Abstract
Virtual ligand screening is an integral part of the modern drug discovery process. Traditional ligand-based, virtual screening approaches are fast but require a set of structurally diverse ligands known to bind to the target. Traditional structure-based approaches require high-resolution target protein structures and are computationally demanding. In contrast, the recently developed threading/structure-based FINDSITE-based approaches have the advantage that they are as fast as traditional ligand-based approaches and yet overcome the limitations of traditional ligand- or structure-based approaches. These new methods can use predicted low-resolution structures and infer the likelihood of a ligand binding to a target by utilizing ligand information excised from the target’s remote or close homologous proteins and/or libraries of ligand binding databases. Here, we develop an improved version of FINDSITE, FINDSITEfilt, that filters out false positive ligands in threading identified templates by a better binding site detection procedure that includes information about the binding site amino acid similarity. We then combine FINDSITEfilt with FINDSITEX that uses publicly available binding databases ChEMBL and DrugBank for virtual ligand screening. The combined approach, FINDSITEcomb, is compared to two traditional docking methods, AUTODOCK Vina and DOCK 6, on the DUD benchmark set. It is shown to be significantly better in terms of enrichment factor, dependence on target structure quality and speed. FINDSITEcomb is then tested for virtual ligand screening on a large set of 3,576 generic targets from the DrugBank database as well as a set of 168 Human GPCRs. Excluding close homologues, FINDSITEcomb gives an average enrichment factor of 52.1 for generic targets and 22.3 for GPCRs within the top 1% of the screened compound library. Around 65% of the targets have better than random enrichment factors. The performance is insensitive to target structure quality, as long as it has a TM-score ≥ 0.4 to native. Thus, FINDSITEcomb makes the screening of millions of compounds across entire proteomes feasible. The FINDSITEcomb web service is freely available for academic users at http://cssb.biology.gatech.edu/skolnick/webservice/FINDSITE-COMB/index.html
Keywords: Virtual ligand screening, FINDSITE, FINDSITEX, FINDSITEcomb
INTRODUCTION
Virtual ligand screening has become an integral part of modern drug discovery processes for lead identification1. It utilizes computational techniques, is easily automated, and, in principle, can be high throughput. It is attractive to the drug discovery community because experimental high throughput screening has bottlenecks in data analysis and assay development2. Traditionally, there are two broad categories of virtual ligand screening: (a) ligand-based and (b) structure-based. Ligand-based virtual screening is fast, but it requires a set of ligands that are known to bind to the target; this limits its large-scale application. Here, compounds are ranked by their similarity to known binding ligands. Molecular similarity can be computed using 1D, 2D or 3D molecular descriptors such as fingerprints3–5. The most popular similarity measure for comparing chemical structures represented by means of fingerprints is the Tanimoto Coefficient6. Structure-based virtual screening utilizes the structure of the target, docks drug molecules to potential binding pocket/sites and evaluates the binding likelihood using physics-based or knowledge-based scoring functions7. The advantage of structure-based methods is the ability to discover novel active compounds without prior knowledge of known active ligands. Disadvantages are the requirement for high-resolution structures of the target protein that are not always available, as is the case for G-protein coupled receptors (GPCRs) and ion-channels. Structure-based virtual screening is also computationally expensive. This precludes their application to screen millions of compounds across thousands of proteins even when protein structures of requisite quality are available.
To overcome the shortcomings of traditional ligand-based and structure-based methods for virtual ligand screening, recently, novel threading/structure-based approaches that eliminate the prerequisites for known actives and/or high-resolution structure of a given target have been developed8–17. The basic assumption of these methods is that evolutionarily related proteins have similar functions and thus bind similar ligands. It was shown that this assumption is useful even for evolutionarily remote proteins8, 14. Threading/structure is used to detect a possible evolutionary relationship between a target and those proteins that have known binding ligands. If the target protein does not have an experimentally solved structure, threading followed by structure refinement will also provide a model. Subsequently, the structures of the threading detected holo PDB18 templates (structures with bound ligands), along with their bound ligands, are aligned onto the target structure by structural alignment methods19, 20. Template ligand positions are then clustered to infer the binding pocket location and pose of the target’s ligands, and the ligands of the top-ranking cluster (best predicted pocket) are utilized for compound similarity search against a ligand library in a similar way as in traditional ligand-based methods. Thus, threading/structure-based methods inherit the advantages of the speed and lack of requirement of a high-resolution protein structures of ligand-based approaches, and yet, like structure-based methods, do not need known binders to the target protein. Other methods that overcome the need for high-resolution structures and computational demanding of docking approaches have also been developed21–23. These methods utilize predicted target structures and sample binding conformations in coarse-grained protein and ligand representations. The scoring functions for ranking binding conformations are usually knowledge-based21, 23. Their accuracy for virtual ligand screening is comparable to traditional structure-based docking approaches with all-atom representations and scoring functions21.
Since threading/structure-based approaches eliminate the prerequisite for a known set of binders and a high-resolution target structure, they open up the possibility of proteomic-scale drug discovery, since 75% of the sequences in a typical proteome can be reliably modeled24. Proteomic-scale virtual ligand screening is attractive because it could contribute to the understanding of the molecular basis of diseases25. However, threading/structure-based methods for functional analysis have to a large extent focused mainly on protein function and/or binding site predictions, with just a few applications to virtual ligand screening that involve kinases and HIV-1 protease inhibitors9, 10, 26. Large-scale benchmarking tests of these methods for virtual ligand screening of generic targets and systematic comparison to traditional structure–based approaches have not yet been carried out.
An obvious limitation of previous threading/structure-based methods is the requirement that for the protein target of interest, the PDB must contain a significantly number of, at worst, evolutionary distant holo PDB18 templates structures. This makes them inapplicable to membrane proteins, as well as any other class of proteins, (e.g. ion channels) for which an insufficient number of PDB holo templates exist. To address this significant limitation, we recently developed FINDSITEX 26. FINDSITEX utilizes experimental ligand binding databases such as the ChEMBL27 and DrugBank28 databases and does not require experimental holo structures; rather, the structures of the templates are modeled and virtual holo templates are constructed. It is thus useful for targets such as GPCRs and other membrane proteins.
In this work, we improve FINDSITE8 for virtual ligand screening by developing an approach that selects better ligands from the threading identified PDB templates. The improved method, FINDSITEfilt, is then combined with FINDSITEX into a composite method, FINDSITEcomb that will generalize the threading/structure-based FINDSITE approach for generic targets. Here, FINDSITEX utilizes two publicly available protein-small molecule binding databases: ChEMBL27 and DrugBank28. In the Methods section, we describe how these ideas are implemented. Then, in the Results section, we compare the performance of FINDSITEcomb with two freely available traditional docking approaches, AUTODOCK Vina29 and DOCK 630, on the DUD-A Directory of Useful Decoys set31. We then benchmark FINDSITEcomb for virtual ligand screening on a large set of generic drug targets from DrugBank28 and Human GPCR targets from the GLIDA database32. Finally, in the Discussion section, we discuss current and future work.
