Abstract
Purpose
Protein kinase D (PKD) mediates diverse biological responses including cell growth and survival. Therefore, PKD inhibitors may have therapeutic potential. We evaluated the in vitro cytotoxicity of two PKD inhibitors, kb-NB142-70 and its methoxy analog, kb-NB165-09, and examined their in vivo efficacy and pharmacokinetics.
Methods
The in vitro cytotoxicities of kb-NB142-70 and kb-NB165-09 were evaluated by MTT assay against PC-3, androgen independent prostate cancer cells, and CFPAC-1 and PANC-1, pancreatic cancer cells. Efficacy studies were conducted in mice bearing either PC-3 or CPFAC-1 xenografts. Tumor-bearing mice were euthanized between 5 and 1440 min after iv dosing, and plasma and tissue concentrations were measured by HPLC-UV. Metabolites were characterized by LC-MS/MS.
Results
kb-NB142-70 and kb-NB165-09 inhibited cellular growth in the low-mid μM range. The compounds were inactive when administered to tumor-bearing mice. In mice treated with kb-NB142-70, the plasma Cmax was 36.9 nmol/mL and the PC-3 tumor Cmax was 11.8 nmol/g. In mice dosed with kb-NB165-09, the plasma Cmax was 61.9 nmol/mL while the PANC-1 tumor Cmax was 8.0 nmol/g. The plasma half-lives of kb-NB142-70 and kb-NB165-09 were 6 and 14 min, respectively. Both compounds underwent oxidation and glucuronidation.
Conclusions
kb-NB142-70 and kb-NB165-09 were rapidly metabolized, and concentrations in tumor were lower than those required for in vitro cytotoxicity. Replacement of the phenolic hydroxyl group with a methoxy group increased the plasma half-life of kb-NB165-09 2.3-fold over that of kb-NB142-70. Rapid metabolism in mice suggests that next-generation compounds will require further structural modifications to increase potency and/or metabolic stability.
Keywords: Protein Kinase D (PKD) inhibitors, pharmacokinetics, prostate cancer, pancreatic cancer, kb-NB142-70, kb-NB165-09
Introduction
Recent research has revealed that protein kinase D (PKD) plays an important role in the regulation of a wide array of fundamental biological processes, including; cell survival, migration, differentiation, proliferation, membrane trafficking and gene expression [1-10]. The PKD family of serine-threonine kinases consists of PKD1, PKD2 and PKD3, which share a similar modular structure [11]. PKDs are expressed in multiple tissues and cell types, and different expression patterns and functions of PKD depend on the cell type and external stimuli [12-18]. Because of their interaction with multiple signaling pathways and their various intracellular locations, PKDs appear to play a critical role in cancer cell proliferation and invasion [11-17].
Prostate cancer is the leading cancer in men and is the 2nd highest cause of cancer related deaths [18]. Pancreatic cancer is the 10th most common cancer diagnosed and is the 4th highest cause of cancer related deaths [18]. Due to disappointing treatment results and the dismal outcome of pancreatic cancer with an average survival rate of less than 5 years, there is an urgent need for better understanding of tumor pathogenesis and pathways involved in disease progression, in order to develop efficacious targeted small-molecules. The expression of multiple PKD isoforms has been reported in pancreatic and prostate cancer cells [19-32], where PKD enzymes are involved in DNA synthesis and cellular proliferation as well as inhibiting apoptosis [30]. The benzoxoloazepinolone CID755673 was the first reported potent and specific pan PKD inhibitor with IC50 values of 200-300 nM against all 3 PKD isoforms. Given its specificity, CID755673 revealed and validated specific PKD-mediated cell functions in prostate cancer cells and was effective at blocking cellular proliferation, cell migration and invasion [33].
kb-NB142-70 and its methoxy-analogue kb-NB165-09 are structural analogues of CID755673, and are a least 2-fold more potent and showed improved selectivity. LaValle et al. analyzed these two CID755673 analogs and showed that in addition to being more potent in inhibiting all PKD isoforms in vitro, in cells these compounds also inhibited PKD1 autophosphorylation in the low μM range, drastically inhibited cell proliferation by induction of cell cycle arrest at G2/M phase, and significantly inhibited wound healing and tumor cell invasion [34]. Because of the enhanced selectivity and potency of kb-NB142-70 and its analogue kb-NB165-09 against PKD, we evaluated their in vitro cytotoxicity, and their in vivo pharmacokinetics, metabolism and efficacy in mice bearing human prostate or pancreatic xenografts.
Materials and Methods
Cell Lines and Reagents
PC-3 human androgen independent prostate cancer cells were obtained from ATCC (Manassas, VA) and CFPAC-1 and PANC-1 pancreatic cancer cells were obtained from the NCI Tumor Repository (Frederick, MD) and were MAP (murine antibody profile) test negative. The cells were cultured in RPMI 1640 medium (BioWhittaker Inc, Wakersville, MD) supplemented with 10% heat inactivated fetal bovine serum (Invitrogen) and 100 units of penicillin/mL and 100 μg/mL of streptomycin (Biofluids Biosource, Rockville, MD) in a humidified atmosphere of 5% CO2.
Acetonitrile (HPLC-grade) and water (HPLC-grade) were purchased from Thermo-Fisher Scientific (Fairlawn, NJ). Nitrogen gas and liquid nitrogen were purchased from Valley National Gases Inc. (Pittsburgh, PA). Formic Acid (99%) was purchased from Sigma-Aldrich (St. Louis, MO). kb-NB142-70 and kb-NB165-09 (batch synthesized lot# kb-NB184-43) were synthesized according to published procedures [35,36] and were greater than 97% pure (Fig. 2d and 2h). 17-Allyl-amino-(17-demethoxy) geldanamycin (NSC 330507, 17-AAG, internal standard) was obtained from the Developmental Therapeutics Program of the NCI (Rockville, MD). Dextrose (5%) for injection, saline (0.154 M NaCl) and sterile water were purchased from Baxter Healthcare Corporation (Deerfield, IL). β-Glucuronidase (10,000 units/mg from bovine liver, G0501) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma Aldrich (St. Louis, MO).
