement data with a theoretical expression that models cell migration as a biased random walk [78]. Directionality, defined as the ratio of net migration distance to the total distance traveled for a given period of time, is a commonly used metric that is more straightforward to compute [79].
The stochasticity of cell migration and the fact that direction and speed can be modulated independently complicates the use of end-point migration assays. For example, migration with high speed and low persistence cannot be distinguished from low speed, highly persistent migration. This issue is compounded in Transwell assays, where only the number of cells reaching and crossing the membrane is measured, and increased migration speed with random direction could be confused with directionally biased migration. Furthermore, there is often significant cell–cell heterogeneity in cell migration characteristics, and migration statistics are often averaged over a cell population. These bulk, or population-averaged, metrics can obscure aspects of the migratory response that may be important for understanding the response of tumor cells to various stimuli. These disadvantages can be overcome by the use of live-cell imaging in microfluidic systems. For example, Haessler et al. [80] used population histograms and scatter plots to identify subpopulations of cells that respond uniquely to interstitial flow, with downstream migrating cells moving with high directional persistence and upstream migrating cells moving faster but with less directional persistence, supporting the hypothesis that flow imparts simultaneous and competing stimuli to tumor cells [32].
Mechanical signals
ECM stiffness and density
Traditionally, extracellular stimuli to migration are divided into mechanical and chemical signals, and the ECM imparts a variety of mechanical signals to migrating cells. The relationship between the density and organization of stromal tissue and tumor progression is bilateral and dynamic, with tumor cell phenotype dictated by the stromal properties and tumor cells modifying the chemical and mechanical profile of the stroma [81]. In particular, the density and stiffness of the stromal ECM has been shown to influence the invasion of tumor cells. Mammographically dense breast tissue has long been known to be a risk factor for breast cancer [82], but recently it was demonstrated that increased matrix density can promote invasion of tumor cells even in the absence of stromal cells [83]. Collagen crosslinking [84] and lysyl oxidase (LOX) overexpression [85] contribute to increased ECM stiffness in the tumor stroma [38], and stiffness of the ECM is a critical regulator of mammary gland function and morphology [86]. The surrounding stromal tissue in a normal mammary gland is characterized by an elastic modulus of ~200 Pa, while the elastic modulus of mammary tumor-associated stroma is ~4 kPa (Table 2) [12].
The stiffness and density of the substrate are sensed by the cell through adhesion molecules, primarily integrins [87], a class of tension-sensitive transmembrane molecules that can initiate the formation of a continuous mechanical link between the ECM fibers and cellular cytoskeleton [88]. Integrins are both substrate and tension dependent [89], and activation and clustering of integrins initiates a mechanosensitive signaling cascade [90]. The mechanical strength of cell–ECM adhesions is dependent on integrin engagement [91]. Vinculin recruitment, a measure of matrix adhesion maturation and an integrin signaling component, is dependent both on externally applied force [92] and internally generated myosin-mediated force [93]. Integrins are overexpressed in cancer cells, and a drug that blocks αVβ3-integrins has been shown to increase apoptosis and the efficacy of radiation treatment of mice with mammary adenocarcinoma [94].
Advances in techniques to engineer substrate stiffness in vitro have allowed investigation of the effect of ECM stiffness on tumor cells. Similar to the mechanosensing by fibroblasts and other cell types [95], prostate tumor cells are sensitive to substrate stiffness, increasing their elastic modulus with an increase in the substrate stiffness [96]. Interestingly, breast cancer cells that overexpress the receptor tyrosine kinase ErbB2 are more sensitive to substrate stiffness [97]. Furthermore, ECM stiffening contributes to altered integrin and ERK signaling, resulting in high Rho-dependent cellular contractile stress, disrupting the breast epithelial phenotype and promoting the malignant phenotype [12].
Rho-mediated contractile stress and integrin signaling are key regulators of cell migration [98], and integrin signaling is altered in tumors (see review of integrins in cancer in [99]). Recently, a technique involving crosslinking collagen in vitro enabled Levental et al. [38] to demonstrate that the increased stiffness of tumor-associated stroma increases levels of β1 integrin and FAKpY397 colocalization, and that these signals contribute to increased breast tumor cell invasion through enhanced PI3K signaling. The rigidity of the substrate alters invadopodia function of breast cancer cells, with enhanced matrix degradation and more functional invadopodia per cell on stiffer substrates [100].
Cellular contractile stress generation is required for rigidity sensing [13], and interfering with myosin motor activity attenuates the effects of stiffness on migration. Myosin light chain kinase inhibitor ML-7 impedes invadopodia maturation and decreases FAKpY397 and CaspY165 localization within breast carcinoma invadopodia [100], and inhibition of myosin II or Rho-associated kinase (ROCK) attenuates the ability of glioma cells to sense substrate rigidity [101]. Stiffness sensing is also cell-type specific, and the stiffness-dependent invasion of breast cancer cells into 3D matrices is dictated by the specific genetic profile of the metastatic cells. Single cell subpopulations of MDA-MB-231 have been shown to demonstrate tissue-specific metastatic lesions, tissue tropisms [102], and these subpopulations have shown varying stiffness dependence when invading 3D tissues. For example, breast cancer cells that metastasize to bone do not show stiffness dependent invasiveness; however, cells that metastasize to the lung invade less with increasing stiffness [103].
Although most experiments to date investigate cell migration as a function of stiffness on substrates or within matrices of uniform stiffness, stiffness gradients can guide cell migration on 2D substrates [40], and in 3D [104, 105]. Furthermore, patterning stiffness on substrates has been shown to lead to self-aggregation of fibroblasts [106]. The tumor microenvironment is heterogeneous, characterized by abrupt changes in the mechanical properties of the tissue [107], and the role this mechanical heterogeneity plays in guiding cell migration has yet to be determined. However, improved resolution in measuring the mechanical properties of tumors in vivo, multi-scale models that address substrate and cell mechanics, and improved techniques for creating artificial ECMs with customized mechanical properties will provide insight into the relationship between stiffness gradients and cell migration.
MMPs and matrix synthesis
As mentioned above, stiffness and stiffness gradients have been shown to influence cell migration on 2D substrates, and recently there has been much interest in determining the role of stiffness of 3D matrices on cell invasion. In 2D, mechanical properties can be easily quantified using a variety of rheometric techniques, and these bulk mechanical properties agree well with microscale mechanical properties measured with techniques such as atomic force microscopy (see review of 2D vs. 3D for cancer cell invasion in [36]). However, 3D matrices that mimic in vivo ECM are fibrous and the material is structured on cellular length scales, so assessing the mechanical properties relevant to cell migration is challenging. For example, the storage modulus of a 0.5 mg/ml collagen hydrogel is less than 10 Pa [108], while the extensional modulus of a fiber is on the order of GPa [109]. The coupling of matrix ligand density, pore size, and matrix stiffness in 3D migration assays employing collagen type I hydrogels further complicates investigation of the role of mechanical properties on 3D tumor cell invasion. For example, increasing hydrogel matrix protein density increases stiffness but also decreases pore size and increases ligand density. Tumor cell invasion is regulated both by pore size and stiffness [110], and crosslinking techniques provide orthogonal control of matrix ligand density and stiffness, though pore size and stiffness remain inversely coupled [108].
Cells in matrices with subcellular pore sizes implement a combination of force and proteolytic matrix degradation to overcome the structural barrier to migration imposed by the matrix fibers. Transformed breast carcinoma and fibrosarcoma cells apply force to collagen fibers to reorganize and reorient local matrix, while MMPs focally degrade matrix to reduce steric hindrance [111]. MMPs are key regulators of carcinogenesis, and their function extends beyond degrading matrix proteins [112]. As tumor cells secrete MMPs to degrade the interstitial matrix, this provides microtracks of weakened and digested matrix enabling migration of subsequent cells [113]. MMPs diffuse from tumor or stromal cells into the surrounding interstitium [112], and the expression, secretion, and action of MMPs couples the tumor cell chemical and mechanical microenvironment. For example, MT1-MMP expression is required for carcinoma cells to digest matrix proteins and invade into the 3D matrix [114], and MT1-MMP activity is also required to activate surface-bound TGF-β in carcinoma cells, which affects cell growth and matrix secretion [115]. Furthermore, MMP cleavage of matrix materials can release soluble cytokines that are bound to the ECM in inactive, precursor form [116], and the transport environment within the tissue can induce gradients in these soluble signals, providing directional cytokine cues to cells [117].