METHODS
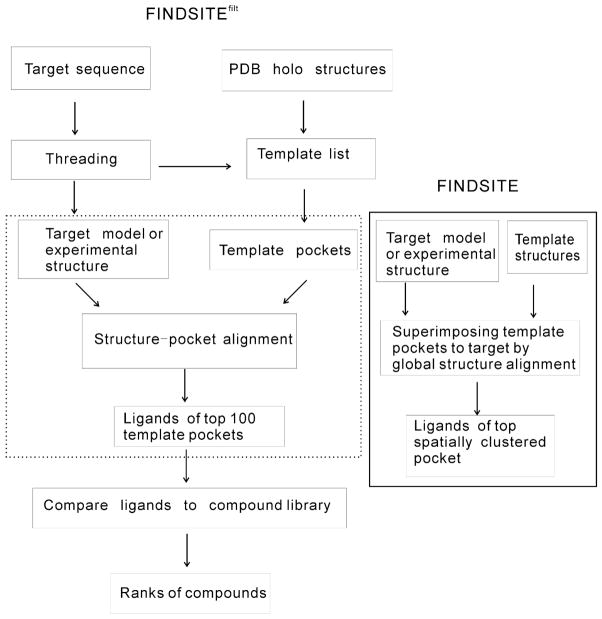
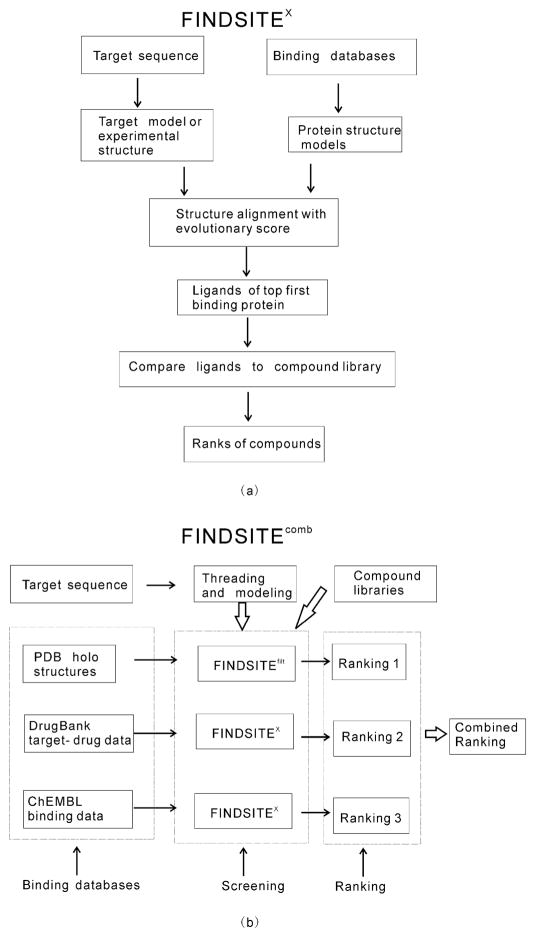
Figure 1 shows the flowchart of the improved version of the FINDSITE methodology, FINDSITEfilt. Figure 2 (a) shows the flowchart of FINDSITEX, and Figure 2 (b) shows an overview of FINDSITEcomb. We describe these methods in what follows.
Figure 1.
Flowcharts of FINDSITEfilt, replacing the steps in dotted-line box with those in the solid-line bordered box gives the original FINDSITE approach.
Figure 2.
(a) Flowchart of FINDSITEX and (b) Overview of FINDSITEcomb.
Improving FINDSITE for ligand virtual screening using heuristic structure-pocket alignment
The flowchart of original FINDSITE8 approach can be found in Figure 1 by replacing the steps in the dotted-line box with those in the solid-line box. The original FINDSITE employs template identification, structure superimposition and binding site clustering as follows: First, for a given target sequence, structure templates are selected from the PDB template library18 by the threading procedure PROSPECTOR_333. Templates are ranked by their Z-score (score in standard deviation units relative to the mean of the structure template library) of the sequence mounted in a given template structure using the best alignment as given by dynamic programming. Only those templates with a Z-score ≥4 and a TM-score ≥0.4 to the target structure/model are used. The TM-score34 is a structural similarity measure that lies between 0 and 1, with a value of 1.0 for identical structures. For a pair of randomly related proteins its average value is around 0.15, with the best average random value of 0.30. A TM-score ≥0.4 means two structures are significantly similar, with a P-value of 3.4 × 10−5. Subsequently, template structures bound to ligands are identified and superimposed onto the target protein structure using the global structure alignment algorithm TM-align19. Then, the centers of mass of ligands bound to threading templates are clustered according to their spatial proximity, using an 8Å cutoff distance. This cutoff maximizes ranking accuracy and accommodates some structural distortions. The geometrical center of each cluster corresponds to the center of a putative binding pocket. Finally, the predicted binding pockets are ranked according to the number of threading templates that share the common binding pocket (cluster multiplicity). For virtual ligand screening, FINDSITE selects ligands that occupy the top ranked binding pocket from the identified ligand-bound threading templates. Hereafter, these ligands will be designated as “template ligands”. The 1,024-bit version of Daylight fingerprints35 is used to represent the ligands and compounds in libraries. Then, the Tanimoto Coefficient (TC)6 of two 1,024-bit fingerprints is used to evaluate the chemical similarity between the two compounds, and compounds in libraries are ranked accordingly (the larger their TC is, the better is the rank).
In the original FINDSITE, the position of the target pocket is determined by global structure alignment (global alignment of two full length protein structures) and the alignment depends only on geometric properties (Cα coordinates). Based on the observation that there are similar pockets in globally different structures and between globally similar structures that have no evolutionary relationship36, the original version of FINDSITE could miss some true positive and include some false positive template ligands. The objective of our improved approach is to filter out these false positive and negative template ligands by a better alignment procedure and by including amino acid type dependent information about binding site similarity between the target and template structures.