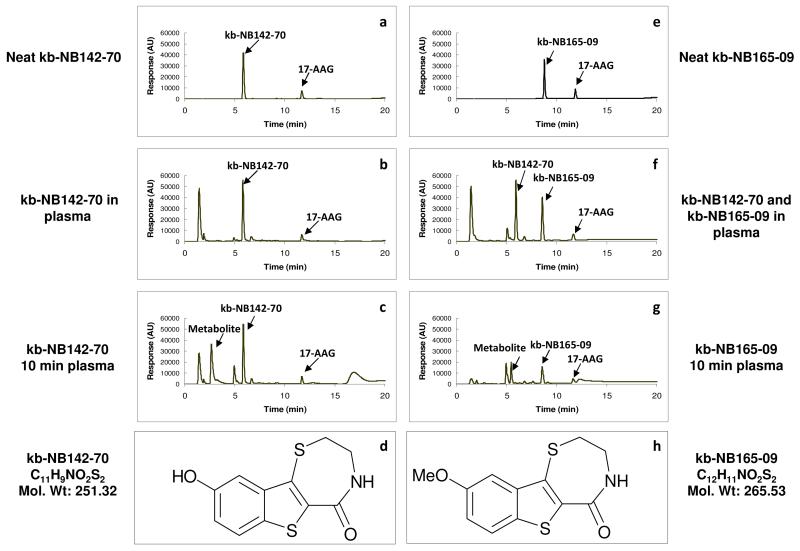
Fig. 2.
Structures and HPLC chromatography of kb-NB142-70 and kb-NB165-09 and their metabolites in plasma. (a) Neat kb-NB142-70 with internal standard (17-allylamino-17-demethoxygeldanamycin, 17-AAG) (b) the chromatogram of kb-NB142-70 added to plasma as a quality control assay standard (c) the chromatogram of kb-NB142-70 and metabolites in plasma obtained 10 min after treating a mouse with 25 mg/kg of kb-NB142-70 iv (d) Structure of kb-NB142-70 (e) Neat kb-NB165-09 with internal standard (17-AAG) (f) Chromatogram of kb-NB165-09 and kb-NB142-70 added to plasma as a quality control assay standard (g) Chromatogram of kb-NB165-09 and metabolites in plasma obtained 10 min after treating a mouse with 25 mg/kg of kb-NB165-09 iv (h) Structure of kb-NB165-09
Mice
Specific-pathogen-free, adult, female C.B.-17 SCID mice (5-6 weeks of age) were obtained from Charles River Laboratories (Wilmington, MA). Mice were allowed to acclimate to the University of Pittsburgh Cancer Institute Animal Facility for at least 1 week before studies were initiated. To minimize exogenous infection, mice were maintained in microisolator cages and handled in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 1996) and on a protocol approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Ventilation and airflow in the animal facility were set to 12 changes/h. Room temperature was regulated at 22 ± 1°C. The mice received Prolab ISOPRO RMH 3000, Irradiated Lab Diet (PMI Nutrition International, Brentwood, MO) and water ad libitum except on the evening prior to dosing for the pharmacokinetic studies, when all food was removed and withheld until 4 h after dosing.
MTT assay
PC-3 cells (3000 cells/well), PANC-1 cells (5000 cells/well), or CFPAC-1 cells (3000 cells/well) in logarithmic growth were plated in a 96-well culture plate. kb-NB142-70 or kb-NB165-09 was added such that the final concentrations in the wells were 1-100 μM in medium containing 0.3% DMSO. After 72 h, 50 μL of 1 mg/mL MTT was added to each well and allowed to incubate for 4 h. At the end of the incubation, medium containing drug and MTT were removed from each well and 100 μL DMSO was added, followed by shaking for 5 min. The absorbance at 570 nm was read on DYNEX NRX Revelation microplate reader (Dynex, Vienna, VA). Results were compared to wells containing vehicle-treated cells and expressed as % inhibition. The IC50 was calculated from triplicate experiments using the Hill equation and the program ADAPT II [37].
Tumor implantation
Cells in logarithmic phase growth were harvested, and 5 × 106 cells in 0.1 mL of medium were implanted subcutaneously on the right flanks of passage C.B.-17 SCID mice. When the tumors reached approximately 500 mm3, the passage mice were euthanized with CO2, and the tumors harvested aseptically. The harvested tumors were cut into approximately 25 mm3 fragments and implanted on the right flanks of study mice. When the tumors in the study mice were approximately 400 mm3, the animals were stratified into time point groups, such that there were no differences in mean body weights or tumor volumes between the groups (ANOVA and Kruskal-Wallis, p ≥0.95).
Efficacy Study
Mice bearing PC-3 xenografts were stratified into 5 mice per treatment group and treated with 25 mg/kg/dose kb-NB142-70 for 5 consecutive days. Mice bearing CFPAC-1 xenografts were stratified into 10 mice per treatment group and received either: kb-NB165-09 (37.5 mg/kg/dose, qdx4); kb-NB165-09 (25 mg/kg/dose, qdx4); gemcitabine (Gemzar, 50 mg/kg/dose, q3dx3), or vehicle (10% hydroxypropyl β-cyclodextrin, 0.01 ml/g body weight, qdx4). Mice were dosed by lateral tail vein injection. Body weights and tumor diameters were recorded twice weekly and tumor volumes were calculated using the formula: TV= L × W2/2 where L is the longest tumor diameter and W is the shortest tumor diameter perpendicular to L. %T/C was calculated as the mean and median tumor volumes divided by the mean and median tumor volumes of the control treated groups × 100%. The time to tumor doubling was the mean or median number of days it took each treatment group to double from its starting tumor volume on day 0, the first day of dosing.