Intercellular mechanical signals
Fibroblasts are stromal cells that remodel the ECM through secretion of type I, type II, and type V collagen and fibronectin and contribute to basement membrane formation through secretion of collagen type IV and laminin (see review of fibroblasts in ECM remodeling in [118]). In tumors, stromal fibroblasts acquire an activated phenotype similar to fibroblast activation in wound healing [119], with 80 % of stromal fibroblasts achieving the activated phenotype in breast carcinomas [120]. Along with myofibroblasts [121], these cancer-associated fibroblasts (CAFs) are characterized by increased deposition of matrix, known as desmoplasia, which contributes to the increased stiffness of tumor-associated ECM (see review of CAFs in [122]), and CAFs demonstrate altered gene expression from normal stromal fibroblasts including upregulation of genes governing ECM deposition [123]. CAFs dynamically interact with tumor cells, sensing changes to the matrix through integrin-dependent adhesions, and further regulating the ECM by modulating matrix and MMP secretion. The dynamic evolution of the tumor stroma is known as stromagenesis (see review of stromagenesis in [124]), and altered stroma is a key feature of carcinogenesis [125].
CAFs alter the mechanical landscape of the tumor stroma and contribute to increased invasiveness of carcinoma cells [126]. In addition to the desmoplasia-associated matrix stiffening, fibroblasts generate high levels of contractile stress and can thus apply tensile stresses to the surrounding ECM. Fibroblasts seeded in 3D collagen gels increase the mechanical stiffness and resistance to fracture of tissue constructs [127], and TGFβ-activated fibroblasts seeded in 3D collagen gels can induce Rho-dependent contraction of these constructs [128]. These CAF-mediated alterations to the mechanical properties of the tumor stroma promote the invasive phenotype, and fibroblasts can directly promote the invasion of carcinoma cells. Migrating fibroblasts locally degrade matrix and secrete fibronectin and tenascin-C when seeded in collagen type-I and Matrigel hybrid tissue constructs. The matrix degradation is dependent on integrin engagement and Rho- and ROCK-mediated regulation of myosin light chain activity, suggesting that cellular tension generated by the fibroblasts is necessary to degrade the matrix. These microtracks, created through matrix degradation, are sufficient to promote carcinoma cell migration, even when Rho and ROCK are blocked in the migrating tumor cells; however, the tumor cell migration requires Cdc42 and MRCK to modulate myosin light chain accumulation and activity [129]. The effects of the microtracks on tumor cell migration suggest fibroblast-mediated local mechanical and matrix architectural stimuli guide tumor cell migration, and advances in micropatterning technology and computational models for investigating pericellular matrix organization [130] will provide further insight into the mechanisms by which fibroblasts influence migration of local tumor cells.
ECM topography
Bulk reorganization of the tumor ECM during cancer progression promotes cell invasion, and mammary tumor explants cultured ex vivo within collagen hydrogels reorganize the peripheral collagen fibers [131]. In mouse models, mammary tumor stroma is characterized by structured, dense collagen type I. The collagen fibers are strained around the tumor and oriented normally to the tumor surface at the tumor margin. The aligned collagen fibers correlate with local cell invasion, and both collective and single cell invasion are observed along the radially oriented collagen fibers [131], consistent with the observation that metastatic carcinoma cells migrate in the direction of collagen fibers in vivo [132]. Questions remain, however, regarding the mechanism by which the ECM is altered in tumor development. For example, it remains to be seen whether dense collagen regions promote carcinogenesis and precede tumor growth, or whether collagen alignment is induced by increased matrix contraction by the epithelial tumor cells and altered matrix deposition by activated fibroblasts during tumor growth. Furthermore, questions remain as to whether the radial collagen fibers are formed by invading cells at the tumor periphery or if the aligned fibers precede and promote cell invasion.
Just as fibroblasts can reorganize the stromal matrix, tumor cells can induce topographical stimuli that guide migration. ROCK-mediated epithelial cell contractility can locally deform the ECM, even in the absence of protease activity and stromal fibroblasts [133], and, in vitro, Rho/ROCK and myosin-dependent contractility of carcinoma cells can lead to alignment of collagen fibers normal to the tumor periphery [134]. Alignment of collagen fibers has long been known to influence fibroblast migration through increased migration velocity along the fibers and suppressed migration normal to the fiber alignment direction, a mechanism known as contact guidance [135], and fibroblasts migrate rapidly along micropatterned lines of matrix [43]. Interestingly, fiber alignment promotes carcinoma cell invasion in the direction of the fibers even when Rho and ROCK activity is blocked in the migrating cells, suggesting that tension generated in the stroma could promote invasion through collagen fiber alignment [134]. These results, in conjunction with the data mentioned above from Gaggioli et al. [129] on fibroblast-generated microtracks, suggest that restriction of cell–ECM adhesions, either by fiber alignment or cell confinement, provides a strong directional stimulus for tumor cell migration.
Recently, advances in experimental techniques have provided insight into the role of mechanical confinement on cell migration. Irimia and Toner [45] developed a microfluidic platform to confine cells in channels of varying cross-sectional area, and they found that, in channels with cross-sectional dimensions comparable to a cell diameter, tumor cells, including breast, lung, prostate, and colorectal adenocarcinoma, migrated rapidly with high directional persistence. These channels, however, were rigid and nonporous. Recently, Ilina et al. [136] implemented a two-photon laser ablation technique to generate microtracks within collagen matrices and found that not only do the microtracks guide the migration of breast cancer cells even in the absence of MMP activity but the cells use an MMP-independent pushing mechanism to expand the track diameter during migration. One limitation of these studies is that the channels were synthesized from uniform hydrogels, whereas in vivo fibroblasts create microtracks and secrete matrix proteins, producing spatial heterogeneity in the chemical and mechanical signals within the tissue. Advances in micropatterning techniques could provide insight into how the altered chemical, mechanical, and steric properties of the microtrack influence cell migration in vivo.
Solid stress
Growth of a tumor within a confined tissue compresses the surrounding matrix and generates compressive stresses, which, in combination with elevated interstitial fluid pressure [137], contributes to high levels of solid stress within the tumor [138]. To investigate the role of the compressive stresses on tumor progression, Helmlinger et al. [139] developed a technique for growing tumor spheroids within agarose gels and demonstrated that solid stress generated by the growing spheroid limited tumor growth. Computational models accurately captured the experimentally observed growth curves and verified that increasing agarose density further limits growth by increasing the radial and circumferential stresses generated by growing spheroids [140]. Subsequent work has demonstrated that compressive stresses limit the growth of tumor spheroids by increasing apoptosis while decreasing proliferation, with the spatial regions of highest compressive stress corresponding to the highest rates of apoptosis [141]. High compressive stress within tumors, which can collapse intratumoral blood vessels [142], can also influence the invasive properties of cells within the tumor by creating hypoxia and high interstitial pressure. Compressive stress increased the motility, measured by the rate of migration in a wound-healing assay, of transformed and partially transformed breast carcinoma cells, while it suppressed the migration of non-malignant breast epithelial cells. The increased level of invasion was dependent on the development of leader cells [143], characterized by high directional persistence and directionality in their migration, and myosin light chain kinase inhibitor ML-7 blocked the migration of leader cells, suggesting a role for intracellular tension in the migration response to compressive stress [144].
Chemical signals
Chemotaxis
Traditionally, chemical migratory stimuli, mediated primarily by soluble chemical signals, have been considered independently of mechanical stimuli. Gradients in soluble molecules can guide the migration of tumor and stromal cells and modulate cancer progression [145]. Tumor cells and stromal cells sense the local concentration of chemokines and growth factors through surface receptors, and a recent review highlights the key factors and pathways involved in chemotaxis in cancer progression [14]. Chemokine receptors are often upregulated in cancer cells in vivo [146], and both tumor cells and stromal cells migrate in response to chemokine gradients. Chemokines can be secreted autologously, released from the matrix, or secreted by stromal cells, and cells respond to the magnitude of the transcellular chemokine gradient as well as the mean chemokine concentration [147].
Despite the growing body of work on the molecular mechanisms behind directional sensing and migration in cancer cells, little is known about the chemo-mechanical dynamics of chemotaxis in 3D environments. Furthermore, tumor cells secrete chemokines, and autocrine chemokine gradients can guide cell migration in the absence of an externally applied chemokine gradient [148]. Tumor cells in vivo are likely exposed to multiple, competing chemotactic stimuli, and it is unknown how tumor cells respond to simultaneous growth factor gradients. As discussed below, there is crosstalk and cooperation between growth factor receptors and integrins, and the mechanical environment must be considered when evaluating tumor cell chemotaxis. The advent of new, microfluidic platforms for investigating chemotaxis [149] will help to elucidate the mechanisms by which cells integrate mechanical and chemical signals in vivo.