The improvements to FINDSITE for ligand virtual screening are shown in the dotted-line box of Figure 1. After threading by SP3 37 as employed in the TASSERVMT-lite structure modeling approach26, for each ligand bound to the threading selected template, a template pocket structure is extracted from the holo template PDB structure. The template pocket structure consists of the Cα atoms of the template residues, any of whose backbone and/or side chain heavy atoms are within 4.5 Å of the bound ligand’s heavy atoms as well as the template residues’ Cα atoms that are within 8 Å of the bound ligand’s heavy atoms. The pocket usually has several dozen Cα atoms scattered along the protein’s sequence. We shall re-label the Cα atoms sequentially for the following alignment. Next, we apply a heuristic structure (of the target) -pocket (of the template) alignment method that effectively determines where the putative target pocket should be and measures its evolutionary closeness to the template pocket. Given the target structure (either modeled or experimental, if available) and a PDB template pocket, the heuristic structure-pocket alignment is carried out as follows: (1) initial alignment: three Cα atoms (consisting of three consecutive I1=I, I2=I+1, I3=I+2, re-labeled residues) of the template pocket are compared to three Cα atoms of the target (residues J1, J2, J3 with J3>J2>J1); if the lengths of all corresponding sides of the two triangles are within 1Å (i.e. |d(I1,I2)-d(J1,J2)|≤1, |d(I2,I3)-d(J2,J3)|≤1, |d(I1,I3)-d(J1,J3)|≤1), the whole template pocket will be superimposed on to the target using the alignment I1 aligned to J1, I2 to J2, I3 to J3. Otherwise, the next pair of triplets is tested. (2) Extension of the alignment based on the superimposed structure: For each template pocket Cα atom, if its nearest target Cα atom in the superimposed structure is within 1 Å, the pocket residue is defined as aligned to the target residue; (3) Superimpose the whole pocket to the target using the alignment in (2) and repeat (2) until the alignment does not change; (4) Calculate the SP- score (Structure-Pocket alignment score) of the alignment in (2) using:
| (1) |
where BLOSUM62(a,b) is the BLOSUM62 substitution matrix38; (5) Repeat steps (1)–(4) for all possible I1, I2, I3, and J1, J2, J3, and the alignment with the largest SP-score is saved as the final alignment. Notice that current implementation of the structure-pocket alignment is sequence order dependent (thus, circularly permuted pockets will be missed). Template pockets are ranked by their SP-scores and the ligands corresponding to the top 100 template pockets selected as template ligands for ligand virtual screening using the following compound similarity score:
| (2) |
where TC stands for the Tanimoto Coefficient6, Nlg is the number of template ligands from the putative evolutionarily related proteins; Ll and Llib stand for the template ligand and the ligand in the compound library, respectively; w is a weight parameter. w=1 gives the average TC in the original FINDSITE screening score. The second term is the maximal TC between a given compound and all the template ligands. Here, we empirically choose w=0.1 to give more weight to the second term so that when the template ligands are true ligands of the target, they will be favored. This new threading/structure based virtual screening approach is called FINDSITEfilt. In contrast to the original FINDSITE, FINDSITEfilt does not cluster the selected top (up to)100 ligands for virtual screening. However, for binding site prediction, spatial clustering is needed. This issue will be addressed elsewhere.
FINDSITEcomb for ligand virtual screening
In order for our FINDSITE based approach to be applicable to all protein classes including membrane receptors, ion-channels, etc., we combine FINDSITEfilt that uses ligand-bound complex structures in the PDB with the FINDSITEX approach that utilizes binding data without complex structures. The original version of FINDSITEX 26 that uses the GLIDA binding database32 was originally developed for GPCR targets. Here, we extend it to treat all protein targets. The FINDSITEX flowchart is shown in Figure 2(a). Given a binding database, the structures of all the target proteins in the database are modeled using the fast version of the latest variant of the TASSER39 based method, TASSERVMT40, TASSERVMT-lite26. If a ligand binding database protein has an experimental structure in the PDB18, TASSERVMT-lite will automatically produce a model very close to the experimental structure (usually having a root-mean-square-deviation of its Cαs <2 Å). The structure of the target protein can also be modeled with TASSERVMT-lite if it’s not available experimentally. Proteins in the binding database that are potentially evolutionarily related to the target are detected by the fr-TM-align20 structure alignment method supplemented with an evolutionary score26: The target structure and the structure of protein in the binding database are aligned by fr-TM-align. Then, an evolutionary score is calculated over the aligned residues as . This score is used to rank the database proteins. The larger the score is, the closer is the database protein to the target evolutionarily. The ligands of the top ranked database protein will be used as template ligands in Eq. (2) for searching against the compound library. As with FINDSITEfilt, mTC given in Eq. (2) is used. Again, this is slightly different from the compound similarity score in our original FINDSITEX 26; this is equivalent to the first term in Eq.(2).
In this work, we shall utilize the DrugBank28 targets and associated drugs as one binding database for FINDSITEX. The DrugBank28 database (http://www.drugbank.ca) has 4,227 non-redundant protein targets and 6,711 drug entries. For our current purpose, we use 3,576 targets and their 6,507 drugs because some targets are too large for TASSERVMT-lite26 to model (Currently, TASSERVMT-lite is applicable to proteins up to 1000 residues in length). Another binding database employed by FINDSITEX is ChEMBL27 (version 12, https://www.ebi.ac.uk/chembl/) that has binding data for broad categories of targets across various species, and thus is helpful for targets such as GPCRs and ion-channels. From ChEMBL, we downloaded data for 593 kinases, 395 proteases, 69 phosphatases, 57 phosphodiesterases, 54 cytochrome P450s, 546 membrane receptors, 325 ion-channels, 134 transporters, 101 transcription factors, 92 cytosolic, 56 secreted, 25 structural, 17 surface antigen, 14 adhesion, 13 other membrane, and 10 nuclear proteins (total 2,501 proteins). The total number of non-redundant ligands binding to these targets is 409,703. We are able to model 2,449 (98%) of the protein targets using TASSERVMT-lite26 and employ these predicted structures in FINDSITEX. The ones we cannot model are too large for our current modeling method. All structural models are provided on our website at http://cssb.biology.gatech.edu/skolnick/webservice/FINDSITE-COMB/index.html.
Figure 2(b) shows the overview of the combined approach FINDSITEcomb that combines the three FINDSITE based virtual screening approaches: FINDSITEfilt using the PDB database, FINDSITEX using the DrugBank database and FINDSITEX using the ChEMBL database. Given a target, for each compound in the compound library, the combined screening score is the maxima of the three mTC scores (see Eq. (2)). The combined screening score gives the final combined ranking.
RESULTS
In what follows, for the evaluation of the performance in DUD, large scale testing of drug targets and GPCRs, we report the performance of a given approach to virtual screening by the Enrichment Factor within the top x fraction (or 100×%) of the screened library compounds defined as:
| (3) |
A true positive is defined as an experimentally known binding ligand/drug or one that has a TC=1 to an experimentally validated binding ligand/drug. For x=0.01, EF0.01 ranges from 0 to 100 (100 means that all true positives are within the top 1% of the compound library).
Comparison to traditional docking methods
We compare FINDSITEcomb in benchmarking mode, (all proteins with > 30% sequence identity to target in the binding databases are excluded from template ligand selection), to two freely available traditional docking methods AUTODOCK Vina29 (http://vina.scripps.edu/) and DOCK 630 (http://dock.compbio.ucsf.edu/DOCK_6/) using the 40 target DUD benchmark set31 (http://dud.docking.org/). The DUD set is designed to help test docking algorithms by providing challenging decoys. It has a total of 2,950 active compounds and a total of 40 protein targets. For each active, there are 36 decoys with similar physical properties (e.g. molecular weight, calculated LogP) but dissimilar topology. AUTODOCK Vina is an open source drug discovery program29 that was tested on the DUD set and shown to be a strong competitor against some commercially distributed docking programs (http://docking.utmb.edu/dudresults/). DOCK 6 is an update of the DOCK 4 program30 and is free for academic users. It has relatively more complicated inputs than AUTODOCK Vina and its performance depends on the input preparation protocols41. AUTODOCK Vina, however, depends on random number generation for the specific target-ligand docking score. In this work, we apply default options for AUTODOCK Vina and use only rigid body docking in DOCK 6 with the default input parameters/options in the examples provided with the program.