Statistics
Mean tumor volumes and body weight were evaluated using ANOVA with pairwise comparisons using Dunnett’s T-test, Tukey’s and Fisher’s exact test. Nonparametic analysis of median data was performed using Kruskall-Wallis, and pairwise comparisons were conducted using the Mann-Whitney test. Significance was set at p ≤ 0.05. All statistics were performed using Minitab statistical software (Minitab, State College, PA).
Pharmacokinetic studies
C.B.-17 SCID mice bearing PC-3 xenografts were dosed 25 mg/kg of kb-NB142-70 iv and mice bearing PANC-1 xenografts were treated with kb-NB165-09 iv and po at a dose of 25 mg/kg. The mice (3/time point) were euthanized at the following times: 5, 10, 15, 30, 60, 120, 240, 360, 960 and 1440 min after administration iv or 5 min and 1440 min after iv vehicle administration. Mice were euthanized at 5, 15, 30, 60 and 120 min after po administration of kb-NB165-09. Mice were euthanized by CO2 inhalation and blood was collected by cardiac puncture using heparinized syringes and needles. Blood was centrifuged at 12,000 × g for 4 min to obtain plasma and packed red blood cells. The following tissues were collected, weighed and snap frozen in liquid nitrogen after iv administration of kb-NB142-70: liver, kidney, spleen, skeletal muscle, brain, heart, lungs, peritoneal fat and tumor. After iv administration of kb-NB165-09; liver, kidneys and tumor were collected. After oral administration of kb-NB165-09, only plasma and red blood cells were collected. All samples were stored at −70 °C until analysis.
Western blot analysis of tissue samples
Samples of tumor, liver and kidneys from the pharmacokinetic studies of kb-NB142-70 and kb-NB165-09 were weighed and snap frozen in liquid nitrogen and then processed for Western blot analysis of protein kinase D 1, 2, and 3 and their phosphorylated forms. Western blot analysis was carried out as previously reported [34]. Briefly, tissues were homogenized in lysis buffer containing 200 mM Tris-HCl, pH 7.4, 100 μM 4-(2-aminoethyl) benzenesulfonyl fluoride, 1 mM EGTA, and 1% Triton X-100. Protein concentration was determined using the BCA Protein Assay kit (Pierce, Invitrogen, Grand Island, NY) and then equal amounts of protein were subjected to SDS-PAGE followed by electrotransfer to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk in Tris-buffered saline and then probed with primary antibodies for PKD1 and PKD3 (Cell Signaling Technology, Danvers, MA), PKD2 (Millipore, Billerica, MA), pS916-PKD1 (Cell Signaling Technology, Danvers, MA), pS742-PKD1 (Invitrogen, Grand Island, NY), or GAPDH, followed by anti-mouse or anti-rabbit secondary antibodies conjugated to horseradish peroxidase (Bio-Rad, Hercules, CA). The enhanced chemiluminescence (ECL) Western blotting detection system (Amersham Biosciences, Piscataway,NJ) was used to facilitate detection of protein bands.
Pharmacokinetic sample processing
Plasma was extracted directly. Tissues were thawed and homogenized using an Omni Tissuemizer (OMNI International, Marietta, GA) in three volumes (g/v) of phosphate buffered saline (PBS) (Invitrogen). To 200 μL of plasma or tissue homogenate, 30 μL of internal standard (17-AAG, 10 μg/mL) was added, and the samples were vortexed for 15 sec at setting 4 (Vortex Genie, Scientific Industries, Inc. Springfield, MA). Acetonitrile (1 mL) was added to each sample followed by vortexing for 1 min and centrifugation at 12,000 × g for 6 min. Supernatants were transferred to clean 12 × 75 mm culture tubes and evaporated to dryness under a gentle stream of nitrogen gas. Residues were resuspended in 300 μL of initial mobile phase and 100 μL was injected into the HPLC or resuspended in 100 μL mobile phase, and 25 μL was injected onto the HPLC-MS.
HPLC-UV
The HPLC system consisted of a Beckman Coulter System Gold with a 568 auto-sampler, 126 solvent module, and a 166 detector. Separation was performed on a Phenomenex Luna C18 (5 μm, id 4.6 × 100 mm) column (Torrence, CA) and a Brownlee C18 guard column (PerkinElmer, Shelton, CT). The mobile phase was a gradient of acetonitrile and water, each containing 0.1% formic acid: acetonitrile from 20-80% over 10 min, held at 80% for 5 min, reduced to 20% (the initial condition) over 5 min followed by re-equilibration for 5 min. The flow rate was 1 mL/min, and the absorption was monitored at 310 nm. The assay was linear between 0.1 and 10 μg/mL. The retention times of kb-NB142-70 and kb-NB165-09 were 6 min (Fig. 2a) and 8.5 min (Fig. 2e), respectively. The major metabolite’s retention time for kb-NB142-70 was 3 min (Fig. 2c) and kb-NB165-09 was 5.4 min (Fig. 2g). Standard curves of kb-NB142-70 or kb-NB165-09 were constructed by plotting the internal standard ratio versus the known concentration of kb-NB142-70 (Fig. 2b) or kb-NB165-09 (Fig. 2f) in the sample. kb-NB142-70 was added to control plasma in kb-165-09 standard curve because kb-NB142-70 was initially expected to be a metabolite of kb-NB165-09 in vivo (Fig. 2f). Triplicate standard curves containing kb-NB142-70 or kb-NB165-09 concentrations of 0, 0.03, 0.1, 0.3, 1, 3 μg/mL were included with each analytic run. Standard curves were fit by linear regression followed by back calculation of concentrations. The lower limit of kb-NB142-70 and kb-NB165-09 quantification was 0.03 μg/mL. Coefficients of variation of kb-NB142-70 and kb-NB165-09 in plasma at a low mid-range concentration (0.3 μg/mL) and high mid-range concentration (3 μg/mL) were less than 10%. Recoveries of kb-NB142-70 and kb-NB165-09 were higher than 93%.