Heterotypic tumor-stromal cell paracrine signaling
Chemical signal gradients that regulate tumor cell chemotaxis can be established through secretion of soluble chemokines by non-cancerous stromal cells and diffusion through the ECM in the tumor microenvironment. This cell–cell communication through paracrine factors is critical during tumor progression and invasion [150], and involves a variety of different cell types in the tumor microenvironment, such as fibroblasts, immune cells, and endothelial cells [151]. Recent progress in characterization of these cells showed that stromal cells isolated from mammary tumors have altered gene expression patterns [152] compared to their normal counterparts. These stromal cells have been characterized as “cancer-associated stromal cells,” such as cancer-associated fibroblasts [122] and tumor-associated macrophages [153]. The study of these interactions in vivo is difficult because of the challenges with precisely dissecting the underlying molecular pathways. Traditionally, Transwell assays have been employed to study the migration or invasion of tumor cells in response to unidirectional conditioned medium or bi-directional signaling with stromal cells, while in recent years a large number of investigators have also developed new microsystems-based assays [28, 35, 46, 154]. Stromal–tumor cell interactions may also include juxtacrine signaling involving direct physical contact [150] and through secretion and binding of ECM molecules. Our focus here is on how tumor cell migration is regulated through stromal cell-secreted factors; however, an important direction for further investigation is the mechanisms by which tumor cells modulate stromal cell phenotypes and how these changes can subsequently result in feedback loops further promoting tumor cell invasion.
Tumor-macrophage and tumor-fibroblast interactions
In vivo studies utilizing a microneedle chemotaxis invasion assay and multiphoton imaging showed that carcinoma cells and macrophages interact via a paracrine loop involving colony stimulator factor 1 (CSF-1) secreted by the tumor cells to signal EGF secretion by the macrophages leading to tumor cell chemotaxis. Involvement of this paracrine loop in promoting tumor cell invasion has also been demonstrated in vitro through studies of macrophage––tumor cell signaling [155]. De Wever et al. [156] demonstrated that myofibroblasts isolated from cancer stroma, or TGF-β transformed fibroblasts induce proinvasive activity in colon cancer cells through production of the extracellular matrix glycoprotein tenascin-C and secretion of scatter factor/hepatocyte growth factor (HGF). In their study, however, they did not address the question of whether physical contact between the two cell types was required to establish the proinvasive effects on cancer cells. A microfluidic model incorporating 3D co-culture of carcinoma cells and fibroblasts was utilized to demonstrate that cancer cell invasion was upregulated by cancer-associated fibroblasts compared to their normal counterparts [157]. Sung et al. [154] developed a microfluidic 3D compartmentalized system of co-cultured epithelial cells and fibroblasts to study the transition from a localized carcinoma into an invasive carcinoma. By modulating the intercellular distance between the two cell types, the authors demonstrated that morphology of the mammary clusters changes with direct contact between the two cell types, leading to a more invasive phenotype with long protrusions.
Tumor-endothelial cell interactions
Using a Transwell-based co-culture model of human glioblastoma and microvascular endothelial cells, Kenig et al. [17] demonstrated that endothelial cells increased the invasiveness of cancer cells. Enhanced tumor cell invasion was driven by endothelial cell SDF-1 expression resulting in increased expression of MMP-9 by the tumor cells. Kaji et al. [46] used a micropatterned system to co-culture HeLa and HUVEC cells, which included fluid flow control to direct transport of soluble factors from the tumor to the endothelial cells or vice versa. Interestingly, tumor-conditioned medium resulted in retraction of the endothelial monolayer, while endothelial-conditioned medium did not influence the migration of HeLa cells. Recently, Franses et al. [158] investigated the role of endothelial cells in tumor cell proliferation and invasion, and the authors showed that dysfunctional endothelial cells, which expressed lower levels of perlecan, secreted higher levels of interleukin-6 (IL-6), resulting in increased invasion of human breast and lung carcinoma cell lines. Apart from blood vessel endothelial cells, lymphatic endothelial cells have also been implicated in cancer metastasis. Using a co-culture Transwell assay, Issa et al. [159] demonstrated that lymphatic endothelial cells promoted melanoma cell proteolytic activity and motility via a paracrine loop involving tumor cell-secreted VEGF-C and lymphatic endothelial cell-secreted CCL21.
Interactions of tumor cells with multiple stromal cell types
Although most studies have investigated the interaction between tumor cells and a single stromal cell type, recent experiments employed three different cell types to more closely mimic the tumor microenvironment. Interestingly, different stromal cell types may also communicate directly via paracrine signals and secrete factors that inhibit or promote tumor cell migration factors. Hsu et al. [34] employed a microfluidic model with three chambers to demonstrate that macrophage-conditioned medium reduced the ability of myofibroblasts to promote lung adenocarcinoma cell migration. Walter-Yohrling et al. [160] showed that tumor cells of different origin, including breast, ovarian, prostate, and lung carcinoma, have positive or negative effects on the induction of endothelial/myofibroblast invasion in a tri-culture organotypic assay. Clearly, multiple cell type interactions are implicated in tumor cell invasion, and further studies are needed that incorporate a greater variety of tissue-specific cells to better mimic the in vivo microenvironment.
Chemo-mechanical crosstalk
Interstitial flow
Interstitial fluid provides a transport medium for nutrients and signaling molecules [161, 162]. Jain and Baxter [163] identified that elevated interstitial fluid pressure (IFP) within the tumor interstitium inhibits drug delivery to tumor tissue, and subsequent work established a mathematical framework for determining the critical parameters that govern transport of soluble species from the vasculature into neoplastic tissue [57]. High blood vessel permeability, elevated oncotic pressure [164], and abnormal lymphatic vessels contribute to high IFP within the tumor, and high intratumoral pressure has been correlated with poor prognosis in vivo [165]. IFP within tumors can be nearly as high as the microvascular pressure (up to 30 mmHg; [137]), while IFP in normal tissue is close to zero [166], and this difference in pressure between normal and neoplastic tissue leads to high IFP pressure gradients at the tumor margin. IFP gradients drive convection of interstitial fluid, and Chary and Jain [167] determined typical interstitial flow (IF) speeds to be on the order of 0.1–2.0 µm/s. The magnitude of IF velocity depends on the magnitude of the IFP gradient and the hydraulic conductivity of the tumor (Table 2), and subsequent work has indicated that the velocity can reach up to 4.0 µm/s [168]. Computational models demonstrate that IF velocity is a function of the ratio of interstitial to vascular flow resistance [57], and when the resistance to IF transport is much greater than resistance to vascular transport, IF velocity at the tumor periphery can reach as high as 10.0 µm/s [166].
The convection of soluble signals by interstitial flow provides a host of migration signals to invading tumor cells by transporting both autocrine and paracrine chemokines (see review of IF and effects on tumor cell invasion in [169]). In recent work by Shields et al. [170], IF was found to increase the metastatic potential of breast cancer and melanoma tumor cells through binding of self-secreted chemoattractant via the CCR7 receptor. This autocrine signaling, termed “autologous chemotaxis,” arises when IF transports secreted ligand downstream, and the balance between convection and diffusion establishes a Peclet number-dependent pericellular gradient, which provides a chemotactic signal in the downstream direction [117]. IF also influences fibroblast function, promoting matrix reorganization, differentiation into myofibroblast phenotype [171], and fibroblast migration [172], and these mechanisms prime the stromal environment through MMP secretion and Rho-dependent matrix contraction to promote tumor cell invasion [173].
Computational models have helped elucidate the effects of IF on the mechanical microenvironment, and the simulations demonstrate that IF applies stress to matrix fibers, which can be sensed by the cell through matrix adhesions [174], and IF imparts shear stress on the cell membrane, the magnitude of which is determined by the arrangement of matrix fibers [175]. Recent work demonstrates that, when the CCR7 receptors are blocked, breast cancer cells migrate in the upstream direction, and the data suggest that the upstream stimulus is mechanically mediated. This flow mechanotaxis arises primarily from pressure gradients across the cell, resulting in a net force on the cell in the flow direction, which is balanced by a net force on matrix adhesions in the upstream direction (Fig. 4) [32]. Originally, flow mechanotaxis was thought to be mediated by tension in integrins, supported by the increase FAKpY397 in cells exposed to flow, but recently a potential role for the glycocalyx has been identified in the cellular response to interstitial flow. Interstitial flow increases the migration of vascular smooth muscle cells through increased MMP13 expression, mediated by heparan sulfate proteoglycans [176]. Although this work was performed with vascular smooth muscle cells, syndecan expression in breast cancer has been demonstrated [177], though it remains to be seen whether proteoglycans play a role in the tumor cell response to interstitial flow.
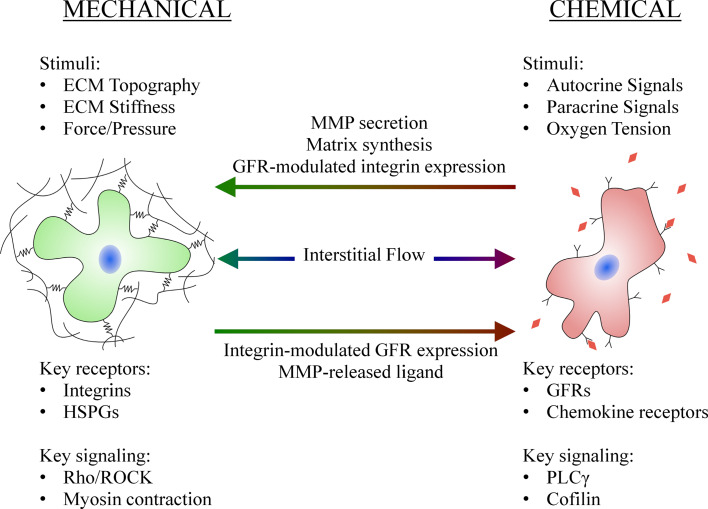
Fig. 4.
