SUMMARY
Inhibitors of Apoptosis Proteins (IAPs) are guardian ubiquitin ligases that keep classic pro-apoptotic proteins in check. Systematic identification of additional IAP substrates is challenged by the heterogeneity and sheer number of ubiquitinated proteins (>5000). Here we report a powerful catalytic tagging tool, the NEDDylator, which fuses a NEDD8 E2 conjugating enzyme, Ubc12, to the ubiquitin ligase, XIAP or cIAP1. This permits transfer of the rare ubiquitin homolog NEDD8 to the ubiquitin E3 substrates allowing them to be efficiently purified for LC/MS/MS identification. We have identified >50 potential IAP substrates of both cytosolic and mitochondrial origin that bear hallmark N-terminal IAP binding motifs. These substrates include the recently discovered protein phosphatase, PGAM5, which we show is proteolytically processed, accumulates in cytosol during apoptosis, and sensitizes cells to death. These studies reveal mechanisms and antagonistic partners for specific IAPs, and provide a powerful technology for labeling binding partners in transient protein-protein complexes.
INTRODUCTION
The Inhibitors of Apoptosis Protein (IAPs) are cellular guardians that are critical for controlling pro-death proteins such as caspases, Smac and HtrA2. IAPs contain both a RING ubiquitin ligase domain and characteristic baculoviral IAP repeat (BIR) domains that recognize substrates and promote their ubiquitination (Vaux and Silke, 2005; Vucic et al., 2011). Biochemical studies have shown that X chromosome-linked IAP (XIAP) can directly bind to and inhibit caspases, proteases that execute apoptotic cell death. Smac and HtrA2 are allies of caspases and antagonize XIAP using a characteristic N-terminal four residue IAP-binding motif (IBM) to prevent XIAP from binding caspases. However, these pro-apoptotic collaborators are also victims of XIAP as they become ubiquitinated and degraded in this mutually antagonistic relationship (Khan et al., 2008; MacFarlane et al., 2002). Cellular IAPs (mainly cIAP1 and cIAP2) have similar roles as XIAP in regulating caspases activity but also regulate NF-κB pathways by targeting TRAF2, RIP1 and NIK for ubiquitination (Vucic et al., 2011). cIAP1 also mediates the ubiquitination and degradation of other IAP proteins, including XIAP and cIAP2, and thus fine tunes overall IAP levels in the cell (Silke et al., 2005). Given the central importance of XIAP and cIAP1 in regulating cell death and inflammatory signaling, we sought an unbiased proteomics approach to search for additional substrates for the IAPs.
Identification of substrates for specific E3 ligases is extremely challenging because of the weak transient interaction between E3s and their substrates, the tremendous heterogeneity of modifications with over 5000 ubiquitinated proteins in cells (Kim et al., 2011), and the rapid proteasomal degradation of ubiquitinated proteins. To facilitate E3 substrate identification we produced a catalytic tagging device, called the NEDDylator, that covalently marks E3 substrates with the rare and stable ubiquitin homolog, NEDD8 (Figure 1). NEDD8, like ubiquitin, is covalently conjugated to its substrate proteins through lysines. It uses an orthogonal system consisting of a NEDD8 E1 activating enzyme, E2 conjugating enzymes, and E3 ligases. NEDD8 is highly homologous to ubiquitin, but unlike ubiquitin, has a very limited number of endogenous protein substrates (Xirodimas, 2008), mainly the cullins which are the scaffolding subunits of the cullin-RING ubiquitin ligase family (Deshaies et al., 2010). NEDD8 modifications are known to regulate protein activity, and do not target proteins for proteasomal degradation.
Here, we generated a chimeric protein with a NEDD8 E2 (Ubc12) fused to the substrate binding domain of IAP ubiquitin ligases, termed the NEDDylator. This fusion protein permits robust, stable and simple-patterned NEDDylation of IAP substrate proteins that are readily purified and identified by mass spectrometry. These studies revealed >50 IAP substrate candidates. Almost all of these have classic IBM motifs of both cytosolic and mitochondrial origin. One of the strongest hits was PGAM5, a protein phosphatase located in the outer mitochondrial membrane. We show PGAM5 is cleaved by a non-caspase protease, and rapidly released from the mitochondria during apoptosis. The released form sensitizes cells to death, and cleavage uniquely generates a neo-IBM motif that can be bound to IAPs to mediate its ubiquitination. These studies both reveal important mutually antagonistic IAP substrates and a powerful approach for hunting substrates for E3 ligases or other protein-protein partners.
RESULTS
Engineering the NEDDylator for Tagging IAP Substrates
The RING domains in E3 ubiquitin ligases function to bind ubiquitin E2s and place them in close proximity to the bound substrate so they can catalyze the transfer of ubiquitin from the E2 to amino groups on the substrate bound to its E3 (Figure 1A). The NEDDylator for human XIAP (NEDDylatorXIAP) was engineered by removing the RING domain (residues 435-497) from XIAP to prevent its association with ubiquitin E2 (Figure 1B). The N-terminus (residues 1-434) of XIAP was then fused via a flexible Gly-Gly-Ser-Gly linker to the NEDD8 E2, Ubc12. This construct was thus designed to ablate the ability to ubiquitinate, but empower the ability to NEDDylate the ubiquitin ligase substrates.
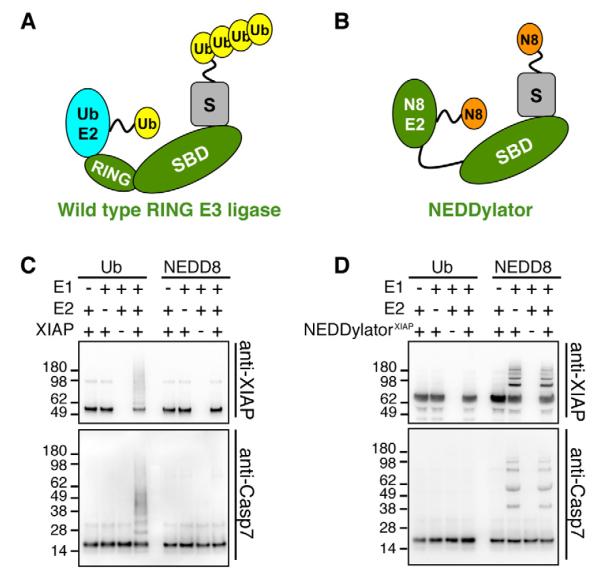
Figure 1. Design and Validation of NEDDylator.