Pharmacokinetic analysis
The terminal half-life, clearance, volume of distribution at steady state, and areas under the concentration (AUC) versus time curves for plasma, and the AUC and terminal half-life for tissues were estimated by noncompartmental analysis using the program LAGRAN and the Lagrange function [38, 39]. The AUC’s were confirmed using the trapezoidal method with the software PK Solutions (Summit PK, Montrose, CO).
Partial Mass Balance of kb-NB142-70
The % of dose in each tissue or organ was estimated by calculating the exact mean dose of kb-NB142-70 that each mouse received, e.g. 1.827 μmoles for an average 18.41 g mouse. For tissues and organs where we have the exact tissue weight the % dose in that tissue at each time point was calculated accordingly: the concentration/g was multiplied by the organ/tissue weight and divided by the dose administered and multiplied by 100%. For tissues or organs for which we did not have total weight, but only the weight that we collected at euthanasia, the % dose in that organ was calculated based on Table 21 in a “Tissue Mass Balance in Physiological Parameter Values for PBPK Models” A Report Prepared by the International Life Science Institute Risk Science Institute under a Cooperative Agreement with the U.S. Environmental Protection Agency, Office of Health and Environmental Assessment, Office of Health and Environmental Assessment, December 1994 [40]. For example, muscle represents 38.4% of body weight for a mouse. Tissues that were not measured, such as: skin, GI tract and contents, bone, and rest of the body, can account for approximately 39.6% of body weight and we do not know the concentrations of kb-NB142-70 or metabolites in these tissues or organs.
Plasma protein binding
Plasma protein binding of kb-NB142-70 was conducted using the Rapid Equilibrium Dialysis (RED) Device (Pierce Biotechnology, Inc., Rockford, IL) since kb-NB142-70 bound to the filter from Amicon Centrifree® Ultrafiltration Devices (Millipore, Billerica, MA). kb-NB142-70 at concentrations of 0.3 and 3 μg/mL were added to separate 500 μL samples of plasma or phosphate buffered saline (PBS containing 100 mM sodium phosphate and 150 mM sodium chloride) and 500 μL of each sample was added into the sample chamber and dialyzed against 750 μL of phosphate buffered saline for 4 h at 37 °C with shaking following the manufacturer’s instructions. Complete contents of each chamber were removed and processed as described above for plasma samples. Samples were analyzed by HPLC.
Plasma protein binding of kb-NB165-09 was conducted using Amicon Centrifree® Ultrafiltration Devices (Millipore, Billerica, MA) according to the manufacturer’s instructions.
Metabolite analysis by LC-MS/MS
Metabolic identifications were performed on plasma samples obtained 15 min after iv dosing of mice, and on urine samples collected 0-6 h after dosing (see above for details). Control murine samples were analyzed as a negative control for endogenous signals.
Sample preparation (200 μL) consisted of protein precipitation with 1 mL of acetonitrile. The clear supernatant was evaporated to dryness under a gentle stream of nitrogen. The dried residue was reconstituted in 100 μL of 10% acetonitrile. A sample volume of 30 μL was injected onto the LC system.
The HPLC system consisted of an Agilent (Palo Alto, CA) 1200 autosampler and binary pump, a Phenomenex (Torrance, CA, USA) Luna C18 (5 mm, 100 × 4.6 mm) column kept at ambient temperature, and a gradient mobile phase. Mobile-phase solvent A consisted of 0.1% (v/v) formic acid in water, and mobile-phase solvent B consisted of 0.1% (v/v) formic acid in acetonitrile. The initial mobile phase composition was 10% solvent A and 90% solvent B pumped at a flow rate of 1.0 mL/min. From 0 to 15 min, solvent A was increased linearly from 10% to 50%. From 15 to 16 min, the flow was increased to 2.0 mL/min, and the gradient was increased to 90% A. These conditions were maintained until 19 min. From 19 to 20 min, the flow rate was maintained at 2.0 mL/min and the gradient was decreased to 10% A. These conditions were maintained until 25 min, after which the next sample was injected.
HPLC eluent was split with an ASI (El Sobronte, CA, USA) model 620-P010 flow splitter with 90% of the eluent flowing through the UV detector and 10% of the eluent being introduced into the mass spectrometer.
The UV detector was an Agilent 1200 DAD series G1315C diode array detector operated in spectral mode between 190 and 400 nm, to allow comparison of the peaks identified by the LC-MS/MS system with those identified by the HPLC-UV system.
Mass spectrometric detection was carried out using an ABI 4000 QTRAP (MDSSciex, Concord, ON, Canada) mass spectrometer with electrospray ionization in positive mode. The settings of the mass spectrometer were as follows: CEM, 2500 V; GS1, 40 L/h; GS2, 30 L/h; CAD, high; CUR, 20 L/h; IS, 5500 V; DP, 40 V; CES, 10 V; CE, 40 V. The mass spectrometer was operated in the enhanced product ion (EPI) mode, with CE operated at 30, 40, and 50 V. The HPLC system and mass spectrometer were controlled by Analyst software (version 1.4.2), and data were collected with the same software.
Based on the structure of the parent compounds, we screened for masses corresponding to expected metabolic transformations, such as hydroxylation, glucuronidation, etc.
β-glucuronidase treatment of urine
Sample of 0-6 h urine (200 μL) from mice treated with 25 mg/kg kb-NB142-70 or kb-NB165-09 were incubated for 24 h with 10,000 units of β-glucuronidase in a shaking water bath at 37 °C. Samples were then collected, extracted and processed as described above for HPLC-UV analysis.