Chemical factors influence the cellular response to mechanical factors and vice versa. MMP and matrix secretion modulates the stiffness and pore size of the surrounding matrix, while growth factor receptor (GFR) activation influences integrin expression and activation. MMP-mediated matrix degradation releases chemical signals, while integrin activation and clustering can alter GFR expression and activation. GFRs and integrins can form macromolecular complexes, and elements of the intracellular signaling pathway are shared. Interstitial flow induces mechanical signals through fluid shear and pressure stresses, while simultaneously inducing chemical signals through convection of autocrine and paracrine signaling factors (HSPGs are heparan sulfate proteoglycans and PLCγ is phospholipase C-gamma)
Although the details of the mechanism guiding cells to migrate upstream have yet to be verified, recent evidence supports the hypothesis that IF imparts competing stimuli on migrating breast cancer cells [80]. The flow-induced expression of MMPs in fibroblasts [172] further complicates investigation of IF as a migration cue as the MMPs degrade the matrix, altering the flow field and pressure distribution around the cell. In glioma cells, fluid stress activates MMP1 [178], and secreted soluble MMPs could be convected downstream, degrading the matrix on the downstream side of the cell, further activating adhesion molecules on the upstream side, though that remains to be demonstrated.
Focal adhesion kinase and Src kinase as mediators for crosstalk
IF applies chemical and mechanical stimuli to cells, but the signals are further coupled intracellularly through shared signaling pathways from chemical and mechanical receptors (Fig. 4). Focal adhesion kinase (FAK) is an important regulator of tumor progression, with elevated FAK expression found in invasive human cancers [179], and FAK has been identified as a potential target for cancer therapeutics [180]. Autophosphorylation of FAK at Y397 requires the cell to be bound to a rigid substrate, while clustering of integrins can induce phosphorylation at other sites in FAK [181], and autophosphorylation at Y397 increases linearly with stiffness of the substrate [91]. FAKpY397 increases the affinity of FAK for the Src homology 2 domain of many proteins, including Src kinase [182], which can further phosphorylate FAK at other tyrosines, resulting in full activation of FAK enzymatic activity [183].
FAK further regulates downstream signals crucial to cell migration, and FAKpY397 is required for cell motility [184, 185]; however, the role of FAK in 2D cell migration and 3D invasion seems to be different. Expression of v-Src restores 2D migration defects in FAK−/− fibroblasts, but FAK expression and the formation of a FAK-Src signaling complex was required for 3D cell invasion [186]. The discrepancy between 2D and 3D is presumably due to the fact that the FAK-Src-p130cas signaling construct leads to activation of MMP-2 and MMP-9 and matrix degradation for invasion [187]. Src further contributes to mechanotransduction through selective modulation of integrin-dependent traction forces [188].
FAK and Src are also key regulators of chemokine-induced migration. FAK can be transphosphorylated by activated EGFR, and FAK function is required for EGF-stimulated chemotaxis and motility [184]. Src can also be directly phosphorylated by EGFR [187], and as FAK is recruited to sites of integrin clustering and is required for growth factor-stimulated motility, Src is likely involved in the cooperativity or crosstalk between growth factor and integrin signaling [184]. The intracellular coupling of chemical and mechanical stimuli through FAK/Src complicates cell migration assays because testing for phosphorylation of FAK, Src, or downstream components does not distinguish between chemical and mechanical activation. Thus, assays that allow independent modulation of chemical and mechanical stimuli, or assays for further upstream signals, such as vinculin, are required to test the mechanism of activation within the cell.
Growth factor and integrin crosstalk
Integrins regulate the activity and downstream signaling of receptor protein tyrosine kinases (RPTKs) such as EGFR, coupling adhesion and growth factor signaling [189]. Matrix adhesion induces the formation of integrin-EGFR macromolecular signaling complexes [190], and integrin clustering can induce phosphorylation and activation of EGFR and downstream signals (see review of integrin and growth factor cooperation in [191]). The association of integrins and growth factor receptors introduces crosstalk between adhesion and growth factor pathways, and this crosstalk necessitates consideration of the ECM when investigating chemokines and vice versa (Fig. 4). For example, when breast cells are cultured in 3D, blocking of β1 integrin or EGFR reduces expression of both receptors, restoring malignant breast cancer cells to the normal epithelial phenotype, an effect not seen when the cells are cultured on 2D substrates [192]. Integrins can also act to amplify growth factor signals influencing chemokine-induced invasion. In carcinoma cells, α6β4 integrin associates with and is phosphorylated by the HGF-specific receptor tyrosine kinase c-Met, and functional α6β4 integrin is required for HGF-dependent invasion [193]. The crosstalk between integrins and growth factor receptors results in coupling between the chemical and mechanical microenvironments, and recently it has been shown that stiffening of the ECM can increase the sensitivity of epithelial cells to EGF [194].
This relationship between integrins and growth factor receptors is bi-directional, and growth factor activation can influence integrin function. In prostate carcinoma cells, binding of the chemokine CXCL12 to its receptor CXCR4 increases adhesion to the ECM through α5 and β3 integrin activation [195]. Recent work has demonstrated that α5β1 integrin binds to and activates c-Met, which initiates Src/FAK signaling and promotes migration in ovarian cancer cells. Blocking α5β1 integrin or c-Met decreases Src/FAK activation and cell invasion in vitro and inhibits metastasis formation in vivo; however, activation of c-Met by soluble HGF can restore the ovarian cancer invasive potential even when α5β1 integrin is blocked. Importantly, the integrin-mediated activation of c-Met was substrate-specific, and c-Met phosphorylation by integrin was not observed on collagen substrates [196].
Integrin and cadherin crosstalk
ECM adhesion–growth factor crosstalk is further complicated by crosstalk between cell-–ECM and cell–cell adhesion molecules. A variety of mechanisms contribute to matrix and intercellular adhesion crosstalk (see review of adhesion crosstalk in [197]), including feedback in which integrin activation alters cadherin expression and vice versa. The Src/FAK signaling complex is at the heart of this crosstalk [198], and c-Src suppression in breast cancer cells results in E-cadherin upregulation, reversion of the mesenchymal phenotype to an epithelial phenotype, and reduced cell migration [199]. Using a dual-micropipette technique, Martinez-Rico and colleagues [200] demonstrated that the force required to separate cell doublets attached by E-cadherin-mediated adhesion was dependent on specific integrin engagement, and that cells, including carcinoma cells, attached to fibronectin-coated beads required a larger force to separate cell doublets than cells attached to vitronectin- or polylysine-coated beads. Patterning of ECM ligands further allows the regulation of cell–cell junction morphology and force distribution [201]. Recently, Borghi et al. [202] implemented a reductionist micropatterning technique to investigate crosstalk of cadherin- and integrin-mediated adhesions in governing epithelial cell motility. The micropatterning technique allowed cells to simultaneously adhere to collagen IV and the extracellular domain of E-cadherin. Micropatterning directed traction force and migration parallel to the major axis of areas printed with collagen IV. Notably, the presence of E-cadherin stripes decreased lamellipodia activity but did not affect migration rate. Depletion of αE-catenin within the epithelial cells increased lamellipodia activity, migration rate, and decreased migration coordination among epithelial cell sheets. These results suggest E-cadherin engagement influences the direction of migration but not migration rate, consistent with the results of Abercrombie and Heaysman [203], who originally postulated contact inhibition in fibroblasts. Interestingly, contact inhibition is lost in tumor cells [204], and recently it has been shown that matrix stiffening results in the loss of contact inhibition and acquisition of a malignant phenotype [194].
Conclusions and future directions
In this review, we provide an overview of recent experimental studies that investigate the effects of different factors present in the tumor microenvironment on tumor cell migration. These experimental approaches have provided great insight into the factors that guide cell migration in vivo, but the experimental platforms often trade precision for physiologic relevance. Computational models have been developed to supplement experimental data, but cell migration is inherently a multi-scale process, and models developed thus far lack the capability of simulating cancer progression and cell migration at various length scales. Mechanical stimuli such as ECM topography, ECM stiffness, pore size, solid stress, and matrix contraction, and chemical stimuli from autocrine and paracrine sources, influence the direction and speed of cell migration. These various signals signal simultaneously to the tumor cells, and tumor cell migration is an integrated response to multiple signals. Despite significant progress in unraveling the underlying molecular mechanisms and signaling pathways for transducing individual stimuli, the crosstalk between these different factors in the tumor microenvironment at the extracellular and intracellular levels remains largely unknown.