(A) Schematic view of wild type RING ubiquitin ligase. A typical RING ubiquitin ligase (green) contains a RING domain and a substrate-binding domain (SBD). The RING domain binds a ubiquitin E2 (cyan) and facilitates transfer of ubiquitin (yellow) from the catalytic cysteine on the E2 to lysines on substrate protein (grey) recruited by SBD.
(B) Schematic view of the NEDDylator. The RING domain of an ubiquitin ligase is removed and NEDD8 E2 Ubc12 is fused to the substrate binding domains (SBD) of the ligase to generate the NEDDylator (green). This brings the NEDD8 E2 conjugated with NEDD8 (orange) in close proximity to the substrate of the original E3 ligase.
(C) Wild type XIAP has ubiquitin specific ligase activity for self and substrate ubiquitination. In vitro ubiquitination and NEDDylation assays were performed with purified recombinant proteins as described in the supplemental methods. For ubiquitination assays we employed the specific E1 UBE1 and E2 UbcH5b, while for the NEDDylation assay we used the NEDD8 specific E1 NAE1/UBA3 and E2 Ubc12. Immuno-blots against XIAP and caspase-7 were used to detect ubiquitinated proteins.
(D) The NEDDylatorXIAP specifically mediates transfer of NEDD8 and not ubiquitin. The NEDDylatorXIAP, instead of XIAP, was added in the same assay as in (C).
To test this design using purified components, we compared the ability of the native XIAP ubiquitin ligase and the NEDDylatorXIAP to transfer either a ubiquitin or a Hisbiotin tagged NEDD8 (HB-NEDD8) to a known substrate, caspase-7 (Choi et al., 2009; Gray et al., 2010; Schile et al., 2008; Suzuki et al., 2001b), or to XIAP itself. As expected, wild type XIAP produces multiple ubiquitinated species of caspase-7 and itself, but does not NEDDylate either protein (Figure 1C). In sharp contrast, the NEDDylatorXIAP robustly attaches the tagged NEDD8 to itself and caspase-7 without the need for the NEDD8 E2, and does not ubiquitinate caspase-7 or itself (Figure 1D).
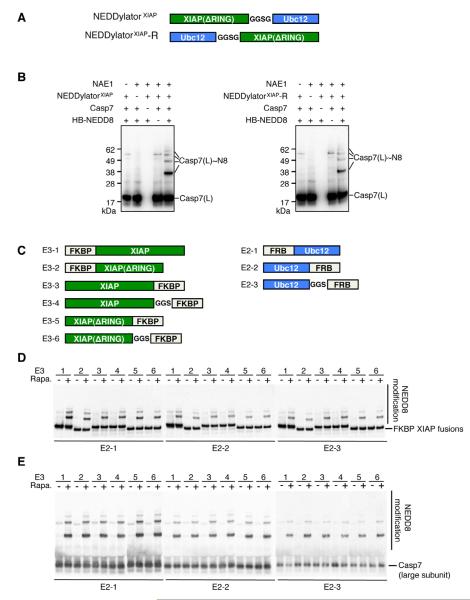
To evaluate the tolerances and flexibility of the NEDDylatorXIAP for efficient NEDDylation we tested a number of other fusion designs. To test how orientation affects the NEDDylation we compared NEDDylatorXIAP with a reversed version NEDDylatorXIAP-R, in which XIAP is fused to the C-terminal of Ubc12 (Figure 2A). Similar NEDD8 modifications of caspase-7 were observed between the two constructs (Figure 2B). To further evaluate the tolerances and to see if the NEDDylatorXIAP would be amenable to small molecule conditional induced activation, we produced a split-NEDDylatorXIAP in which FKBP was fused to XIAP and FRB separately fused to Ubc12. Only in the presence of rapamycin, which induces heterodimerization of FKBP and FRB (Banaszynski et al., 2005), did the split-NEDDylatorXIAP robustly NEDDylate caspase-7. We generated six FKBP-XIAP and three FRB-Ubc12 fusions with various linkers, potentially introducing different flexibility and orientation between XIAP and Ubc12 (Figure 2C). Upon rapamyacin induction, all 18 combinations showed NEDDylation efficiencies that were virtually indistinguishable for two known XIAP substrates, caspase-7 and XIAP itself (Figure 2D, 2E).
Figure 2. The NEDDylatorXIAP Activity is Dependent on Close Proximity between the XIAP Substrate Binding Domain and Ubc12, but not Restricted by the Relative Orientation between XIAP Substrate Binding domain and Ubc12.
(A) Schematic view of the NEDDylatorXIAP and NEDDylatorXIAP-R.
(B) In vitro NEDDylation of caspase-7 by the NEDDylatorXIAP and NEDDylatorXIAP-R produce similar results. Caspase-7 was NEDDylated in the presence of NEDD8 E1 (NAE1) and the NEDDylator fusions. The anti-caspase-7 antibody only recognizes the large subunit Casp-7(L). Casp-7(L)~N8 represents the NEDD8 modified form of the large subunit of caspase-7.
(C) Schematic view of the split NEDDylator, where Ubc12 is fused to FRB, and XIAP is fused to FKBP.
(D) XIAP is NEDDylated in the presence of 1 μM rapamycin with 18 combinations of split NEDDylators. Immuno-blotting was performed with an anti-XIAP antibody.
(E) Caspase-7 is NEDDylated in the presence of 1 μM rapamycin with 18 combinations of split NEDDylators. Immuno-blotting was performed with an anti-caspase-7 antibody. In (D) and (E), DMSO was added to control samples.
See Figure S1.
cIAP1 is believed to have both pro-inflammatory and apoptotic roles (Fulda and Vucic, 2012) and so we sought to compare its substrates with those of XIAP. We generated the NEDDylator for cIAP1 by removing the C-terminal RING domain of cIAP1 (residues 556-618) and fusing it to Ubc12. The NEDDylatorcIAP1 showed robust NEDDylation of caspase-7 (Figure S1A). These results show that the NEDDylator for two different E3 ligases tolerates a wide range of different domain orientations without affecting their abilities to tag target proteins. These data are consistent with the highly flexible nature and structure known for ubiquitin E2-E3 ligase complexes. Such a flexible system evolved to handle a multitude of substrates that vary in size and composition (Duda et al., 2008; Zhuang et al., 2009).