Results
In vitro cytotoxicity
The IC50 values of kb-NB142-70 and kb-NB165-09 were 21 ± 6 μM and 24 ± 7 μM against PC-3, respectively; 34 ± 7 μM and 28 ± 1 μM against PANC-1, respectively, and 5.2 ± 0.1 μM and 13.4 ± 1.7 μM against CFPAC-1, respectively (supplementary Fig 1).
Efficacy
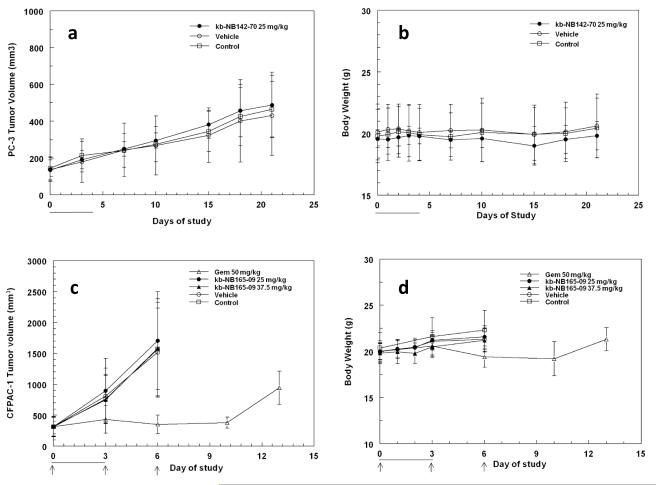
kb-NB142-70 administered qdx5 iv at 25 mg/kg/day to mice bearing PC-3 xenografts had no effect on the growth of the tumor xenografts and did not result in any toxicity or changes in the body weights of the mice. Similarly, kb-NB165-09, administered qdx4 iv at 37.5 and 25 mg/kg/day to mice bearing CFPAC-1 xenografts, had no effect on tumor growth or body weights of the mice. In contrast, the positive control, gemcitabine, administered 50 mg/kg q3dx3 to mice bearing CFPAC-1 xenografts resulted in a significant decrease in CFPAC-1 mean tumor volume on day 6 (T/C, 335 mm3/1520 mm3 = 22%) and an increase in tumor doubling time from 2.2 to 10.4 days (Fig 1).
Fig. 1.
Efficacy studies of kb-NB142-70 and kb-NB165-09 in mice bearing PC-3 or CFPAC-1 xenografts, repectively. (a) Mean tumor volume ± std and (b) body weight of mice dosed 25 mg/kg kb-NB142-70 iv qdx5. (c) Mean tumor volume ± std and (d) body weight of mice dosed either 37.5 or 25 mg/kg kb-NB165-09 iv qdx4. Lines indicate days of dosing for kb-NB142-70 and kb-NB165-09; arrows indicate days of dosing for gemcitabine.
Pharmacodynamic response
When PC-3 or Panc-1 tumors from the pharmacokinetic studies were analyzed for expression of protein kinase D 1, D2, and D3 using Western blot analysis, there were no consistent changes in expression of these proteins and their phosphorylated forms were not detected (data not shown). These data are in agreement with the lack of inhibition of tumor growth in the PC-3 efficacy study. Further, there were no consistent changes in protein kinase D expression in liver or kidney samples of the mice treated with either compound. This could be due to the heterogeneity of tissue samples collected and/or the low immunoreactivity of the phospho-specific antibodies.
Pharmacokinetics
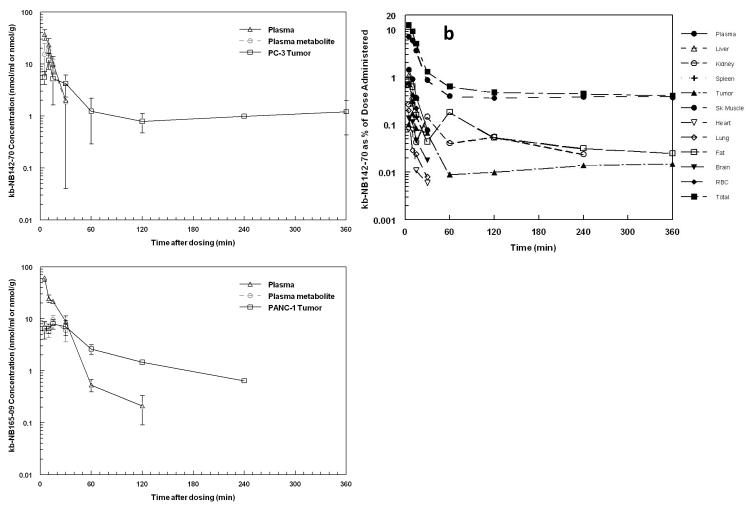
HPLC chromatograms of kb-NB142-70 and kb-NB165-09 are shown in Figure 2. The retention times of kb-NB142-70 and kb-NB165-09 were 6 and 8.5 min, respectively. Plasma kb-NB142-70 Cmax of 37 ± 8 nmol/mL was observed at 5 min after intravenous administration of 25 mg/kg to mice bearing PC-3 xenografts. Plasma disappearance of kb-NB142-70 was rapid and accompanied by the appearance of a metabolite (Fig. 3a). Neither kb-NB142-70 nor its metabolite was detectable in plasma beyond 30 min. The plasma half-life of kb-NB142-70 was 6 min, clearance was 243 mL/min/kg and the volume of distribution for kb-NB142-70 was approximately 2744 mL/kg. Highest tissue Cmax of 63 and 27 nmol/g were observed in kidney and lung, respectively, and occurred at the earliest time point sampled, 5 min (Table 1a). Tissue concentrations of kb-NB142-70 were detectable for a longer period than plasma concentrations with fat concentrations detectable out to the last time point sampled (24 h after dosing). kb-NB142-70 appeared to cross the blood brain barrier, although brain concentrations were lower than plasma concentrations. PC-3 xenograft Cmax was 12 nmol/g of kb-NB142-70 and was observed at 10 min (Fig. 3a, Table 1a). Because tumor concentrations were detectable longer than plasma concentrations, the AUC for tumor was 731 nmol min mg−1 compared to the AUC for plasma, 409 nmol min mL−1. The largest AUC was in fat at 1681 nmol min g−1. Plasma kb-NB165-09 Cmax of 62 nmol/mL was observed at 5 min after iv dosing (Fig. 3c, Table 1b). The plasma half-life of kb-NB165-09 was 14 min, clearance was 121 mL/min/kg and the volume of distribution was approximately 1943 mL/kg. The highest tissue Cmax, 95 and 43 nmol/g were observed in kidney and liver, respectively, and occurred at the earliest time point sampled, 5 min (Table 1b). A Cmax of 8 nmol/g of kb-NB165-09 in PANC-1 xenografts was observed at 15 min (Table 1b). After po dosing of kb-NB165-09, the compound was poorly bioavailable at 1.7%. Tissue Cmax of kb-NB165-09 after iv administration were higher than tissue Cmax of kb-NB142-70, likely reflective of the higher lipophilicity.