Integrating computational and experimental approaches will allow for a more detailed understanding of these mechanisms and their interplay, and this strategy has proven to be successful regarding the treatment of pancreatic ductal adenocarcinoma. Computational modeling has demonstrated that IFP acts as a barrier for drug delivery to tumors [57], and these models suggest that altering vascular and interstitial permeability could increase the efficacy of anti-tumor therapy [166]. In vitro experiments, coupled with in vivo data, have further demonstrated that IFP and IF alter the migration properties of migrating tumor cells and suggest that IF might increase the formation of metastases [32, 170]. Recently, Provenzano et al. [205] demonstrated that, by enzymatically degrading matrix elements in the tumor stroma, IFP is significantly reduced within the tumor allowing increased transport of small molecule therapeutic drug, which resulted in increased mouse survival and decreased formation of metastases. Furthermore, a growing body of work, motivated in part by computational models, suggests that targeting the tumor microenvironment in combination with anti-angiogenic therapy, increases drug transport and improves cancer therapy [58].
To this end, development of the next generation functional migration assays that enable integration of spatiotemporal control of mechanical and chemical signals with high-resolution, real-time imaging, and the ability to conduct biochemical assays, is crucial. The investigation of tumor cell migration at the single cell level will allow investigation of the diverse migration strategies employed by individual tumor cells inside the complex and heterogeneous tumor microenvironment. Progress in this direction will help to develop more accurate diagnostic techniques to quantify and predict cell migration in patients, thus informing methods for constraining tumor cell dissemination and halting metastasis formation.
Acknowledgments
The authors would like to thank members of the Kamm lab for critically reading the manuscript. Funding from The National Cancer Institute (R21CA140096), The Charles Stark Draper Laboratory University Research and Development Program (N.DL-H-550151) is greatly appreciated. W.J.P. was supported by a National Science Foundation Graduate Research Fellowship.
Abbreviations
- CAF
Cancer-associated fibroblast
- ECM
Extracellular matrix
- EGF
Epidermal growth factor
- EMT
Epithelial to mesenchymal transition
- FAK
Focal adhesion kinase
- IF
Interstitial flow
- IFP
Interstitial fluid pressure
- LEGI
Local excitation, global inhibition
- LIMK1
LIM domain kinase 1
- LOX
Lysyl oxidase
- MMP
Matrix metalloproteinase
- ROCK
Rho-associated kinase
- RPTK
Receptor protein kinase
References
- 1.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 3.Mueller MM, Fusenig NE. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4(11):839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 4.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Bristow RG, Hill RP. Hypoxia and metabolism: hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8(3):180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 6.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197(4306):893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 7.Weigelt B, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 8.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369(9574):1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almog N. Molecular mechanisms underlying tumor dormancy. Cancer Lett. 2010;294(2):139–146. doi: 10.1016/j.canlet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Paez D, et al. Cancer dormancy: a model of early dissemination and late cancer recurrence. Clin Cancer Res. 2011;18(3):645–653. doi: 10.1158/1078-0432.CCR-11-2186. [DOI] [PubMed] [Google Scholar]
- 11.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19(2):194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326(5957):1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zigmond SH. Orientation chamber in chemotaxis. Methods Enzymol. 1988;162:65–72. doi: 10.1016/0076-6879(88)62064-7. [DOI] [PubMed] [Google Scholar]
- 19.Zicha D, Dunn GA, Brown AF. A new direct-viewing chemotaxis chamber. J Cell Sci. 1991;99(Pt 4):769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 20.Gundersen RW, Barrett JN. Neuronal chemotaxis: chick dorsal-root axons turn toward high concentrations of nerve growth factor. Science. 1979;206(4422):1079–1080. doi: 10.1126/science.493992. [DOI] [PubMed] [Google Scholar]
- 21.Soon L, et al. Description and characterization of a chamber for viewing and quantifying cancer cell chemotaxis. Cell Motil Cytoskelet. 2005;62(1):27–34. doi: 10.1002/cm.20082. [DOI] [PubMed] [Google Scholar]
- 22.Yarrow JC, et al. A high-throughput cell migration assay using scratch wound healing, a comparison of image-based readout methods. BMC Biotechnol. 2004;4:21. doi: 10.1186/1472-6750-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong MK, Gotlieb AI. The reorganization of microfilaments, centrosomes, and microtubules during in vitro small wound reendothelialization. J Cell Biol. 1988;107(5):1777–1783. doi: 10.1083/jcb.107.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Horssen R, ten Hagen TL. Crossing barriers: the new dimension of 2D cell migration assays. J Cell Physiol. 2011;226(1):288–290. doi: 10.1002/jcp.22330. [DOI] [PubMed] [Google Scholar]
- 25.Toley BJ, et al. Micrometer-scale oxygen delivery rearranges cells and prevents necrosis in tumor tissue in vitro. Biotechnol Prog. 2012;28(2):515–525. doi: 10.1002/btpr.1510. [DOI] [PubMed] [Google Scholar]
- 26.Simpson KJ, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10(9):1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 27.Murrell M, Kamm R, Matsudaira P. Tension, free space, and cell damage in a microfluidic wound healing assay. PLoS One. 2011;6(9):e24283. doi: 10.1371/journal.pone.0024283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung S, et al. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9(2):269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 29.von Philipsborn AC, et al. Growth cone navigation in substrate-bound ephrin gradients. Development. 2006;133(13):2487–2495. doi: 10.1242/dev.02412. [DOI] [PubMed] [Google Scholar]
- 30.Salieb-Beugelaar GB, et al. Latest developments in microfluidic cell biology and analysis systems. Anal Chem. 2010;82(12):4848–4864. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 31.Saadi W, et al. A parallel-gradient microfluidic chamber for quantitative analysis of breast cancer cell chemotaxis. Biomed Microdevices. 2006;8(2):109–118. doi: 10.1007/s10544-006-7706-6. [DOI] [PubMed] [Google Scholar]
- 32.Polacheck WJ, Charest JL, Kamm RD. Interstitial flow influences direction of tumor cell migration through competing mechanisms. Proc Natl Acad Sci USA. 2011;108(27):11115–11120. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang CW, et al. Electrotaxis of lung cancer cells in a multiple-electric-field chip. Biosens Bioelectron. 2009;24(12):3510–3516. doi: 10.1016/j.bios.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Hsu TH, et al. The migration speed of cancer cells influenced by macrophages and myofibroblasts co-cultured in a microfluidic chip. Integr Biol (Camb) 2011;4:177–182. doi: 10.1039/c2ib00112h. [DOI] [PubMed] [Google Scholar]
- 35.Zervantonakis IK, et al. Microfluidic devices for studying heterotypic cell–cell interactions and tissue specimen cultures under controlled microenvironments. Biomicrofluidics. 2011;5(1):13406. doi: 10.1063/1.3553237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 2011;3(4):267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 37.Kim HD, Peyton SR. Bio-inspired materials for parsing matrix physicochemical control of cell migration: a review. Integr Biol (Camb) 2012;4(1):37–52. doi: 10.1039/c1ib00069a. [DOI] [PubMed] [Google Scholar]
- 38.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185(1):11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo CM, et al. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaji H, et al. Engineering systems for the generation of patterned co-cultures for controlling cell–cell interactions. Biochim Biophys Acta. 2011;1810(3):239–250. doi: 10.1016/j.bbagen.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polio SR, et al. A micropatterning and image processing approach to simplify measurement of cellular traction forces. Acta Biomater. 2012;8(1):82–88. doi: 10.1016/j.actbio.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle AD, et al. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184(4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X, et al. Directing cell migration with asymmetric micropatterns. Proc Natl Acad Sci USA. 2005;102(4):975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irimia D, Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integr Biol (Camb) 2009;1(8–9):506–512. doi: 10.1039/b908595e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaji H, et al. Controlled cocultures of HeLa cells and human umbilical vein endothelial cells on detachable substrates. Lab Chip. 2009;9(3):427–432. doi: 10.1039/b812510d. [DOI] [PubMed] [Google Scholar]