Before virtual screening comparison, we compared the relative speed of FINDSITEcomb, AUTODOCK Vina and DOCK 6. On a single CPU node in our cluster, for a typical 325 amino acid protein screened against 100,000 compounds, FINDSITEcomb takes ~10 hours for modeling, ~20 hours for structure comparison and 3 minutes for the compound similarity search, for a total of ~30 hours; AUTODOCK Vina takes around 1,000 hours and DOCK 6 around 5,000 hours. Thus, for screening against 100,000 compounds, FINDSITEcomb is ~ 30 times faster than AUTODOCK Vina and ~160 times faster than DOCK 6, respectively.
Cross docking using experimental and modeled target structures
“Cross docking” means docking all ligands and decoys of all targets to a given target. This scenario is closest to the realistic situation when we do not have much information about which molecule is a true active or decoy to which target. A total of 97,974 non-redundant compounds have been screened for each target. Here, we use both experimental structures and homology-modeled structures for the detection of evolutionary relationships in FINDSITEcomb and for docking methods. Since all DUD targets have crystal structures in the PDB, straightforward modeling will produce models that are very close to their crystal structures. We thus use remote homology modeling by excluding templates in the threading library whose sequence identity > 30% to a given target. However, models for some targets are too extended because a large portion of their sequence is not aligned to a template. Although this is not an issue for FINDSITEcomb (provided that the ligand binding site is in the modeled region), the size of these models is too large for the traditional docking methods to produce output within a tractable time. Therefore, only 30 DUD targets (denoted as DUD-30) are examined. The average actual (predicted) model TM-scores26, 34 to native of these 30 targets are 0.84/0.76. All, but one, model has an actual TM-score to native > 0.4 (hivpr has actual/predicted TM-scores of 0.38/0.48).
The results of this scenario are given in Table 1. Using experimental structures, FINDSITEcomb has an average EF0.01 (27.69) that is 3 times that of AUTODOCK Vina (8.92) and 9 times that of DOCK 6 (3.14). For these 40 DUD targets, the main contribution to FINDSITEcomb is from the PDB, whereas DrugBank and ChEMBL contribute equally. A Student-t test between FINDSITEcomb and the two docking methods indicates that the differences are significant (two sided p-value < 0.05). We note that any of the individual components of FINDSITEcomb is better than the two other docking methods. When modeled structures are used, FINDSITEcomb performs as well as with experimental structures and is significantly better than the two traditional docking methods (EF0.01 of 23 vs. 2–3). Table 1 shows that AUTODOCK Vina performs much worse when modeled structures (EF0.01 ~2) than when experimental structures are used (EF0.01 ~9). The performance of DOCK 6 does not seem to be affected greatly by target structure quality. However, it shows a significant change in performance for EF0.1 in non-cross docking (see below).
Table 1.
Performance of methods on DUD using experimental and modeled structures in cross docking
| Experimental Structures | Modeled Structuresa | |||
|---|---|---|---|---|
| Method (binding database) | Average EF0.01 | P-valueb | Average EF0.01 | P-value |
| FINDSITEX (DrugBank) | 16.89 | 20.05(21.76) | ||
| FINDSITEX (ChEMBL) | 13.78 | 12.69(11.28) | ||
| FINDSITEfilt (PDB) | 22.32 | 21.26(22.44) | ||
| FINDSITEcomb | 27.69 | 23.10 (24.60) | ||
| AUTODOCK Vina | 8.92 | 1.3×10−3 | 2.17 | 1.3×10−4 |
| DOCK 6 | 3.14 | 6.7×10−5 | 3.05 | 1.2×10−3 |
Results are the average of DUD-30 targets; numbers in brackets are results for 40 DUD targets.
Two-sided p-values of Student-t test between FINDSITEcomb and docking methods.
Non-cross docking using experimental target structures
In this scenario, each target’s ligands and decoys (36 times the number of actives) are docked onto itself. The number of screened compounds thus differs between targets. Here, due to fewer compounds screened for each target, we assess the enrichment factors, within the top 5% & 10% as well as 1% of the screened compounds. Another quantity assessed is the area under the accumulation curve (AUAC) of the fraction of actives vs. the fraction of screened compounds.
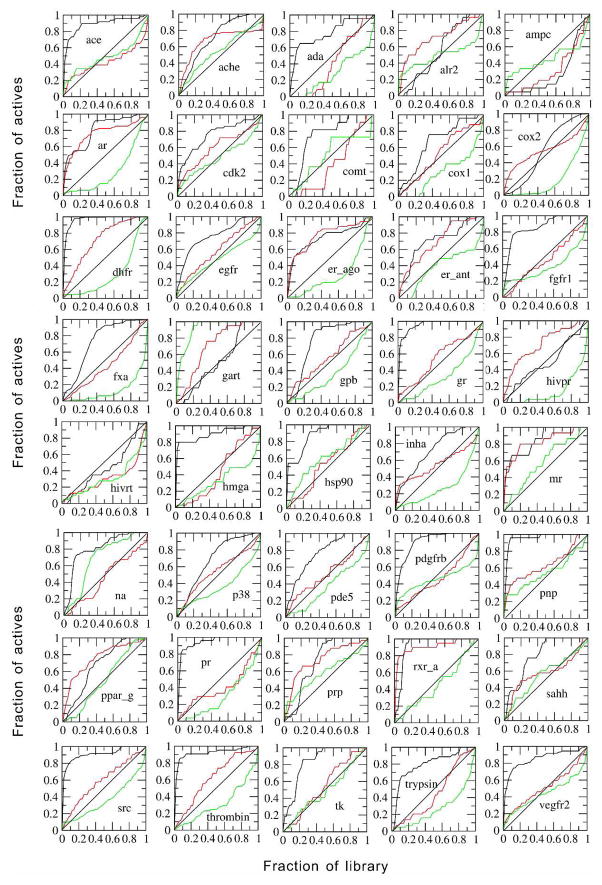
Table 2 shows the performance of different methods in this scenario. Consistent with above results, FINDSITEcomb and its individual components are all better than AUTODOCK Vina and DOCK 6 in terms of enrichment factor. Assessed by the AUAC, DOCK 6 is worse than random and AUTODOCK Vina is better than random (the random AUAC=0.5). Both are significantly worse than FINDSITEcomb. FINDSITEcomb has 38 targets having an AUAC > 0.5, whereas AUTODOCK Vina and DOCK 6 have 28 and 11 targets having an AUAC > 0.5, respectively. For the two FINDSITEcomb failed targets (ampc, hivrt), the two other docking methods also failed. The reason for FINDSITEcomb’s failure is the overwhelming number of false positive, selected template ligands at the lower template sequence identity cutoff (30%). If the sequence identity cutoff is set to 95% to allow the inclusion of ligands from closely homologous templates, the AUACs will be 0.88 and 0.64 for ampc and hivrt, respectively. In Figure 3, we present plots of the fraction of actives vs. the fraction of screened compounds for all 40 targets. Table 3 shows the statistics of targets that: (a) are always above the random diagonal line; (b) start above and go under the random diagonal line; (c) start under and go above the random diagonal line; (d) are always under the random diagonal line. For FINDSITEcomb, the majority (27) of targets are always above the random diagonal line; whereas, AUTODOCK Vina and DOCK 6 have a majority of targets (19 & 22) that start from above and go under the random diagonal line. This latter property could be a typical memory effect of some trained approaches.