Fig. 3.
(a) Mean plasma concentration, plasma metabolite concentration (expressed as nmol/ml equivalent of parent) and tumor concentration of kb-NB142-70 versus time in C.B-17 SCID mice bearing PC-3 xenografts treated iv with 25 mg/kg kb-NB142-70 (b) mass balance of kb-NB142-70 expressed as the % of dose in each tissue or organ (c) Mean plasma concentration, plasma metabolite concentration (expressed as nmol/mL equivalent of parent) and tumor concentration of kb-NB165-09 versus time in C.B-17 SCID mice bearing PANC-1 xenografts treated iv with 25 mg/kg kb-NB165-09.
Table 1 (a).
Concentrations and AUCs of kb-NB142-70 in plasma and tissues after administration of 25 mg/kg kb-NB142-70 iv to C.B-17 SCID female mice bearing PC-3 xenografts
| Time (Min) |
Plasma (nmol/mL) |
Tumor (nmol/g) |
Liver (nmol/g) |
Kidney (nmol/g) |
Lung (nmol/g) |
Spleen (nmol/g) |
Heart (nmol/g) |
Brain (nmol/g) |
Skeletal Muscle (nmol/g) |
RBC (nmol/mL) |
Fat (nmol/g) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 36.9±8.3 | 5.62±1.63 | 23.4±1.1 | 63.4±13.7 | 26.8±3.3 | 21.2±3.8 | 14.5±2.0 | 6.34±1.72 | 18.4±4.6 | 23.6±1.7 | 9.92±0.52 |
| 10 | 23.4±7.4 | 11.8±4.0 | 6.41±0.56 | 48.9±15.8 | 16.7±1.2 | 12.6±2.5 | 13.1±2.5 | 5.07±0.57 | 15.5±4.0 | 13.2±2.7 | 8.93±1.71 |
| 15 | 10.3±3.7 | 5.22±3.60 | 8.41±0.29 | 27.0±12.4 | 10.4±4.9 | 8.03±3.39 | 6.23±1.96 | 2.40±0.40 | 9.45±2.47 | 6.94±2.26 | 6.65±0.74 |
| 30 | 2.0±1.9 | 4.22±1.92 | ND | 10.1±2.0 | 2.95±1.69 | ND | 2.60±0.53 | 0.84±0.11 | 2.28±0.56 | 2.31±1.70 | 2.80±0.45 |
| 60 | ND | 1.24±0.95 | ND | 2.87±0.56 | ND | ND | ND | ND | 1.05±0.02 | ND | 2.62±1.35 |
| 120 | ND | 0.79±0.32 | ND | 3.76±0.82 | ND | ND | ND | ND | 0.95±0.01 | ND | 0.77±0.13 |
| 240 | ND | 0.57±0.60 | ND | 1.78±0.29 | ND | ND | ND | ND | 1.00±0.01 | ND | 0.45±0.06 |
| 360 | ND | 1.21±0.78 | ND | ND | ND | ND | ND | ND | 0.97±0.06 | ND | 0.36±0.03 |
| 960 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0.85±0.42 |
| 1440 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0.44±0.07 | |
| AUC0→∞ (nmol·min/mL) or (nmol·min/g) |
409 | 731 | 266 | 1712 | 377 | 288 | 236 | 85 | 748 | 288 | 1681 |
Table 1 (b).
Concentrations and AUCs of kb-NB165-09 in plasma and tissues after administration of 25 mg/kg kb-NB165-09 iv to C.B-17 SCID female mice bearing PANC-1 xenografts
| Time (Min) |
Plasma (nmol/mL) |
Tumor (nmol/g) |
Liver (nmol/g) |
Kidney (nmol/g) |
|---|---|---|---|---|
| 5 | 61.9±9.4 | 6.46±2.39 | 42.6±6.4 | 94.8±1.3 |
| 10 | 24.8±4.2 | 6.52±1.31 | 25.7±3.6 | 56.4±8.0 |
| 15 | 21.9±1.8 | 8.01±1.72 | 22.6±3.6 | 30.5±1.5 |
| 30 | 8.94±2.45 | 7.00±2.28 | 11.3±0.8 | 22.4±6.1 |
| 60 | 0.54±0.14 | 2.61±0.54 | 2.77±0.14 | 5.03±0.15 |
| 120 | 0.21±0.12 | 1.45±0.04 | 0.20 | ND |
| 240 | ND | 0.64±0.02 | ND | ND |
| AUC0→∞ (nmol·min/mL) or (nmol·min/g) |
782 | 604 | 854 | 1724 |
Urinary and fecal excretion
kb-NB142-70 0-6 h urinary excretion accounted for only 2.5% of dose and only 0.1% was excreted in 6-24 h urine. Fecal excretion accounted for only 0.67% of the dose. Assuming equivalent UV absorption, its metabolite accounted for approximately 7.6% and 1.2% of the dose excreted in the 0-6 h and 6-24 h urine samples, respectively. Only 0.6% of kb-NB165-09 could be accounted for in the 0-6 h urine and an additional 0.9% of the dose was excreted between 6 and 24 h. Metabolites were detected, but not quantified. Fecal excretion represented less that 0.17% of the administered dose of kb-NB165-09.