We next tested the NEDDylatorXIAP in cell lysates as well as in cells. The N-terminal acetylation of Ubc12 is important for specific NEDDylation of cullins (Scott et al., 2011). Thus, we used the NEDDylatorXIAP that has XIAP fused to the N-terminus of Ubc12 to mask the N-terminus of Ubc12 and thereby reduce the background from cullin NEDDylation. We produced apoptotic Jurkat cell lysates by addition of staurosporin (STS) so that both procaspase-7 (a non-substrate) and active caspase-7 (a known substrate) were present. Addition of the NEDDylatorXIAP and free HB-NEDD8 induced NEDD8 modification of mature caspase-7 but not procaspase-7 as expected (Figure S1B). We further tested the NEDDylator in Jurkat cells, which were transiently transfected with NEDDylatorXIAP and HB-NEDD8. Immuno-blotting showed NEDD8 modifications of the known XIAP substrates, caspase-7 and SMAC are significantly increased in the presence of NEDDylatorXIAP (Figure S1C).
We then determined which lysines were modified on caspase-7 by the wild type XIAP or the NEDDylatorXIAP. We identified two lysines (K38 and K80) out of 25 on caspase-7 that can be both ubiquitinated by XIAP and modified with NEDD8 by the NEDDylatorXIAP (Figure S2). These data suggest that NEDDylator mediated NEDDylation resembles the native ubiquitination and indicates that fusion of the substrate binding domain of XIAP to Ubc12 in the NEDDylator did not bias the sites that were modified.
Screening for XIAP and cIAP1 Substrates by NEDDylator
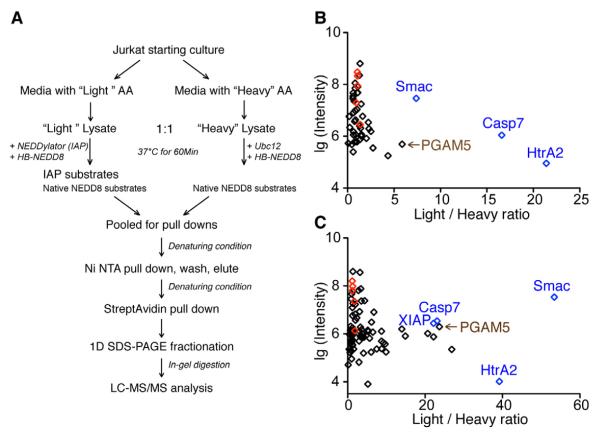
To globally identify the substrates of XIAP and cIAP1 using their respective NEDDylators, we applied stable isotope labeling by amino acid in cell culture (SILAC) (Ong and Mann, 2006) – based mass spectrometry strategy outlined in Figure 3A. Jurkat cells were grown in media containing either light or heavy isotopes of lysine and arginine and induced to undergo apoptosis with STS. Light cell extracts were incubated with the respective NEDDylator for each of the two IAPs and the heavy extracts labeled only with the wild-type NEDD8 E2, Ubc12. The NEDDylated proteins were isolated under denaturing conditions via the HB-NEDD8 tag using a tandem affinity pull down procedure (Tagwerker et al., 2006) (Figure S3). Purified NEDDylated proteins were trypsinized and analyzed with LC-MS/MS. Endogenous NEDD8 substrates, and not substrates of either IAP, are expected to have Light/Heavy (L/H) ratios close to 1. In contrast the IAP substrates labeled by the respective NEDDylators should have high L/H ratios. In both experiments, the five cullins that are well-known NEDD8 substrates were identified with low L/H ratios ranging from 0.9 to 1.8. In stark contrast, three known IAP substrates, including caspase-7, Smac and HtrA2, had L/H ratios that were greater than 7 (Figure 3B, 3C, Table S1).
Figure 3. Identification of Potential XIAP and cIAP1 Substrates, Using Respective NEDDylators and SILAC-based Mass Spectrometry.
(A) Workflow for comparing proteins that are NEDDylated with (“Light”) or without (“Heavy”) the NEDDylator.
(B) Distribution of potential XIAP substrate protein Light/Heavy ratios obtained by the SILAC experiments. Each dot represents a protein, from which at least two unique peptides were identified. Light/Heavy ratios of each protein are plotted against the log scale of summed intensity. Known XIAP substrates caspase-7, HtrA2 and Smac are shown in blue; native NEDD8 substrate cullins, including Cul1, 2, 3, 4A, and 5 are shown in red.
(C) Distribution of potential cIAP1 substrate protein Light/Heavy ratios obtained by a similar SILAC experiment. Labels are the same as in (B).
To evaluate the quality of the SILAC data, we calculated the L/H ratio standard deviation (SD) from all the cullin peptides identified in each dataset (Figure S3D, S3E). The observed SD was <0.35 for both datasets, which give an upper L/H ratio cutoff of 2.2 and 2.8, respectively, for the XIAP and cIAP1 datasets. Here we use a more stringent L/H ratio cutoff of 4 as the criteria for candidate substrates.
With the NEDDylatorXIAP, we identified 52 proteins, each having two or more unique tryptic peptides (Figure 3B); 39 of these proteins had L/H ratios <2, probably representing the background from native NEDD8 modification. Five proteins had high L/H ratios >4, among which were the three known XIAP ubiquitin ligase substrates, caspase-7, Smac and HtrA2. With the NEDDylatorcIAP1, we identified 87 proteins, each having two or more tryptic peptides (Figure 3C); 37 of which have background-level L/H ratios <2, while 29 proteins had L/H ratios >4, indicating a larger group of potential substrates for cIAP1.