Table 2.
Performance of methods on DUD using experimental structures in non-cross docking
| Method (Binding database) | Average EF0.01 | Average EF0.05 | Average EF0.1 | Average AUAC |
|---|---|---|---|---|
| FINDSITEX (DrugBank) | 6.26 | 3.77 | 3.11 | |
| FINDSITEX (ChEMBL) | 7.03 | 4.49 | 3.13 | |
| FINDSITEfilt (PDB) | 11.2 | 5.54 | 3.86 | |
| FINDSITEcomb | 13.4 | 6.56 | 4.37 | 0.774 |
| AUTODOCK Vina | 4.80 (5.3×10−4)a | 3.01 (9.4×10−4) | 2.40 (7.7×10−4) | 0.586 (3.0×10−7) |
| DOCK 6 | 3.72 (1.5×10−4) | 1.79 (1.8×10−5) | 1.24 (9.9×10−7) | 0.426 (1.3×10−12) |
Numbers in brackets are two-sided p-values of Student-t test between FINDSITEcomb and docking methods.
Figure 3.
Fraction of actives vs. fraction of screened compounds curves for the DUD set using experimental structures in non-cross docking. Black line: FINDSITEcomb, red line AUTODOCK Vina, green line DOCK 6.
Table 3.
Behavior of the curves showing the fraction of actives versus the fraction of screened compoundsa
| Method | Always above diagonal | Above to under | Under to above | Always under |
|---|---|---|---|---|
| FINDSITEcomb | 27 | 4 | 9 | 0 |
| AUTODOCK Vina | 9 | 19 | 12 | 0 |
| DOCK 6 | 2 | 22 | 6 | 10 |
Under/over refers to whether when/if the ROC curve crosses the random, diagonal line.
In Ref. 42, several commercially available docking programs including the DOCK 6 are compared on the DUD set for virtual screening accuracy using experimental structures. The results of DOCK 6 were generated using flexible docking and expertise in input preparation and is thus better than what we have in this work. FINDSITEcomb with mean AUAC=0.77 is as good as the best performing program GLIDE (v4.5)43, 44 (mean AUAC=0.72) and therefore is better than all other compared methods: DOCK 6 (mean AUAC=0.55), FlexX45 (mean AUAC=0.61), ICM46, 47 (mean AUAC=0.63), PhDOCK48, 49 (mean AUAC=0.59) and Surflex50–52 (mean AUAC=0.66) 42.
Non-cross docking using modeled versus experimental target structures
Table 4 shows the comparison of different methods using modeled and experimental target structures for the 30 DUD targets. FINDSITEcomb has almost identical EF0.1 and close EF0.01 values for modeled and experimental target structures. All of its component methods have no significant differences (p-value > 0.05) between using experimental and modeled target structures. In contrast, AUTODOCK Vina and DOCK 6 have significantly worse (p-value <0.05) performance for EF0.1 when modeled structures are used. FINDSITEcomb is insensitive to model quality as long as the model’s TM-score to native ≥ 0.4 (see below). However, it should be emphasized that this finding is correct only in a statistical sense (e.g. average EF0.1 or EF0.01). For a particular target, it might not be true.
Table 4.
Comparison of methods for DUD-30 using experimental and modeled structures in non-cross docking
| Method (binding database) | Ave. EF0.01 (expt. structure) | Ave. EF0.01a (modeled structure) | Ave. EF0.1 (expt. structure) | Ave. EF0.1 (modeled structure) |
|---|---|---|---|---|
| FINDSITEX (DrugBank) | 5.92 | 8.28(0.13) | 3.08 | 3.47(0.27) |
| FINDSITEX (ChEMBL) | 8.68 | 8.99(0.86) | 3.55 | 3.09(0.33) |
| FINDSITEfilt (PDB) | 11.0 | 11.3(0.85) | 3.88 | 3.93(0.90) |
| FINDSITEcomb | 14.1 | 13.3(0.58) | 4.54 | 4.53(0.97) |
| AUTODOCK Vina | 5.45 | 2.39(0.037) | 2.48 | 1.40(4.0×10−3) |
| DOCK 6 | 3.82 | 3.05(0.40) | 1.29 | 0.87(0.049) |
Numbers in brackets are two-sided p-values of Student-t test between experimental and modeled structures.
Large scale benchmarking test on generic drug targets
We next tested FINDSITEcomb on all the 3,576 DrugBank targets that we can model. The other targets in the database are too large for our current TASSER-based modeling methods. This issue will be addressed in the future. To test our method under challenging conditions, we exclude all proteins in all three binding databases (PDB, DrugBank, ChEMBL) having sequence identities to the given target > 30%. Target structures are modeled with TASSERVMT-lite26 that is also used for building the structures of the proteins in the binding databases of DrugBank and ChEMBL. The screened compound library consists of all 6,507 drugs (the true binders of all targets) plus 67,871 ZINC8 non-redundant (culled to TC<0.7) compounds53 as background.