Partial mass balance of kb-NB142-70
The total amount of the administered dose that was detectable in plasma and collected tissues at the earliest time point sampled, 5 min, was 13.6% of the dose; 12.5% as the parent compound kb-NB142-70 (the breakdown of % dose of kb-NB142-70 per tissue is shown in Fig. 3b). Several of the metabolites that are detected by LC-MS/MS in the plasma at 15 min (Table 2) are not detectable by HPLC-UV and therefore are not accounted for in this partial mass balance. Muscle represents 38.4% of body weight for a mouse and as is observed in Fig 3b, contains the highest percent dose between 60 and 360 min. Tissues and organs not measured consisted of skin, bone and GI tract as well as the remaining carcass and these organs can account for as much as 40% of the body weight of the mouse.
Table 2.
Metabolites of kb-NB142-70 and its methoxy analogue kb-NB165-09 characterized by LC-MS/MS in urine (U) and plasma (P).
| Peak | NB165-09 | NB142-70 | LC-MS/MS (min) |
[M+H]+ (m/z) |
Product ions | Postulated designation |
|---|---|---|---|---|---|---|
| MS1 | U | U | 5.5 | 268 | 251, 226, 223, 222, 208, 193, 189, 182, 177, 162, 153, 150, 148, 137, 121 |
kb-NB142-70 -sulfoxide-(O) OR -aromatic OH |
| MS2 | U | U, P | 5.8 | 428 | 252, 248, 234, 209, 207, 189, 181, 163, 137 |
kb-NB142-70 -Gluc |
| MS3 | - | U | 6.3 | 268 | 217, 207, 189, 181, 177, 163, 150, 145, 137, 121 |
kb-NB142-70 -alkyl-OH |
| MS4 | U | - | 7.6 | 298 | 280, 263, 238. 235, 234, 220, 209, 207, 203, 192, 177, 166, 165, 160, 151, 150, 121 |
kb-NB165-09 -alkyl-OH -[O] |
| MS5 | U | - | 7.8 | 445 | 322, 296, 282, 277, 251, 239, 237, 225, 211, 205, 195, 177, 168 |
? |
| MS6 | U, P | - | 8.2 | 282 | 265, 240, 222, 203, 196, 191, 177, 164, 153, 134, 121 |
kb-NB165-09 -sulfoxide-(O) OR -aromatic-OH |
| MS7 | - | U | 8.7 | 428 | 252, 248, 234, 209, 207, 189, 181, 163, 137 |
kb-NB142-70 -Gluc |
| MS8 | U, P | - | 9.0 | 282 | 264, 231, 221, 206, 191, 190, 188, 178, 177, 164, 160, 134, 121 |
kb-NB165-09 -alkyl-OH |
| MS9 | U | - | 9.4 | 545 | 298, 248, 231, 204, 171, 153, 143, 135, 107 |
? |
| MS10 | U | U, P | 9.5 | 252 | 503 (di-mer), 234, 209, 207, 189, 181, 163, 153, 145, 137, 109 |
kb-NB142-70 |
| MS11 | U | - | 9.6 | 298 | 280, 256, 253, 238, 221, 204, 196, 189, 161, 148, 140, 115 |
kb-NB165-09 -alkyl-OH -[O] |
| MS12 | U | - | 12.9 | 298 | 253, 237, 235, 223, 209, 207, 195, 192, 177, 165, 164, 163, 150, 135, 121, 108 |
kb-NB165-09 -sulfoxy-(O)2 |
| MS13 | U | U | 13.5-14.5 11.2-13.5 |
583/503/332 | 252, 234, 209, 207, 189, 181, 179, 163, 153, 137 |
kb-NB142-70 multimer? |
| MS14 | U, P | - | 14.8 | 266 | 248, 223, 221, 206, 195, 190, 178, 177, 165, 152, 151, 134, 121, 108 |
kb-NB165-09 |
| MS15 | U | - | 15.8 | 312 | 237, 222, 209, 204, 194, 193, 179, 177, 176, 165, 163, 150, 147, 135, 134, 121 |
kb-NB165-09 -alkyl =O -sulfoxy-(O)2 |
Gluc, glucuronide.
Plasma protein binding
kb-NB142-70 was 78% plasma protein bound at both 0.3 and 3 μg/mL. kb-NB165-09 was 88% and 94% bound to plasma protein at 0.3 and 3 μg/mL, respectively.