- 47.Liu L, et al. Probing the invasiveness of prostate cancer cells in a 3D microfabricated landscape. Proc Natl Acad Sci USA. 2011;108(17):6853–6856. doi: 10.1073/pnas.1102808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3(12):921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 49.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147(5):983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander S, et al. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem Cell Biol. 2008;130(6):1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 51.Stoletov K, et al. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci USA. 2007;104(44):17406–17411. doi: 10.1073/pnas.0703446104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zijlstra A, et al. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13(3):221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyckoff J, et al. High-resolution multiphoton imaging of tumors in vivo. Cold Spring Harb Protoc. 2011;2011(10):1167–1184. doi: 10.1101/pdb.top065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moshitch-Moshkovitz S, et al. In vivo direct molecular imaging of early tumorigenesis and malignant progression induced by transgenic expression of GFP-Met. Neoplasia. 2006;8(5):353–363. doi: 10.1593/neo.05634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyckoff J, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64(19):7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 56.Byrne HM. Dissecting cancer through mathematics: from the cell to the animal model. Nat Rev Cancer. 2010;10(3):221–230. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 57.Baxter LT, Jain RK. Transport of fluid and macromolecules in tumors. 1. Role of interstitial pressure and convection. Microvasc Res. 1989;37(1):77–104. doi: 10.1016/0026-2862(89)90074-5. [DOI] [PubMed] [Google Scholar]
- 58.Chauhan VP, et al. Delivery of molecular and nanoscale medicine to tumors: transport barriers and strategies. Annu Rev Chem Biomol Eng. 2011;2(1):281–298. doi: 10.1146/annurev-chembioeng-061010-114300. [DOI] [PubMed] [Google Scholar]
- 59.Jeon J, Quaranta V, Cummings PT. An off-lattice hybrid discrete-continuum model of tumor growth and invasion. Biophys J. 2010;98(1):37–47. doi: 10.1016/j.bpj.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103(29):10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaman MH, et al. Computational model for cell migration in three-dimensional matrices. Biophys J. 2005;89(2):1389–1397. doi: 10.1529/biophysj.105.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson AR, et al. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127(5):905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 63.Sandersius SA, Weijer CJ, Newman TJ. Emergent cell and tissue dynamics from subcellular modeling of active biomechanical processes. Phys Biol. 2011;8(4):045007. doi: 10.1088/1478-3975/8/4/045007. [DOI] [PubMed] [Google Scholar]
- 64.Schoeberl B, et al. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20(4):370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 65.Painter KJ. Modelling cell migration strategies in the extracellular matrix. J Math Biol. 2009;58(4–5):511–543. doi: 10.1007/s00285-008-0217-8. [DOI] [PubMed] [Google Scholar]
- 66.Ramis-Conde I, et al. Multi-scale modelling of cancer cell intravasation: the role of cadherins in metastasis. Phys Biol. 2009;6(1):016008. doi: 10.1088/1478-3975/6/1/016008. [DOI] [PubMed] [Google Scholar]
- 67.Irimia D, et al. Adaptive-control model for neutrophil orientation in the direction of chemical gradients. Biophys J. 2009;96(10):3897–3916. doi: 10.1016/j.bpj.2008.12.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson EW, et al. Oncogene-induced basement membrane invasiveness in human mammary epithelial cells. Clin Exp Metastasis. 1994;12(3):181–194. doi: 10.1007/BF01753886. [DOI] [PubMed] [Google Scholar]
- 69.Sommers CL, et al. Loss of epithelial markers and acquisition of vimentin expression in adriamycin- and vinblastine-resistant human breast cancer cell lines. Cancer Res. 1992;52(19):5190–5197. [PubMed] [Google Scholar]
- 70.Nieman MT, et al. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147(3):631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 72.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84(3):359–369. doi: 10.1016/S0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 73.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 74.Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis. 2006;23(2):97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 75.Tranquillo RT, Lauffenburger DA, Zigmond SH. A stochastic model for leukocyte random motility and chemotaxis based on receptor binding fluctuations. J Cell Biol. 1988;106(2):303–309. doi: 10.1083/jcb.106.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stoker M, Gherardi E. Regulation of cell movement: the motogenic cytokines. Biochim Biophys Acta. 1991;1072(1):81–102. doi: 10.1016/0304-419x(91)90008-9. [DOI] [PubMed] [Google Scholar]
- 77.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10(8):538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dickinson RB, Tranquillo RT. Optimal estimation of cell-movement indexes from the statistical-analysis of cell tracking data. AIChE J. 1993;39(12):1995–2010. doi: 10.1002/aic.690391210. [DOI] [Google Scholar]
- 79.Ware MF, Wells A, Lauffenburger DA. Epidermal growth factor alters fibroblast migration speed and directional persistence reciprocally and in a matrix-dependent manner. J Cell Sci. 1998;111(Pt 16):2423–2432. doi: 10.1242/jcs.111.16.2423. [DOI] [PubMed] [Google Scholar]
- 80.Haessler U, et al. Migration dynamics of breast cancer cells in a tunable 3D interstitial flow chamber. Integr Biol (Camb) 2012;4(4):401–409. doi: 10.1039/c1ib00128k. [DOI] [PubMed] [Google Scholar]
- 81.Bissell MJ, et al. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70(9–10):537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyd NF, et al. Mammographic densities and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7(12):1133–1144. [PubMed] [Google Scholar]
- 83.Provenzano PP, et al. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28(49):4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Slot AJ, et al. Elevated formation of pyridinoline cross-links by profibrotic cytokines is associated with enhanced lysyl hydroxylase 2b levels. Biochim Biophys Acta. 2005;1741(1–2):95–102. doi: 10.1016/j.bbadis.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 85.Akiri G, et al. Lysyl oxidase-related protein-1 promotes tumor fibrosis and tumor progression in vivo. Cancer Res. 2003;63(7):1657–1666. [PubMed] [Google Scholar]
- 86.Paszek MJ, Weaver VM. The tension mounts: mechanics meets morphogenesis and malignancy. J Mammary Gland Biol Neoplasia. 2004;9(4):325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- 87.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 88.Alenghat FJ, DE Ingber Mechanotransduction: all signals point to cytoskeleton, matrix, and integrins. Sci STKE. 2002;2002(119):pe6. doi: 10.1126/stke.2002.119.pe6. [DOI] [PubMed] [Google Scholar]
- 89.Jalali S, et al. Integrin-mediated mechanotransduction requires its dynamic interaction with specific extracellular matrix (ECM) ligands. Proc Natl Acad Sci USA. 2001;98(3):1042–1046. doi: 10.1073/pnas.98.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323(5914):642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 92.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159(4):695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 94.Burke PA, et al. Cilengitide targeting of alpha(v)beta(3) integrin receptor synergizes with radioimmunotherapy to increase efficacy and apoptosis in breast cancer xenografts. Cancer Res. 2002;62(15):4263–4272. [PubMed] [Google Scholar]
- 95.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 96.Baker EL, Bonnecaze RT, Zaman MH. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J. 2009;97(4):1013–1021. doi: 10.1016/j.bpj.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baker EL, et al. Cancer cell stiffness: integrated roles of three-dimensional matrix stiffness and transforming potential. Biophys J. 2010;99(7):2048–2057. doi: 10.1016/j.bpj.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moissoglu K, Schwartz MA. Integrin signalling in directed cell migration. Biol Cell. 2006;98(9):547–555. doi: 10.1042/BC20060025. [DOI] [PubMed] [Google Scholar]
- 99.Moschos SJ, et al. Integrins and cancer. Oncology (Williston Park) 2007;21(9 Suppl 3):13–20. [PubMed] [Google Scholar]
- 100.Alexander NR, et al. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18(17):1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gupta GP, et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol. 2005;70:149–158. doi: 10.1101/sqb.2005.70.018. [DOI] [PubMed] [Google Scholar]
- 103.Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4(7):e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hadjipanayi E, Mudera V, Brown RA. Guiding cell migration in 3D: a collagen matrix with graded directional stiffness. Cell Motil Cytoskelet. 2009;66(3):121–128. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- 105.Ehrbar M, et al. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys J. 2011;100(2):284–293. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gray DS, Tien J, Chen CS. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young’s modulus. J Biomed Mater Res A. 2003;66(3):605–614. doi: 10.1002/jbm.a.10585. [DOI] [PubMed] [Google Scholar]
- 107.Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17(5):524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 108.Ulrich TA, et al. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials. 2010;31(7):1875–1884. doi: 10.1016/j.biomaterials.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 109.Sasaki N, Odajima S. Stress-strain curve and Young’s modulus of a collagen molecule as determined by the X-ray diffraction technique. J Biomech. 1996;29(5):655–658. doi: 10.1016/0021-9290(95)00110-7. [DOI] [PubMed] [Google Scholar]
- 110.Yang YL, Motte S, Kaufman LJ. Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials. 2010;31(21):5678–5688. doi: 10.1016/j.biomaterials.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 111.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9(8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 112.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 113.Friedl P, Wolf K. Tube travel: the role of proteases in individual and collective cancer cell invasion. Cancer Res. 2008;68(18):7247–7249. doi: 10.1158/0008-5472.CAN-08-0784. [DOI] [PubMed] [Google Scholar]