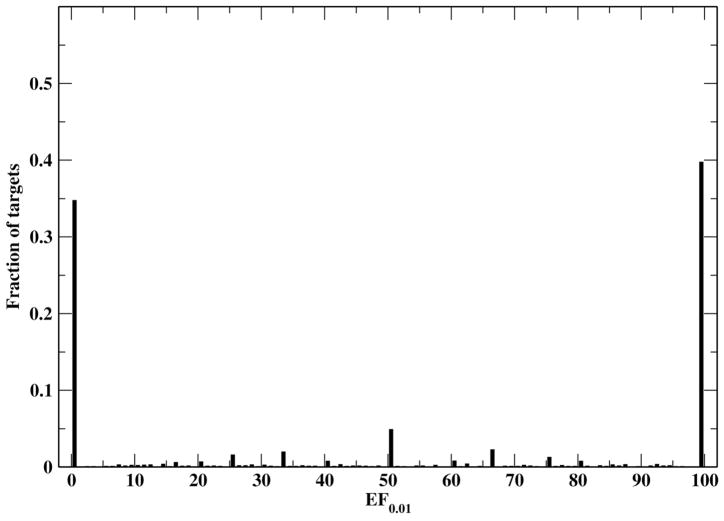
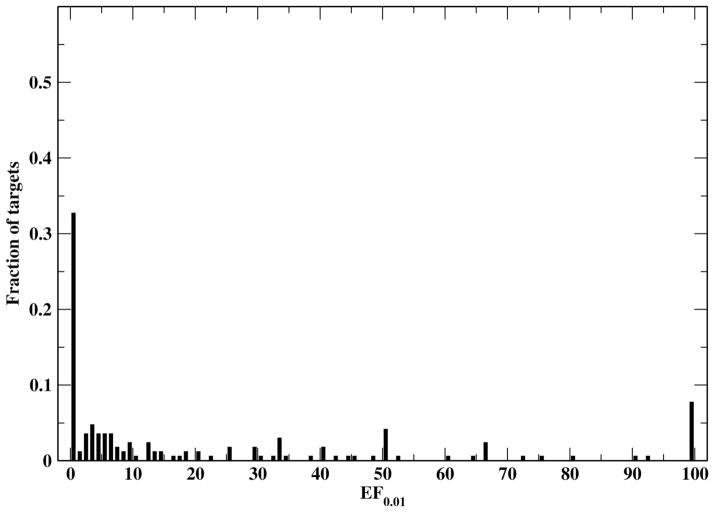
The results of FINDSITEcomb along with its three component methods and the original FINDSITE on this large generic target set are compiled in Table 5. FINDSITEcomb is better than any of its component methods; the major contributions to EF0.01 are from the PDB and DrugBank binding databases. Table 5 also shows that the new FINDSITEfilt is better than the original FINDSITE by a significant ~ 45% for EF0.01 (46.0 vs. 31.7). FINDSITEcomb has an average EF0.01 of 52.1 and is better than random (EF0.01> 1) for 65% of the targets. The histogram of EF0.01 by FINDSITEcomb is shown in Figure 4. Around 40% of the targets have an EF0.01=100. This means that for 40% of the targets, all true drugs can be found within the top 1% (or top ranked 743 ligands) of the screened compounds. FINDSITEcomb fails for ~ 35% targets (EF0.01 < 1). Here we examine two of them. Target Prolyl endopeptidase has predicted TM-score of 0.92 that means its model is very close to experimental structure. It has an EF0.01=0 because the selected template (satisfying sequence identity cutoff < 30%) inside the binding data libraries has no ligands close to that of the target (DB03535) and the templates having close ligands to the target protein all have TM-score < 0.4 to the target (thus are hard to select). The sequence identities of the top ranked ligand binding templates all have <15% sequence identity to the target. Calcium-activated potassium channel subunit beta-3 is a hard target with a predicted TM-score=0.37, indicating that the model is not significantly close to its native structure. Even though in DrugBank alone, there are 16 other targets having the same drug (DB01110), FINDSITEcomb fails to identify them because the target structure is wrong. Thus, FINDSITEcomb could fail because: (1) the binding libraries have no structurally similar templates that have close ligands to the target; (2) the target’s modeled structure is wrong.
Table 5.
Performance of different FINDSITE based methods for the 3,576 drug targets
| Method (binding database) | Average EF0.01 | # (%) of targets having EF0.01 > 1 |
|---|---|---|
| FINDSITE(PDB) | 31.7 | 1526 (43%) |
| FINDSITEX(DrugBank) | 36.6 | 1714 (48%) |
| FINDSITEX(ChEMBL) | 9.5 | 566 (16%) |
| FINDSITEfilt(PDB) | 46.0 | 2080 (58%) |
| FINDSITEcomb | 52.1 | 2333 (65%) |
Figure 4.
Histogram of the FINDSITEcomb enrichment factor EF0.01 for the 3,576 drug targets.
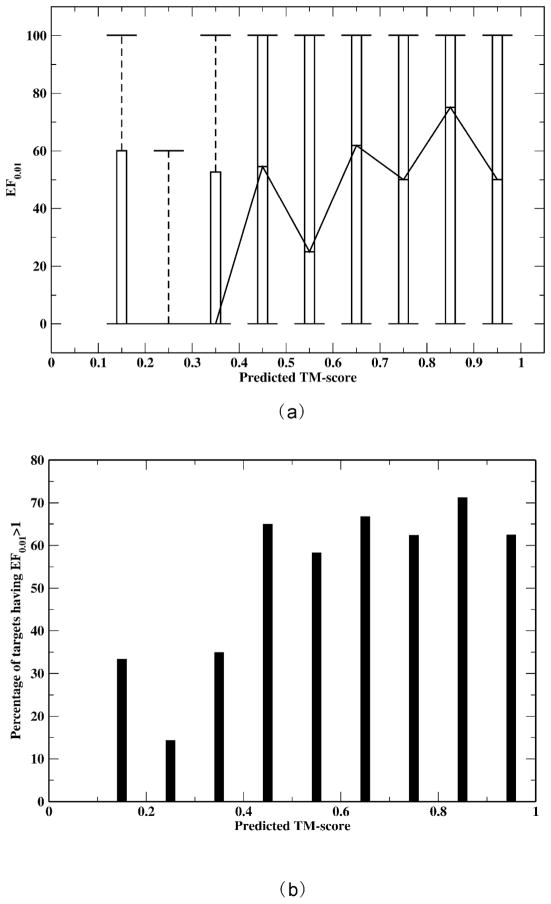
We next examine the relationship between model quality and virtual screening performance. TASSERVMT-lite26 produces a predicted TM-score34 that measures the quality of the model for each target. The predicted TM-score is highly correlated with the actual TM-score of the model to native structure, with a correlation coefficient of 0.86 and a standard deviation of 0.12 over a benchmark set of 690 proteins. A TM-score of 1.0 means that the model is identical to the native structure, and a TM-score of ≥ 0.4 means that the model has significant similarity to the native structure. Figure 5(a) shows box and whisker plots of the EF0.01 within a 0.1 TM-score bin versus the predicted TM-score. Although there is no linear correlation between the median EF0.01 and the predicted TM-score, there is clearly a transition around a TM-score of 0.4. When the predicted TM-score <0.4, all the median EF0.01 are zero; whereas, all the median EF0.01 are at least > 20 when the predicted TM-score >0.4. The transition is also seen for the 75th percentiles (upper box boundaries). The rationale behind this property could be that once the target structure has significant similarity to the native (TM-score ≥ 0.4), the ligands of detected evolutionarily related proteins are roughly similar regardless of how close the target structure is to the native structure. On average, a target with a predicted TM-score ≥ 0.4 has an EF0.01 of 52.8, whereas a target with a predicted TM-score < 0.4 has an EF0.01 of 22.0. Similar results are observed for the percentage of targets having EF0.01 > 1 (better than random) as shown in Figure 5(b) . When the predicted TM-score ≥ 0.4, the probability of EF0.01 > 1 is 66%; this probability drops to around 30% when the predicted TM-score <0.4. Figure 5 demonstrates that as long as the model’s TM-score ≥ 0.4, EF0.01 depends very little on model quality. This feature of FINDSITEcomb was also true for the DUD set (data not shown). Thus, the predicted TM-score can serve as a confidence index of EF0.01 or false positive detection.
Figure 5.