The combination of the NEDDylator with tandem affinity pull down under denaturing conditions has very high specificity. Thus, we tested if we could directly identify substrates in unlabeled lysates without the use of SILAC methods. Indeed, peptides from the three known IAP substrates, caspase-7, Smac and HtrA2, were easily detected and only in the NEDDylatorXIAP or NEDDylatorcIAP1 treated samples but not in the Ubc12 treated control samples. From this experiment, we identified 17 additional potential substrates for XIAP and 20 for cIAP1with ≥2 peptides identified from each protein in the NEDDylator treated sample but not in the control samples (Table S2). Combining the non-labeled and SILAC data sets produced 18 candidate substrates for XIAP and 45 for cIAP1. XIAP and cIAP1 have 11 substrates in common (Table S1 and Table S2), consistent with their partially redundant roles in controlling apoptosis (Harlin et al., 2001). The larger number of cIAP1 substrates likely reflects its broader function in NF-κB and other signaling pathways.
Identification of a Cleaved Form of PGAM5 with a Neo-IBM Motif Generated by a Non-caspase Protease
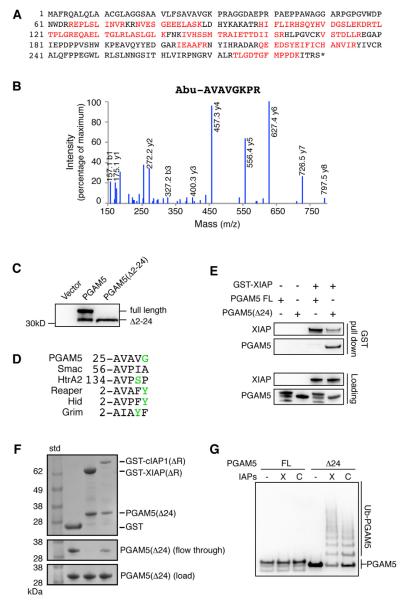
Among the proteins most strongly labeled by the IAP NEDDylators was PGAM5 (phosphoglycerate mutase family member 5), a recently discovered dimeric protein phosphatase (Takeda et al., 2009). PGAM5 is attached to the outer membrane of the mitochondria through an N-terminal mitochondrial membrane peptide (Lo and Hannink, 2008). It is also believed to play roles in mitochondrial fission and fusion (Imai et al., 2010), and in necrotic cell death through interaction with the RIP1/RIP3 kinases (Wang et al., 2012). We identified PGAM5 as a potential substrate for both XIAP and cIAP1 in four independent biological experiments with strong coverage (Figure 4A). It was also identified in SILAC based experiments with very high L/H ratios like those of known IAP substrates (Figure 3B and 3C). Interestingly, we repeatedly identified a cleaved form of PGAM5 lacking the first 24 residues in four different cell lines undergoing apoptosis (Figure 4B, Table S3) using the subtiligase-based N-terminal labeling technique (Mahrus et al., 2008; Shimbo et al., 2012). The cleavage does not derive from a caspase because the P1 residue is serine, not aspartate. We further validated the presence of this truncated PGAM5 in cells by immuno-blotting (Figure 4C).
Figure 4. Identification of the Proteolytic Form of PGAM5 as a XIAP and cIAP1 Substrate In Vitro.
(A) PGAM5 protein sequence is displayed with peptides identified by mass spectrometry highlighted in red. Overall, 15 unique peptides from PGAM5 were recovered and account for 43% of the entire protein sequence.
(B) An example of LC-MS/MS spectrum from endogenous PGAM5 N-terminal peptide after subtiligase labeling and positive enrichment. A non-natural amino acid, 2-aminobutyric acid (Abu), is a designed tag showing characteristic labeling by subtiligase. The same peptide was identified in nine individual experiments, as listed in Table S3.
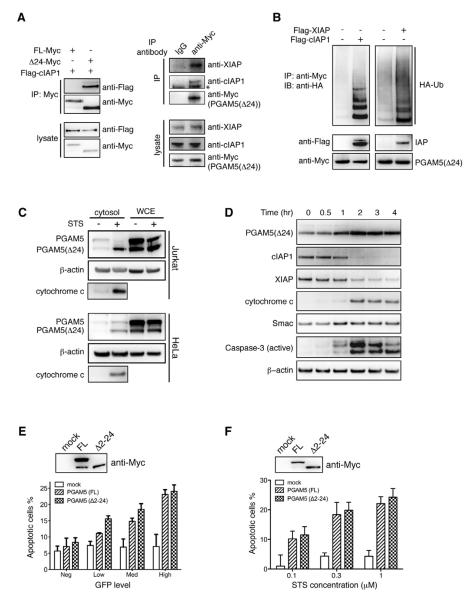
(C) PGAM5 is cleaved into a truncated form in cells. Jurkat cells were transiently transfected with C-terminal Myc tagged full length PGAM5 or PGAM5 (Δ2-24). Immuno-blot with anti-Myc antibody shows an N-terminal cleaved form of PGAM5 with molecular weight identical to PGAM5(Δ2-24).
(D) N-terminal amino acid sequence alignment of PGAM5(Δ24) and other known evolutionarily conserved IBM sequences. Reaper, Hid, and Grim are Drosophila proteins that bind DIAP1, the Drosophila IAP. Amino acids are colored according to their chemical properties.
(E) XIAP binds PGAM5(Δ24) but not the full length PGAM5. Recombinant GST fused XIAP, PGAM5(FL), and PGAM5(Δ24) were used for the in vitro GST pull down experiments.
(F) PGAM5(Δ24) binds to XIAP and cIAP1 independent of the RING domain. GST, GST fused XIAP(ΔR), or GST fused cIAP1(ΔR) were first immobilized on glutathione sepharose beads, then mixed with equal amount to PGAM5(Δ24) for a short time incubation. The resins were washed and proteins were eluted and analyzed with SDS-PAGE and coomassie blue staining.
(G) PGAM5(Δ24) but not the full length protein can be ubiquitinated by both XIAP and cIAP1. PGAM5(FL) and PGAM5(Δ24) containing a C-terminal His tag were expressed and purified from bacteria. The ubiquitination assay was performed in vitro and PGAM5 is detected with an anti-His antibody.
See Figures S4A and Table S3.
Most known IAP substrates have two properties: each is processed to an active form through specific proteolysis, and proteolysis generates a new N-terminus that typically contains a tetra-peptide IBM motif (A-(V/T/I)-(P/A)-(F/Y/I/V)) (Shi, 2002). The IBM, with its invariant amino-terminal alanine, contributes to most of the binding interactions between the IAP and its substrate (Shi, 2002; Verhagen et al., 2001). The truncated PGAM5 we identified contains a putative neo-IBM sequence starting with residue Ala25 (Figure 4D).