- 114.Sabeh F, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167(4):769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mu D, et al. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J Cell Biol. 2002;157(3):493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee S, et al. Processing of VEGF-A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J Cell Biol. 2005;169(4):681–691. doi: 10.1083/jcb.200409115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fleury ME, Boardman KC, Swartz MA. Autologous morphogen gradients by subtle interstitial flow and matrix interactions. Biophys J. 2006;91(1):113–121. doi: 10.1529/biophysj.105.080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tomasek JJ, et al. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 119.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 120.Sappino AP, et al. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41(5):707–712. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- 121.De Wever O, et al. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123(10):2229–2238. doi: 10.1002/ijc.23925. [DOI] [PubMed] [Google Scholar]
- 122.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 123.Sadlonova A, et al. Identification of molecular distinctions between normal breast-associated fibroblasts and breast cancer-associated fibroblasts. Cancer Microenviron. 2009;2(1):9–21. doi: 10.1007/s12307-008-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beacham DA, Cukierman E. Stromagenesis: the changing face of fibroblastic microenvironments during tumor progression. Semin Cancer Biol. 2005;15(5):329–341. doi: 10.1016/j.semcancer.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 125.Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev. 1996;76(1):69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 126.Dimanche-Boitrel MT, et al. In vivo and in vitro invasiveness of a rat colon-cancer cell line maintaining E-cadherin expression: an enhancing role of tumor-associated myofibroblasts. Int J Cancer. 1994;56(4):512–521. doi: 10.1002/ijc.2910560410. [DOI] [PubMed] [Google Scholar]
- 127.Huang D, et al. Mechanisms and dynamics of mechanical strengthening in ligament-equivalent fibroblast-populated collagen matrices. Ann Biomed Eng. 1993;21(3):289–305. doi: 10.1007/BF02368184. [DOI] [PubMed] [Google Scholar]
- 128.Grinnell F, Ho CH. Transforming growth factor beta stimulates fibroblast-collagen matrix contraction by different mechanisms in mechanically loaded and unloaded matrices. Exp Cell Res. 2002;273(2):248–255. doi: 10.1006/excr.2001.5445. [DOI] [PubMed] [Google Scholar]
- 129.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 130.Stevenson MD, et al. Pericellular conditions regulate extent of cell-mediated compaction of collagen gels. Biophys J. 2010;99(1):19–28. doi: 10.1016/j.bpj.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Provenzano PP, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4(1):38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang W, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62(21):6278–6288. [PubMed] [Google Scholar]
- 133.Wyckoff JB, et al. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16(15):1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 134.Provenzano PP, et al. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95(11):5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dickinson RB, Guido S, Tranquillo RT. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann Biomed Eng. 1994;22(4):342–356. doi: 10.1007/BF02368241. [DOI] [PubMed] [Google Scholar]
- 136.Ilina O, et al. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Physical Biol. 2011;8:015010. doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- 137.Boucher Y, Jain RK. Microvascular pressure is the principal driving force for interstitial hypertension in solid tumors: implications for vascular collapse. Cancer Res. 1992;52(18):5110–5114. [PubMed] [Google Scholar]
- 138.Sarntinoranont M, Rooney F, Ferrari M. Interstitial stress and fluid pressure within a growing tumor. Ann Biomed Eng. 2003;31(3):327–335. doi: 10.1114/1.1554923. [DOI] [PubMed] [Google Scholar]
- 139.Helmlinger G, et al. Solid stress inhibits the growth of multicellular tumor spheroids. Nat Biotechnol. 1997;15(8):778–783. doi: 10.1038/nbt0897-778. [DOI] [PubMed] [Google Scholar]
- 140.Roose T, et al. Solid stress generated by spheroid growth estimated using a linear poroelasticity model☆. Microvasc Res. 2003;66(3):204–212. doi: 10.1016/S0026-2862(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 141.Cheng G, et al. Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells. PLoS One. 2009;4(2):e4632. doi: 10.1371/journal.pone.0004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Padera TP, et al. Pathology: cancer cells compress intratumour vessels. Nature. 2004;427(6976):695. doi: 10.1038/427695a. [DOI] [PubMed] [Google Scholar]
- 143.Khalil AA, Friedl P. Determinants of leader cells in collective cell migration. Integr Biol (Camb) 2010;2(11–12):568–574. doi: 10.1039/c0ib00052c. [DOI] [PubMed] [Google Scholar]
- 144.Tse JM, et al. Mechanical compression drives cancer cells toward invasive phenotype. Proc Natl Acad Sci USA. 2012;109:911–916. doi: 10.1073/pnas.1118910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Muller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 146.Ben-Baruch A. The multifaceted roles of chemokines in malignancy. Cancer Metastasis Rev. 2006;25(3):357–371. doi: 10.1007/s10555-006-9003-5. [DOI] [PubMed] [Google Scholar]
- 147.Wang SJ, et al. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300(1):180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 148.Scherber C, et al. Epithelial cell guidance by self-generated EGF gradients. Integr Biol (Camb) 2012;4:259–269. doi: 10.1039/c2ib00106c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Raja WK, et al. A new chemotaxis device for cell migration studies. Integr Biol (Camb) 2010;2(11–12):696–706. doi: 10.1039/c0ib00044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Calvo F, Sahai E. Cell communication networks in cancer invasion. Curr Opin Cell Biol. 2011;23:621–629. doi: 10.1016/j.ceb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 151.Li H, Fan X, Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem. 2007;101(4):805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 152.Schor SL, Schor AM. Phenotypic and genetic alterations in mammary stroma: implications for tumour progression. Breast Cancer Res. 2001;3(6):373–379. doi: 10.1186/bcr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 154.Sung KE, et al. Transition to invasion in breast cancer: a microfluidic in vitro model enables examination of spatial and temporal effects. Integr Biol (Camb) 2011;3(4):439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Green CE, et al. Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization. PLoS One. 2009;4(8):e6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.De Wever O, et al. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004;18(9):1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- 157.Liu T, Lin B, Qin J. Carcinoma-associated fibroblasts promoted tumor spheroid invasion on a microfluidic 3D co-culture device. Lab Chip. 2010;10(13):1671–1677. doi: 10.1039/c000022a. [DOI] [PubMed] [Google Scholar]
- 158.Franses JW, et al. Stromal endothelial cells directly influence cancer progression. Sci Transl Med. 2011;3(66):66ra5. doi: 10.1126/scitranslmed.3001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Issa A, et al. Vascular endothelial growth factor-C and C–C chemokine receptor 7 in tumor cell-lymphatic cross-talk promote invasive phenotype. Cancer Res. 2009;69(1):349–357. doi: 10.1158/0008-5472.CAN-08-1875. [DOI] [PubMed] [Google Scholar]
- 160.Walter-Yohrling J, et al. Identification of genes expressed in malignant cells that promote invasion. Cancer Res. 2003;63(24):8939–8947. [PubMed] [Google Scholar]
- 161.Swartz MA, Fleury ME. Interstitial flow and its effects in soft tissues. Annu Rev Biomed Eng. 2007;9:229–256. doi: 10.1146/annurev.bioeng.9.060906.151850. [DOI] [PubMed] [Google Scholar]
- 162.Jain RK. Transport of molecules in the tumor interstitium: a review. Cancer Res. 1987;47(12):3039–3051. [PubMed] [Google Scholar]
- 163.Jain RK, Baxter LT. Mechanisms of heterogeneous distribution of monoclonal-antibodies and other macromolecules in tumors—significance of elevated interstitial pressure. Cancer Res. 1988;48(24):7022–7032. [PubMed] [Google Scholar]
- 164.Stohrer M, et al. Oncotic pressure in solid tumors is elevated. Cancer Res. 2000;60(15):4251–4255. [PubMed] [Google Scholar]
- 165.Curti BD, et al. Interstitial pressure of subcutaneous nodules in melanoma and lymphoma patients: changes during treatment. Cancer Res. 1993;53(10 Suppl):2204–2207. [PubMed] [Google Scholar]
- 166.Jain RK, Tong RT, Munn LL. Effect of vascular normalization by antiangiogenic therapy on interstitial hypertension, peritumor edema, and lymphatic metastasis: insights from a mathematical model. Cancer Res. 2007;67(6):2729–2735. doi: 10.1158/0008-5472.CAN-06-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chary SR, Jain RK. Direct measurement of interstitial convection and diffusion of albumin in normal and neoplastic tissues by fluorescence photobleaching. Proc Natl Acad Sci USA. 1989;86(14):5385–5389. doi: 10.1073/pnas.86.14.5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dafni H, et al. Overexpression of vascular endothelial growth factor 165 drives peritumor interstitial convection and induces lymphatic drain: magnetic resonance imaging, confocal microscopy, and histological tracking of triple-labeled albumin. Cancer Res. 2002;62(22):6731–6739. [PubMed] [Google Scholar]
- 169.Shieh AC, Swartz MA. Regulation of tumor invasion by interstitial fluid flow. Phys Biol. 2011;8(1):015012. doi: 10.1088/1478-3975/8/1/015012. [DOI] [PubMed] [Google Scholar]
- 170.Shields JD, et al. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11(6):526–538. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 171.Ng CP, Hinz B, Swartz MA. Interstitial fluid flow induces myofibroblast differentiation and collagen alignment in vitro. J Cell Sci. 2005;118(Pt 20):4731–4739. doi: 10.1242/jcs.02605. [DOI] [PubMed] [Google Scholar]