(a) Box and whisker plots of the FINDSITEcomb enrichment factor EF0.01 vs. predicted TM-score for the 3,576 drug targets. The EF0.01s are counted with predicted TM-score within x−0.05 and x+0.05; (b) Percentage of targets having EF0.01 >1 vs. predicted TM-score.
Test on GPCR targets
We developed the FINDSITEX 26 specifically for GPCR proteins by utilizing the GLIDA GPCR binding database32. This early variant of FINDSITEX gives an average enrichment factor EF0.01 of 22.7 for 168 Human GPCRs with known binders in the GLIDA database, when proteins having >30% sequence identity to the target in the binding database (GLIDA) are excluded from template ligand selection. FINDSITEX’s enrichment factor of 22.7 is triple that of the original FINDSITE (7.1). Since FINDSITEcomb does not use the GLIDA GPCR specific database, it is important to test its performance on membrane proteins such as GPCRs, as our goal is to develop a robust and general methodology. Thus, we test FINDSITEcomb using the same 168 Human GPCR set as in Ref. 26 and with the same condition of 30% sequence identity cutoff exclusion for proteins for template ligand selection. Target structures are again modeled with TASSERVMT-lite. The screened compound library consists of all 21,078 true binders of all GPCRs from the GLIDA database (including GPCRs not in this 168 protein set) and the 67,871 ZINC8 TC=0.7 non-redundant compounds.
The results for the 168 Human GPCR set are shown in Table 6. We see that the performance of FINDSITEcomb is almost identical to that of the GPCR specific FINDSITEX that has an average EF0.01 of 22.7 and 114 targets having EF0.01 > 1. Again, for EF0.01 (8.5 vs. 7.1), FINDSITEfilt is better (by ~20%) than the original FINDSITE, and FINDSITEcomb is better than all individual components. In contrast to the above generic targets for which the major contributions of EF0.01 are from the PDB and DrugBank databases, the major contribution to EF0.01 for GPCRs is from the ChEMBL database. Figure 6 shows the distribution of EF0.01. We see that there are few targets having EF0.01=100. For example, for the target TS1R1, FINDSITEcomb has used the drug (DB00168, Aspartame) of the taste receptor type 1 member 2 that has only 23% sequence identity to TS1R1 as the template ligand in Eq. (2). The only active of TS1R1 (L001103) in the GLIDA32 database is identical to DB00168 and is thus ranked top first. Therefore, TS1R1 has an EF0.01 of 100. An example of targets among the 54 (32%) failed ones is SSR3. Its predicted TM-score is 0.68 that is significant (P-value of 3.2 × 10−10). FINDSITEcomb identified these top binding templates: Mu-type opioid receptor, Apelin receptor, CXCR4 from DrugBank, ChEMBL and PDB, respectively. None of these templates has close ligands to those of the SSR3 (TC<0.7). There are, however, 19 templates having at least one identical ligand and sequence identity < 30% to the target in the ChEMBL binding library. All of them have a TM-score < 0.4 to the target.
Table 6.
Performance of different FINDSITE based methods for the 168 Human GPCRs
| Method (binding database) | Average EF0.01 | # (%) of targets having EF0.01 > 1 |
|---|---|---|
| FINDSITE (PDB) | 7.1 | 35 (21%) |
| FINDSITEX (DrugBank) | 10.1 | 76 (45%) |
| FINDSITEX (ChEMBL) | 19.9 | 105 (63%) |
| FINDSITEfilt (PDB)3 | 8.5 | 54 (32%) |
| FINDSITEcomb | 22.3 | 113 (67%) |
Figure 6.
Histogram of the FINDSITEcomb enrichment factor EF0.01for the 168 Human GPCRs.
CONCLUSION and OUTLOOK
We have developed the threading/structure-based approach FINDSITEcomb for virtual ligand screening that utilizes binding information of homologous (remote or close) proteins from publicly available databases such as PDB18, DrugBank28, ChEMBL27. Better accuracy, insensitivity to target structure inaccuracy, and faster speed than traditional docking methods are all attractive features of the current approach. These qualities make proteomic-scale virtual ligand screening possible, since ~75% of the proteins of a typical proteome can be modeled with a predicted TM-score to native ≥0.424. Due to its computational efficiency, we are able to test FINDSITEcomb’s performance across a large variety of protein target classes including GPCRs. We have shown that even in the most challenging condition that only remotely homologous proteins (closest sequence identity of the template protein to the target ≤ 30%) exist in the binding databases, FINDSITEcomb gives an average enrichment factor of 52.1 across all major classes of protein drug targets and 22.3 for GPCRs within the top 1% of screened compound library. More than 65% of targets have better than random enrichment factors when their TM-scores of target structure to native are ≥0.4. Thus FINDSITEcomb is a promising tool for large-scale drug discovery25.
Along with the above-mentioned strengths, the weaknesses of the current methodology are: (a) the inability to treat large proteins (> 1000 amino acids) due to limitations in structure modeling; (b) for around 30% targets, the performance is not better than random (although this ratio might be reduced if closely homologous templates exist in the binding data library); this is mainly due to the failure to accurately model the target structure and the failure to detect structurally different templates that bind to the same ligand. To address, these weaknesses, future improvements of the current method include: (a) extending the modeling approach to large proteins and improving modeling of the 25% of a typical genome’s hard targets where contemporary structure prediction algorithms fail; (b) extending the structure-pocket alignment approach to FINDSITEX using non-PDB libraries; (c) incorporating sequence order independent structure-pocket alignment approaches; (d) combination with low-resolution docking approaches21, 22 to filter out structurally incompatible compounds with respect to binding pockets and to predict binding poses for drug design; and (e) coupling with experimental validation and incorporating feedback from experiment to refine the virtual screening protocol. These efforts are currently underway.
Supplementary Material
Acknowledgments
This work is supported by grant Nos. GM-48835, GM-37408 and GM-08422 of the Division of General Medical Sciences of the National Institutes of Health. The authors thank Dr. Bartosz Ilkowski for managing the cluster on which this work was conducted.