To test if the neo-IBM motif is required for PGAM5 to interact with IAPs, we determined which form of PGAM5 is the substrate of IAP. To express and purify PGAM5(Δ24) in bacteria with the neo-IBM N-terminal sequence, we cloned it as PGAM5(Δ2-24) with N-terminal sequence of Met-Ala-Val-Ala-Val and C-terminal His tag. During protein expression in E. Coli the first Met is found to be fully cleaved, presumably by the endogenous methionyl aminopeptidase (MAP) (Hirel et al., 1989), to generate the authentic PGAM5(Δ24) (Figure S4A). In the in vitro binding experiments, PGAM5(Δ24) and not the full length PGAM5 was pulled down by a GST-XIAP fusion protein (Figure 4E). The interaction between IAPs and PGAM5(Δ24) is dependent on the neo-IBM motif and not the RING domain (Figure 4F). Furthermore, XIAP and cIAP1 ubiquitinates the truncated PGAM5(Δ24) and not the full length protein in the in vitro ubiquitination assay (Figure 4G).
PGAM5(Δ24) Binds to IAPs and is Ubiquitinated in Cells
We next addressed if IAPs can bind and ubiquitinate PGAM5(Δ24) in cells. An N-terminal Ala is an invariant determinant in all known IBM motifs. Thus it was necessary to express PGAM5(Δ24) with its proper neo-IBM motif exposed in mammalian cells without commensurate N-terminal initiator methionine or subsequent acetylation typical seen in cytosolic proteins. To achieve this we used the N-terminal ubiquitin fusion technique, where ubiquitin is co-translationally cleaved at the last Gly residue to expose the desired N-terminal sequence of the fused protein (Varshavsky, 1997). This technique has been used to directly express the Smac containing exposed IBM motif with its transit peptide removed (Hunter et al., 2003). Immuno-blotting confirmed the complete cleavage of the N-terminal ubiquitin and generation of the truncated form of PGAM5(Δ24) in cells (Figure S4B). Consistent with the results from in vitro binding experiments (Figure 4E), cellular cIAP1 specifically binds PGAM5(Δ24) but not the full length protein (Figure 5A, left panels). Moreover, endogenous XIAP and cIAP1 were found to co-immunoprecipitate with PGAM5(Δ24) (Figure 5A, right panels). Ubiquitination of PGAM5(Δ24) was stimulated by over-expression of XIAP or cIAP1 (Figure 5B). In addition, the level of ubiquitination of PGAM5(Δ24) increased in the presence of proteasome inhibitor MG132 indicating the proteasome degrades the ubiquitinated product of PGAM5(Δ24) (Figure S4C).
Figure 5. Functional Studies of PGAM5 in Cells.
(A) PGAM5(Δ24) binds XIAP and cIAP1 in vivo. On the left, HEK293 cells were cotransfected with Flag-cIAP1 and PGAM5(FL)-Myc or PGAM5(Δ24)-Myc. Total cell lysates were analyzed by immune-blotting with anti-Flag and anti-Myc antibodies (bottom two panels). Anti-Myc immune-precipitates (IP) were subjected to immunoblot analysis using anti-Myc and anti-Flag antibodies (top two panels). On the right, PGAM5(Δ24)-Myc was transiently expressed in HEK293 cells. 36 hours after transfection, cells were lysed and subjected for Immuno-precipitation with either anti-Myc antibody or an isotype control antibody (mouse IgG). Proteins in the whole cell lysate (bottom three panels) and proteins that bind to the antibody conjugated sepharose beads (top three panels) were assayed with immuno-blotting. *, indicates the position of the immunoglobulin heavy chain.
(B) XIAP and cIAP1 stimulate PGAM5(Δ24) ubiquitination in vivo. HeLa cells were transiently co-transfected with HA-Ubiquitin, PGAM5(Δ24)-Myc, with or without Flag-IAP construct. Ubiquitination of PGAM5(Δ24)-Myc were addressed by immuno-precipitating with Myc antibody then blotted with an HA antibody.
(C) Cytosolic PGAM5(Δ24) is increased in apoptotic cells. Jurkat and HeLa cells were treated with 1 μM STS for 4 hours. The cytosolic protein extracts were used for immuno-blotting analysis with anti-PGAM5 antibody.
(D) Time course of increasing PGAM5(Δ24) in cytosol. Jurkat cells were treated with 1 μM STS and harvested at different time points to extract cytosolic proteins for immuno-blotting analysis with anti-PGAM5 antibody.
(E) High PGAM5 expression induces apoptosis. Jurkat cells were co-transfected with indicated PGAM5 constructs and GFP at a 5:1 molar ratio. 24 hours after transfection, cell viability was measured by Annexin V staining. Different expression levels of GFP were gated to indicate the expression of PGAM5. Error bars represent SEM calculated from triplicate experiments. Representative anti-Myc immuno-blotting from one of the three experiments shows the relative expression levels of PGAM5(FL) and PGAM5(Δ24).
(F) PGAM5 sensitizes cells to the apoptosis inducer STS. HeLa cells are transiently transfected with indicated PGAM5 constructs in 96 well format. 24 hours after transfection, cells are treated for 2 hours with STS at different concentrations. Cell viability is then immediately measured by CellTiter-Glo luminescent assay. Percentages of apoptotic cells are normalized with DMSO treated samples. PGAM5 levels are analyzed with anti-Myc immune-blotting before STS treatment. Error bars represent SEM calculated from triplicate samples.
See Figure S4.
PGAM5 is a Neo-IBM Class Mitochondrial IAP Substrate
Classic substrates like Smac and HtrA2 are imported into the mitochondrial inter-membrane space where the N-terminal signal sequence is removed to produce the mature forms with IBM motifs. They reside sequestered from the cytosolic IAPs until they are released through changes in mitochondrial outer membrane permeability during apoptosis. In stark contrast the N terminus of full-length PGAM5 masks the internal IBM motif at residue 25 and anchors it to the outer membrane of the mitochondria. We show in non-apoptotic cytosol, there is a low level of PGAM5 that is comparable to Smac. However, upon induction of apoptosis with STS there is a significant increase in PGAM5(Δ24) in the cytosolic level that resembles the kinetics for Smac and cytochrome c release (Figure 5C and 5D). Moreover, the release of PGAM5 proceeds the generation of active caspase-3 and subsequent loss of XIAP and cIAP1 as apoptosis proceeds (Figure 5D). Our data suggest PGAM5 release from the mitochondria is an early event in apoptosis coincident with release of cytochrome c and Smac. In healthy cells by contrast there are significant amounts of cleaved PGAM5 in whole cell extracts but little in the cytosolic fraction (Figure 5C). This suggests a portion of the cleaved PGAM5 is still sequestered on mitochondria. It is not clear whether the large increase of cytosolic PGAM5(Δ24) during apoptosis is due to more proteolysis or from translocation of a pre-cleaved PGAM5(Δ24) from the mitochondria.