- 172.Shi ZD, et al. Interstitial flow promotes vascular fibroblast, myofibroblast, and smooth muscle cell motility in 3-D collagen I via upregulation of MMP-1. Am J Physiol Heart Circ Physiol. 2009;297(4):H1225–H1234. doi: 10.1152/ajpheart.00369.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shieh AC, et al. Tumor cell invasion is promoted by interstitial flow-induced matrix priming by stromal fibroblasts. Cancer Res. 2011;71(3):790–800. doi: 10.1158/0008-5472.CAN-10-1513. [DOI] [PubMed] [Google Scholar]
- 174.Pedersen JA, Lichter S, Swartz MA. Cells in 3D matrices under interstitial flow: effects of extracellular matrix alignment on cell shear stress and drag forces. J Biomech. 2010;43(5):900–905. doi: 10.1016/j.jbiomech.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 175.Pedersen JA, Boschetti F, Swartz MA. Effects of extracellular fiber architecture on cell membrane shear stress in a 3D fibrous matrix. J Biomech. 2007;40(7):1484–1492. doi: 10.1016/j.jbiomech.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 176.Shi ZD, Wang H, Tarbell JM. Heparan sulfate proteoglycans mediate interstitial flow mechanotransduction regulating MMP-13 expression and cell motility via FAK-ERK in 3D collagen. PLoS One. 2011;6(1):e15956. doi: 10.1371/journal.pone.0015956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Beauvais DM, Rapraeger AC. Syndecan-1-mediated cell spreading requires signaling by alpha(v)beta(3) integrins in human breast carcinoma cells. Exp Cell Res. 2003;286(2):219–232. doi: 10.1016/S0014-4827(03)00126-5. [DOI] [PubMed] [Google Scholar]
- 178.Qazi H, Shi ZD, Tarbell JM. Fluid shear stress regulates the invasive potential of glioma cells via modulation of migratory activity and matrix metalloproteinase expression. PLoS One. 2011;6(5):e20348. doi: 10.1371/journal.pone.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McLean GW, et al. The role of focal-adhesion kinase in cancer—a new therapeutic opportunity. Nat Rev Cancer. 2005;5(7):505–515. doi: 10.1038/nrc1647. [DOI] [PubMed] [Google Scholar]
- 180.Shi Q, et al. A novel low-molecular weight inhibitor of focal adhesion kinase, TAE226, inhibits glioma growth. Mol Carcinog. 2007;46(6):488–496. doi: 10.1002/mc.20297. [DOI] [PubMed] [Google Scholar]
- 181.Shi Q, Boettiger D. A novel mode for integrin-mediated signaling: tethering is required for phosphorylation of FAK Y397. Mol Biol Cell. 2003;14(10):4306–4315. doi: 10.1091/mbc.E03-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schlaepfer DD, et al. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372(6508):786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 183.Calalb MB, Polte TR, Hanks SK. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol Cell Biol. 1995;15(2):954–963. doi: 10.1128/mcb.15.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sieg DJ, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2(5):249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 185.Chan KT, Cortesio CL, Huttenlocher A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J Cell Biol. 2009;185(2):357–370. doi: 10.1083/jcb.200809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hsia DA, et al. Differential regulation of cell motility and invasion by FAK. J Cell Biol. 2003;160(5):753–767. doi: 10.1083/jcb.200212114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim LC, Song LX, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6(10):587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- 188.Frame MC. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta. 2002;1602(2):114–130. doi: 10.1016/s0304-419x(02)00040-9. [DOI] [PubMed] [Google Scholar]
- 189.Giancotti FG, Tarone G. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu Rev Cell Dev Biol. 2003;19:173–206. doi: 10.1146/annurev.cellbio.19.031103.133334. [DOI] [PubMed] [Google Scholar]
- 190.Cabodi S, et al. Integrin regulation of epidermal growth factor (EGF) receptor and of EGF-dependent responses. Biochem Soc Trans. 2004;32(Pt3):438–442. doi: 10.1042/BST0320438. [DOI] [PubMed] [Google Scholar]
- 191.Ivaska J, Heino J. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu Rev Cell Dev Biol. 2011;27:291–320. doi: 10.1146/annurev-cellbio-092910-154017. [DOI] [PubMed] [Google Scholar]
- 192.Wang F, et al. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95(25):14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Trusolino L, Bertotti A, Comoglio PM. A signaling adapter function for alpha 6 beta 4 integrin in the control of HGF-dependent invasive growth. Cell. 2001;107(5):643–654. doi: 10.1016/S0092-8674(01)00567-0. [DOI] [PubMed] [Google Scholar]
- 194.Kim JH, Asthagiri AR. Matrix stiffening sensitizes epithelial cells to EGF and enables the loss of contact inhibition of proliferation. J Cell Sci. 2011;124(8):1280–1287. doi: 10.1242/jcs.078394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Engl T, et al. CXCR4 chemokine receptor mediates prostate tumor cell adhesion through alpha5 and beta3 integrins. Neoplasia. 2006;8(4):290–301. doi: 10.1593/neo.05694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mitra AK, et al. Ligand-independent activation of c-Met by fibronectin and alpha(5)beta(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30(13):1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. 2011;124(Pt 8):1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Avizienyte E, et al. Src-induced de-regulation of E-cadherin in colon cancer cells requires integrin signalling. Nat Cell Biol. 2002;4(8):632–638. doi: 10.1038/ncb829. [DOI] [PubMed] [Google Scholar]
- 199.Liu X, Feng R. Inhibition of epithelial to mesenchymal transition in metastatic breast carcinoma cells by c-Src suppression. Acta Biochim Biophys Sin (Shanghai) 2010;42(7):496–501. doi: 10.1093/abbs/gmq043. [DOI] [PubMed] [Google Scholar]
- 200.Martinez-Rico C, et al. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. J Cell Sci. 2010;123(5):712–722. doi: 10.1242/jcs.047878. [DOI] [PubMed] [Google Scholar]
- 201.Tseng Q, et al. Spatial organization of the extracellular matrix regulates cell–cell junction positioning. Proc Natl Acad Sci USA. 2012;109(5):1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Borghi N, et al. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc Natl Acad Sci USA. 2010;107(30):13324–13329. doi: 10.1073/pnas.1002662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Abercrombie M, Heaysman JEM. Observations on the social behaviour of cells in tissue culture. 1. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res. 1953;5(1):111–131. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- 204.Astin JW, et al. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol. 2010;12(12):1194–1204. doi: 10.1038/ncb2122. [DOI] [PubMed] [Google Scholar]
- 205.Provenzano PP, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21(3):418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Song JW, Munn LL. Fluid forces control endothelial sprouting. Proc Natl Acad Sci USA. 2011;108(37):15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Netti PA, et al. Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res. 2000;60(9):2497–2503. [PubMed] [Google Scholar]
- 208.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329(5995):1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cross SE, et al. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2(12):780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 210.Kalluri R. Angiogenesis: basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3(6):422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 211.Pluen A. Role of tumor-host interactions in interstitial diffusion of macromolecules: cranial vs. subcutaneous tumors. Proc Natl Acad Sci USA. 2001;98(8):4628–4633. doi: 10.1073/pnas.081626898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.McKee TD. Degradation of fibrillar collagen in a human melanoma xenograft improves the efficacy of an oncolytic herpes simplex virus vector. Cancer Res. 2006;66(5):2509–2513. doi: 10.1158/0008-5472.CAN-05-2242. [DOI] [PubMed] [Google Scholar]
- 213.Castello-Cros R, et al. Staged stromal extracellular 3D matrices differentially regulate breast cancer cell responses through PI3K and beta I-integrins. BMC Cancer. 2009;9:94. doi: 10.1186/1471-2407-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Montes GS. Structural biology of the fibres of the collagenous and elastic systems. Cell Biol Int. 1996;20(1):15–27. doi: 10.1006/cbir.1996.0004. [DOI] [PubMed] [Google Scholar]
- 215.Yamamura N, et al. Effects of the mechanical properties of collagen gel on the in vitro formation of microvessel networks by endothelial cells. Tissue Eng. 2007;13(7):1443–1453. doi: 10.1089/ten.2006.0333. [DOI] [PubMed] [Google Scholar]
- 216.Thorne RG, Hrabetova S, Nicholson C. Diffusion of epidermal growth factor in rat brain extracellular space measured by integrative optical imaging. J Neurophysiol. 2004;92(6):3471–3481. doi: 10.1152/jn.00352.2004. [DOI] [PubMed] [Google Scholar]
- 217.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 218.Tyska MJ, et al. Two heads of myosin are better than one for generating force and motion. Proc Natl Acad Sci USA. 1999;96(8):4402–4407. doi: 10.1073/pnas.96.8.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guilford WH, et al. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72(3):1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.du Roure O, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102(7):2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kraning-Rush CM, et al. The role of the cytoskeleton in cellular force generation in 2D and 3D environments. Phys Biol. 2011;8(1):015009. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Boucher Y, Baxter LT, Jain RK. Interstitial pressure-gradients in tissue-isolated and subcutaneous tumors—implications for therapy. Cancer Res. 1990;50(15):4478–4484. [PubMed] [Google Scholar]