References
- 1.Reddy AS, Pati SP, Kumar PP, Pradeep HN, Sastry GN. Virtual Screening in Drug Discovery – A Computational Perspective. Current Protein and Peptide Science. 2007;8(3):331–353. doi: 10.2174/138920307781369427. [DOI] [PubMed] [Google Scholar]
- 2.Macarron R, Banks MN, Bojanic D, Burns DJ, Cirovic DA, Garyantes T, Green DVS, Hertzberg RP, Janzen WP, Paslay JW, Schopfer U, Sittampalam GS. Impact of high-throughput screening in biomedical research. Nature Reviews Drug Discovery. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 3.Glen RC, Adams SE. Similarity Metrics and Descriptor Spaces - Which Combinations to Choose? QSAR Comb Sci. 2006;25(12):1133–1142. [Google Scholar]
- 4.Flower DR. On the Properties of Bit String-Based Measures of Chemical Similarity. J Chem Inf Comput Sci. 1998;38(3):379–386. [Google Scholar]
- 5.Nikolova N, Jaworska J. Approaches to Measure Chemical Similarity – a Review. QSAR & Combinatorial Science. 2003;22(9):1006–1026. [Google Scholar]
- 6.Tanimoto TT. An elementary mathematical theory of classification and prediction. IBM Interanl Report. 1958 Nov [Google Scholar]
- 7.Kroemer R. Structure-based drug design: docking and scoring. Curr Protein Pept Sc. 2007;8(4):312–328. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- 8.Brylinski M, Skolnick J. FINDSITE. A threading-based method for ligand-binding site prediction functional annotation. Proc Natl Acad Science. 2008;105:129–134. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brylinski M, Skolnick J. Comprehensive Structural and Functional Characterization of the Human Kinome by Protein Structure Modeling and Ligand Virtual Screening. J Chem Inf Model. 2010;50(10):1839–1854. doi: 10.1021/ci100235n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brylinski M, Skolnick J. Cross-Reactivity Virtual Profiling of the Human kinome by X-ReactKIN: A chemical Systems Biology Approach. Molecular Pharmaceutics. 2010;7(6):2324–33. doi: 10.1021/mp1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brylinski M, Skolnick J. Comparison of structure-based and threading-based approaches to protein functional annotation. Proteins. 2010;78(1):118–34. doi: 10.1002/prot.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy A, Xu D, Poisson J, Zhang Y. A Protocol for Computer-Based Protein Structure and Function Prediction. Journal of Visualized Experiments. 2011:57. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wass MN, Kelly LA, Sternberg MJ. 3DLigandSite. predicting ligand-binding sites using similar structures. Nucl Acid Res. 2010;38(suppl 2):W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brylinski M, Skolnick J. FINDSITELHM. a threading-based approach to ligand homology modeling. PLoS computational biology. 2009;5(6):e1000405. doi: 10.1371/journal.pcbi.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skolnick J, Brylinski M. Novel computational approaches to drug discovery. Proceedings of the International Conference of the Quantum Bio-Informatics III; 2009; 2009. [Google Scholar]
- 16.Roy A, Zhang Y. Recognizing protein-ligand binding sites by global structural alignment and local geometry refinement. Structure. 2012;20:987–997. doi: 10.1016/j.str.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy A, Yang J, Zhang Y. An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Research. 2012;20:W471–W477. doi: 10.1093/nar/gks372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr, MDB, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: A Computer-based Archival File for Macromolecular Structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Skolnick J. TM-align. a protein structure alignment algorithm based on the TM-score. Nucl Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandit S, Skolnick J. Fr-TM-align. a new protein structural alignment method based on fragment alignments and the TM-score. BMC Bioinformatics. 2008;(9):531. doi: 10.1186/1471-2105-9-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brylinski M, Skolnick J. Q-Dock. Low-resolution flexible ligand docking with pocket-specific threading restraints. Journal of Computational Chemistry. 2008;29:1574–88. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brylinski M, Skolnick J. Q-DockLHM. Low-resolution refinement for ligand comparative modeling. Journal of Computational Chemistry. 2010;31:1093–105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HS, Zhang Y. BSP-SLIM. A blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins. 2011;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Gao M, Kumar N, Skolnick J. SUNPRO. Structure and function predictions of proteins from representative organisms. BMC Bioinformatics. 2012 sumitted. [Google Scholar]
- 25.Dean P, Zanders E, Bailey D. Industrial-scale genomics-based drug design and discovery. TRENDS in Biotechnology. 2001;19(8):288–292. doi: 10.1016/s0167-7799(01)01696-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhou H, Skolnick J. FINDSITEX. A Structure-Based, Small Molecule Virtual Screening Approach with Application to All Identified Human GPCRs. Molecular Pharmaceutics. 2012;9(6):1775–1784. doi: 10.1021/mp3000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaulton A, Bellis L, Bento A, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington J. ChEMBL. a large-scale bioactivity database for drug discovery. Nucl Acid Res. 2012;40(D1):D1100–07. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wishart D, Knox C, Guo A, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank. a comprehensive resource for in silico drug discovery and exploration. Nucl Acid Res. 2006;34(Database):D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trott O, Olson AJ. AutoDock Vina. improving the speed and accuracy of docking with a new scoring function efficient optimization multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0. search strategies for automated molecular docking of flexible molecule databases. J Comput-Aided Molec Design. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 31.Huang N, Shoichet B, Irwin J. Benchmarking Sets for Molecular Docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR—ligand database for chemical genomics drug discovery—database tools update. Nucl Acid Res. 2007;36:D907–D912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR 3.0 threading algorithm. Proteins. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. A scoring function for the automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 35.Anonymous. Daylight Theory Manual. Daylight Chemical Information Systems, Inc; Aliso Viejo, CA: 2007. [Google Scholar]
- 36.Skolnick J, Zhou HMB. Further Evidence for the Likely Completeness of the Library of Solved Single Domain Protein Structures. Journal of Physical Chemistry B. 2012;116(23):6654–6664. doi: 10.1021/jp211052j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;(58):321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff JG. Amino Acid Substitution Matrices from Protein Blocks. PNAS. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on genomic scale. Proc Natl Acad Sci (USA) 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Skolnick J. Template-based protein structure modeling using TASSERVMT. Proteins. 2011;80(2):352–361. doi: 10.1002/prot.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brozell S, Mukherjee S, Balius T, Roe D, Case D, Rizzo R. Evaluation of DOCK 6 as a pose generation and database enrichment tool. J Comput Aided Mol Des. 2012;26(6):749–73. doi: 10.1007/s10822-012-9565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of Several Molecular Docking Programs: Pose Prediction and Virtual Screening Accuracy. J Chem Inf Model. 2009;49:1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- 43.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: A new approach for rapid accurate docking andd scoring. 1. Method assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 44.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: A new approach for rapid accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 45.Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 46.Abagyan R, Totrov M, Kuznetsov D. ICM - a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- 47.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1998;(Suppl):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 48.Joseph-McCarthy D, Thomas BEIV, Belmarsh M, Moustakas D, Alvarez JC. Pharmacophore-based molecular docking to account for ligand flexibility. Proteins. 2003;51:172–188. doi: 10.1002/prot.10266. [DOI] [PubMed] [Google Scholar]
- 49.Joseph-McCarthy D, McFadyen IJ, Zou J, Walker G, Alvarez JC. Pharmacophore-based molecular docking: A practical guide. Drug DiscoVery Ser. 2005;1:327–347. [Google Scholar]
- 50.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 51.Pham TA, Jain AN. Parameter Estimation for Scoring Protein-Ligand Interactions Using Negative Training Data. J Med Chem. 2006;49:5856–5868. doi: 10.1021/jm050040j. [DOI] [PubMed] [Google Scholar]
- 52.Jain AN. Surflex-Dock 2.1: Robust performance from ligand energetic modeling ring flexibility, and knowledge-based search. J Comput-Aided Mol Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 53.Irwin JJ, Shoichet BK. ZINC- A Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.