Expression of Smac is known to sensitize cells to apoptosis stimuli (Du et al., 2000), and expression of HtrA2 causes atypical cell death (Suzuki et al., 2001a). We found that high PGAM5 expression levels in cells are similarly correlated with cell death. Exogenous expression of PGAM5(Δ2-24) in cell cytosol alone can induce cell death that is as effective as expression of the full length PGAM5 (Figure 5E, Figure S4D). Like Smac, transient expression of PGAM5 sensitized cells to the apoptosis inducer STS. The cytosolic PGAM5(Δ2-24) is as efficient as the full length PGAM5 at inducing cell death (Figure 5F). Overall, our data suggest an important regulatory role of the cleaved form of PGAM5 in apoptosis.
Additional Mammalian Mitochondrial Proteins as Potential IAP Substrates
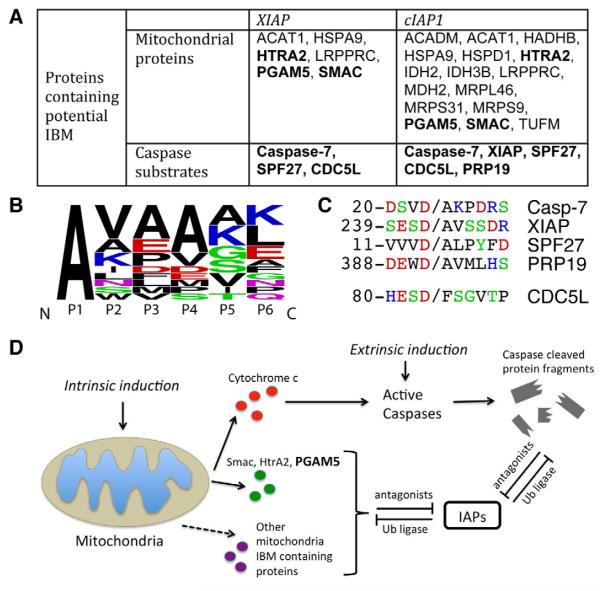
In addition to Smac, HtrA2 and PGAM5, a large portion of the newly identified XIAP and cIAP1 binding proteins are also mitochondrial proteins (Figure 6A). Using our N-terminomics technology (Mahrus et al., 2008), our lab has generated a large experimental degradomic database, where the free N-termini of proteins that result from apoptosis have been identified. By searching against this database, we found that among the 16 mitochondrial proteins labeled by the NEDDylatorcIAP1, 15 are processed into mature forms with the N-terminal mitochondrial targeting transit peptide proteolytically removed, similar to Smac and HtrA2 (Table S4). Interestingly, all the cleaved forms of these mitochondrial proteins contain an N-terminal alanine and an IBM motif-like structure (Figure 6B). The interaction between XIAP and one of these mitochondrial proteins, LRPPR, has been validated in cells (Verhagen et al., 2007). It is tempting to speculate that these other mitochondrial proteins function similarly when released to the cytosol to further antagonize IAPs and relieve caspase inhibition in apoptosis. The IAPs might also mediate the ubiquitination and degradation of these mitochondrial proteins if they are accidentally released to the cytosol by mitochondrial damage.
Figure 6. Proteolytic Cleavage Generates a N-terminal IAP Binding Motif (IBM) that is a Common Feature of IAP Substrates.
(A) Summary table of potential XIAP and cIAP1 substrates containing IBM-like sequences from combined data sets. For simplicity, gene names are used to represent each protein. The full protein names are listed in Table S4.
(B) Sequence logo representation of the frequency of amino acids (Crooks et al., 2004) in the IBM-like cleavage sites identified in 16 mitochondrial proteins listed in table (A). Amino acids are colored according to their chemical properties.
(C) Sequence alignment of amino acid residues surrounding the caspase cleavage sites. Caspases cleave at Asp and generate IBM-like sequences. (D) A model for IAP-IBM interactions.
See Table S4
Common Substrates for Caspases and IAPs
Caspases are the cysteine proteases that specifically cleave at carboxyl side of aspartic acid and drive the final stages of apoptosis. Global proteomics studies have shown that caspases can cleave >1000 substrates during apoptosis (Agard et al., 2012; Dix et al., 2008; Mahrus et al., 2008; Shimbo et al., 2012). Interestingly, five substrates of cIAP1 were identified that are known targets of caspases (Figure 6A): SPF27 (pre-mRNA-splicing factor SPF27), PRP19 (pre-mRNA-processing factor 19), CDC5L (cell division cycle 5-like), XIAP and caspase-7. cIAP1 is known to mediate ubiquitination and degradation of XIAP, through the heterodimerization of RING domains from each protein (Silke et al., 2005). However, the NEDDylatorcIAP1 lacks the RING domain yet robustly tags XIAP. It is known that caspase-mediated proteolysis of XIAP actually generates an IBM-like sequence (Deveraux et al., 1999). Thus, it is reasonable that this neo-IBM motif, not the ring domain, binds cIAP1 during apoptosis when caspases are active without the need for RING-domain assisted heterodimerization with cIAP1. The same binding mechanism may occur for the other two caspase substrates, SPF27 and PRP19, both of which were found cleaved in STS treated Jurkat cells and both exposed an neo-IBM motif upon caspase cleavage (Figure 6C) (Mahrus et al., 2008). Although CDC5L does not contain an IBM motif, it is known to form a complex important for mRNA splicing with IBM-containing SPF27 and PRP19 (Ajuh et al., 2000). Thus, these data suggest the NEDDylator can also indirectly tag bound proteins in complexes, which is known to occur for other ubiquitin ligases (Hao et al., 2005).
Caspase-cleaved protein fragment binding to IAPs may reflect a potential feed forward mechanism to antagonize IAPs and further activate caspases. Indeed, we find >100 proteins that are cleaved by caspases during apoptosis generate an neo-N-terminal alanine (Shimbo et al., 2012), an important characteristic of IBM motifs. This may be an especially important alternative mechanism for enhancing caspase-8 mediated type-I cell death that does not involve immediate release of Smac and HtrA2 from the mitochondria.
DISCUSSION
Applications of NEDDylator
Most ubiquitin substrate identification approaches involve hypothesis-driven candidate approaches, which are slow and heavily biased. Recently, more global substrate identification approaches have been reported using in vitro generated protein microarrays (Gupta et al., 2007), quantitative mass spectrometry (Emanuele et al., 2011; Ota et al., 2008), or global protein stability profiling (GPS) (Emanuele et al., 2011; Yen and Elledge, 2008). We provide an unbiased positive-enrichment method that has several advantages to identifying substrates for specific E3 ubiquitin ligases. Unlike microarrays, the substrates we identify are from native cellular samples and thus contain resident native protein complexes and post-translational modifications. Second, the NEDDylator-based positive enrichment coupled with the dual affinity HB-NEDD8 handle provides a very clean sample that is highly amenable to quantitative mass spectrometry. In addition, modification by the NEDDylator is fully orthogonal to ubiquitination and provides extremely high signal to noise as shown by the SILAC experiments that greatly simplified the data processing. Third, the NEDDylated product is less complex than poly ubiquitination and does not get degraded by the proteasome making it a more stable mark. Lastly, the method is relatively simple to apply and involves only a single genetically encoded enzyme coupled with mass spectrometry that should be easily adapted by other researchers. The fact that the orientation and a variety of split-constructs were all viable NEDDylators speaks further to the robustness and flexibility of the system.
We believe NEDDylator strategy can be used to identify other protein-protein interaction partners, by attaching Ubc12 to the bait portion of the protein. As one test, we made the NEDDylatorPGAM5(Δ24) by fusing Ubc12 to the C-terminus of PGAM5(Δ24) and showed both XIAP and cIAP1 were modified by NEDDylatorPGAM5(Δ24) without the need for the RING domain (Figure S5). This further validates the interactions between PGAM5(Δ24) and IAP proteins, and suggests that NEDDylator can be adapted to identify the interactions in reverse, namely the ubiquitin ligase responsible for a known substrate ubiquitination. Furthermore, given that E3-substrate interactions are considered to be quite transient, the NEDDylator strategy offers an approach for identification of weaker interactions that are very difficult to detect by typical pull-down assays.
PGAM5 is a Pro-apoptotic Regulator that Antagonizes IAPs
Several possible mechanisms can account for the pro-apoptotic function of the cleaved form of PGAM5. The most direct is that PGAM5(Δ24), which contains an IBM motif, would antagonize binding of IAPs to activated capsases and thus foment apoptosis. Second, the release of PGAM5(Δ24) to the cytosol could lead to a significant change in substrate profiles both on the mitochondria and throughout the cell. The mitochondrial PGAM5 was recently proposed as a regulator of induced necrotic cell death (Wang et al., 2012). This was studied by simultaneous addition of TNFα to activate the TNFα receptor, addition of a Smac mimetic compound to block cIAP1 and cIAP2, and by addition of z-VAD-fmk to block caspase activity. Under these conditions it was found that RIP1/RIP3 kinases, critical in driving cellular necrosis, associate with mitochondria bound PGAM5. However during apoptosis our data show that PGAM5 is released early on and could steer the cell away from necrotic cell death by preventing mitochondrial binding of RIP kinases.
A Model for IAP-IBM Interaction
Our studies increase by more than eight-times the number of reported substrates for XIAP and cIAP1 that are generated during apoptosis. Our approach did not fish out all known substrates for cIAP1, such as RIP1 and TRAF2. This is not unexpected because we induced with an intrinsic inducer of apoptosis, staurosporin, as opposed to the extrinsic inducer like TNFα which is needed to activate the TNFα pathway and promote cIAP1 mediated ubiqutination of RIP1 and TRAF2. Additional studies using the NEDDylator should reveal other inducer-specific IAP substrates both for extrinsic apoptosis, necroptosis, inflammosis and perhaps other inducer-specific signaling events mediated by IAPs.
In addition to Smac, HtrA2, and PGAM5, we identified 13 mitochondrial proteins that contain potential IBM motifs. Previous studies using traditional immuno-precipitation coupled with mass spectrometry identified five mitochondrial proteins bearing IBMs (Verhagen et al., 2007). These proteins were shown to bind XIAP including LRPPR that was found here. These studies also showed separate cytosolic expression of each of these mitochondria proteins did not induce apoptosis. The majority of the mitochondrial proteins we identified have functions related to intermediary metabolism such as energy production within mitochondria. Further studies would be important to determine the role of these mitochondrial proteins in modulating the function of IAPs.
In conclusion, we have developed a powerful positive enrichment approach that combines protein engineering and mass spectrometry to stably tag and globally identify >50 candidate substrates for the XIAP and cIAP1. Most candidate substrates contain IBM motifs that are generated by caspase cleavage, or cleavage by other proteases, or upon release from the mitochondria (Figure 6D). These substrates likely oppose the function of the IAPs and we show one of these substrates, PGAM5, is highly pro-apoptotic. These studies validate the NEDDylator as a powerful tool for positive enrichment of substrates for IAPs. We believe the NEDDylator will be generally useful for many more E3 ubiquitin ligase family members now numbering >600. More broadly, this tagging machine may be useful to identify many other transient protein complexes by linking other molecular baits to the NEDDylator and identifying binding partners.
EXPERIMENTAL PROCEDURE
In Vitro Ubiquitination and NEDDylation Assays
Ubiquitination and NEDDylation were assayed at 37°C for 1 hour with purified recombinant proteins in a 20 μl reaction volume with 50 mM Tris pH8.0, 150 mM NaCl, 10 mM MgCl2, 5 mM ATP and 0.1 mM DTT. In Figure 1 and Figure S1A, the ubiquitination reactions contain 50 nM UBE1 (Boston Biochem), 200 nM UbcH5b (Boston Biochem), 200 nM NEDDylator, 200 nM active caspase-7 (purified as previously described in (Wolan et al., 2009), 50 μM ubiquitin (Boston Biochem) and 1 mg/ml bovine serum albumin (BSA, Sigma); the NEDDylation reactions contain 50 nM GST-NAE1/UBA3, 200 nM Ubc12, 200 nM NEDDylator, 200nM active caspase-7, 10 μM HB-NEDD8 and 1 mg/ml BSA. All the reactions were stopped by addition of 4x LDS loading buffer (Invitrogen). Caspase-7 and NEDDylator were detected by western blotting using anti-caspase-7 (Cell Signaling, 9494), anti-XIAP (Cell Signaling, 2045) and anti-cIAP1 (Cell Signaling 4952) antibodies. PGAM5 ubiquitination (Figure 4G) was assayed in the presence of 50 nM UBE1, 200 nM UbcH5b, 700 nM XIAP or cIAP1, and 1.5 μM PGAM5-His6.
Sample Preparation for LC-MS/MS Analysis
For SILAC experiments, Jurkat cells were cultured separately in “Light” (supplemented with regular Lys and Arg) or “Heavy” (supplemented with 13C6 15N2 Lys and 13C6 Arg) medium for 7 days to allow for thorough isotope incorporation. Apoptotic “Light” and “Heavy” cell extracts were prepared individually. The lysates were cleared by spinning and then diluted in 50 mM HEPES pH 7.4, 150 mM NaCl to 5 mg/ml and supplemented with 5 mM ATP, 10 mM MgCl2. Endogenous NEDD8 E1 is cell lysate was used to catalyze the reaction. HB-NEDD8 was added to both lysate at mass ratio 1:500; the NEDDylator was added to the “Light” lysate at mass ratio 1:1000. Equal molar of Ubc12 was added to the “Heavy” lysate. The reactions proceeded at 37 °C for 1 hour, then stopped by addition of iodoacetamide at 20 mM and solid urea to a final concentration of 8 M. “Light” and “Heavy” lysates were pooled and subjected for tandem affinity purification as described (Tagwerker et al., 2006). NEDDylated proteins were separated by one-dimensional SDS-PAGE, and trypsinized for LC-MS/MS analysis.
Supplementary Material
Highlights.
An orthogonal ligase was designed to tag ubiquitin ligase substrate with NEDD8
NEDDylator based proteomic screen identified >50 potential IAP substrates
Most IAP substrate candidates bear hallmark N-terminal IAP binding motifs
PGAM5 is a pro-apoptotic neo-IBM class mitochondrial IAP substrate
ACKNOWLEDGMENTS
We would like to thank David Maltby and the UCSF Mass Spectrometry for equipment assistance, Rebecca Levin for technical assistance, Dr. Emily Crawford for constructing the Degrabase, Drs. Julie A. Zorn and David J. Stanley for providing constructs, Dr. Nicholas J. Agard for invaluable discussions. This work has been supported by R01 GM097316 (J.A.W.) and R01 GM081051 (J.A.W.). Mass spectrometry was performed at the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (ALB, Director) supported by NIH NCRR P41RR001614 and 1S10RR026662.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agard NJ, Mahrus S, Trinidad JC, Lynn A, Burlingame AL, Wells JA. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci U S A. 2012;109:1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh P, Kuster B, Panov K, Zomerdijk JC, Mann M, Lamond AI. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. Embo J. 2000;19:6569–6581. doi: 10.1093/emboj/19.23.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Choi YE, Butterworth M, Malladi S, Duckett CS, Cohen GM, Bratton SB. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase-3 and -7 via unique mechanisms at distinct steps in their processing. J Biol Chem. 2009;284:12772–12782. doi: 10.1074/jbc.M807550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Emberley ED, Saha A. Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem. 2010;54:41–56. doi: 10.1007/978-1-4419-6676-6_4. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. Embo J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Harlin H, Reffey SB, Duckett CS, Lindsten T, Thompson CB. Characterization of XIAP-deficient mice. Mol Cell Biol. 2001;21:3604–3608. doi: 10.1128/MCB.21.10.3604-3608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AM, Kottachchi D, Lewis J, Duckett CS, Korneluk RG, Liston P. A novel ubiquitin fusion system bypasses the mitochondria and generates biologically active Smac/DIABLO. J Biol Chem. 2003;278:7494–7499. doi: 10.1074/jbc.C200695200. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FS, Fujioka M, Datta P, Fernandes-Alnemri T, Jaynes JB, Alnemri ES. The interaction of DIAP1 with dOmi/HtrA2 regulates cell death in Drosophila. Cell Death Differ. 2008;15:1073–1083. doi: 10.1038/cdd.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane M, Merrison W, Bratton SB, Cohen GM. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J Biol Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Ota K, Kito K, Okada S, Ito T. A proteomic screen reveals the mitochondrial outer membrane protein Mdm34p as an essential target of the F-box protein Mdm30p. Genes Cells. 2008;13:1075–1085. doi: 10.1111/j.1365-2443.2008.01228.x. [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. A conserved tetrapeptide motif: potentiating apoptosis through IAP-binding. Cell Death Differ. 2002;9:93–95. doi: 10.1038/sj.cdd.4400957. [DOI] [PubMed] [Google Scholar]
- Shimbo K, Hsu GW, Nguyen H, Mahrus S, Trinidad JC, Burlingame AL, Wells JA. Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proc Natl Acad Sci U S A. 2012;109:12432–12437. doi: 10.1073/pnas.1208616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, Huang DC, Vaux DL. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci U S A. 2005;102:16182–16187. doi: 10.1073/pnas.0502828102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001a;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001b;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009;106:12301–12305. doi: 10.1073/pnas.0901823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Mol Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Coulson EJ, Vaux DL. Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-7-reviews3009. REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen AM, Kratina TK, Hawkins CJ, Silke J, Ekert PG, Vaux DL. Identification of mammalian mitochondrial proteins that interact with IAPs via N-terminal IAP binding motifs. Cell Death Differ. 2007;14:348–357. doi: 10.1038/sj.cdd.4402001. [DOI] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Chen S, Du F, Wang X. The Mitochondrial Phosphatase PGAM5 Functions at the Convergence Point of Multiple Necrotic Death Pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.