LC-MS/MS analysis of metabolites
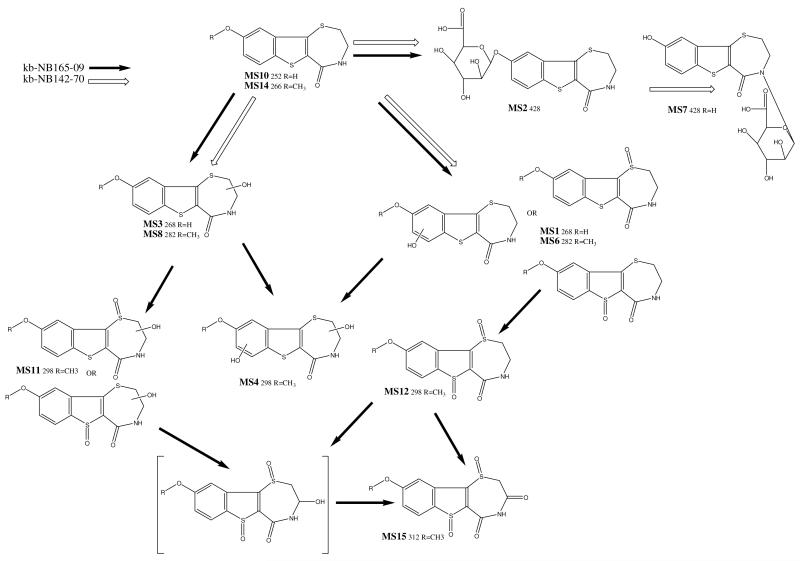
The short plasma half-lives of kb-NB142-70 and kb-NB165-09 prompted us to examine plasma and urine for potential metabolites that were not detectable with our HPLC-UV assay. The m/z of 252 and 428 were detected in the LC-MS ESI positive spectrum chromatogram of plasma obtained 15 min after dosing (Table 2). The m/z 252 presents the parent compound, kb-NB142-70. The m/z 428 was consistent with a glucuronide conjugate (Fig. 4, Table 2). More metabolites were detected in the 0-6 h urine of kb-NB165-09 in the LC-MS ESI positive spectrum chromatogram (Table 2). Based on the masses observed, we propose the kb-NB142-70 and kb-NB165-09 metabolic scheme as depicted in Figure 4. MS1 is a hydroxy analogue of kb-NB142-70 and was observed as metabolic product of both kb-NB142-70 and kb-NB165-09. The hydroxyl moiety is likely positioned on the aromatic end of the molecule or on the sulfur atom. The analogous hydroxy kb-NB165-09 was observed as a metabolic product of kb-NB165-09 as MS6.
Fig. 4.
Metabolic scheme of the postulated metabolites of kb-NB142-70 (open arrows) and its methoxy analogue kb-NB165-09 (solid arrows).
MS2 is a glucuronide analogue of kb-NB142-70 and was observed as metabolic products of both kb-NB142-70 and kb-NB165-09.
MS3 is a hydroxy analogue of kb-NB142-70 and was observed as metabolic product of kb-NB142-70, but not kb-NB165-09. The hydroxyl moiety is likely positioned on one of the two aliphatic carbons as suggested by fragment m/z 207, also observed as a product ion of kb-NB142-70 and represents kb-NB142-70 after loss of CO and NH3. The analogous hydroxy kb-NB165-09 was observed as a metabolic product of kb-NB165-09 as MS8.
MS4 is a di-hydroxy analogue of kb-NB165-09 and observed as a metabolic product of kb-NB165-09. One of the hydroxyl moieties is likely positioned on the aliphatic end of the molecule as suggested by the fragment m/z 280 (elimination of H2O). MS4 is very similar to MS11, apart from the retention time. Because of a phenolic oxide is more polar than a sulfoxide, MS4 likely has a phenolic moiety, whereas MS11 has a sulfoxide. The analogous di-hydroxy kb-NB142-70 was not observed as a metabolic product of kb-NB142-70.
MS5 has an m/z ratio of 445 and was observed as a metabolic product of kb-NB165-09. kb-NB142-70 did not yield this metabolite, nor an analogue 14 mass units lower. The odd m/z ratio suggests an even number of nitrogens as opposed to the single nitrogen in the parent compound. No structure could be postulated.
MS6 is a hydroxy analogue of kb-NB165-09 and was observed as metabolic product of kb-NB165-09. The hydroxyl moiety is likely positioned on the aromatic end of the molecule or on the sulfur atom. The corresponding kb-NB142-70 analogue MS1, 14 mass units lower, was observed as a metabolic product of kb-NB142-70.
MS7 is a glucuronide analogue of kb-NB142-70 and was observed as a metabolic product only of kb-NB142-70.
MS8 is a hydroxy analogue of kb-NB165-09 and was observed as metabolic product of kb-NB165-09. The hydroxyl moiety is likely positioned on one of the two aliphatic carbons as suggested by fragments m/z 264 (elimination of H2O) and m/z 221, also observed as a product ion of kb-NB165-09 and represents kb-NB165-09 after loss of CO and NH3. The corresponding kb-NB142-70 analogue MS3, 14 mass units lower, was observed as a metabolic product of kb-NB142-70.
MS9 has an m/z ratio of 545 and was observed as a metabolic product of kb-NB165-09. kb-NB142-70 did not yield this metabolite, nor an analogue 14 mass units lower. The product ion spectrum of MS9 includes a high m/z ratio of 298, which corresponds to several di-oxygenated metabolites of kb-NB165-09. However, the product ion spectrum of MS9 does not agree with the spectrum of any of the metabolites with m/z 298.
MS10 is kb-NB142-70 and was observed in urine and plasma after administration of kb-NB142-70, but only a minute signal could be observed in urine as a metabolic product of kb-NB165-09.
MS11 is a di-hydroxy analogue of kb-NB165-09 and observed as a metabolic product of kb-NB165-09. One of the hydroxyl moieties is likely positioned on the aliphatic end of the molecule as suggested by the fragment m/z 280 (elimination of H2O). The analogous di-hydroxy kb-NB142-70 was not observed as a metabolic product of kb-NB142-70.
MS12 is a di-oxy analogue of kb-NB165-09 and observed as a metabolic product of kb-NB165-09. The absence of a mass at m/z 280 (elimination of H2O) suggests there is no aliphatic hydroxylation. The presence of m/z 108, 121, and 134 suggests an unsubstituted methoxy phenyl ring. Together, this suggests oxidation of the sulfur atom(s). The analogous di-hydroxy kb-NB142-70 was not observed as a metabolic product of kb-NB142-70.
MS13 eluted as a broad peak and appears to be a metabolic product of both kb-NB142-70 and kb-NB165-09. Masses at 583, 503 (kb-NB142-70 dimer), and 332 all fragmented to the identical set of product ions. The late retention time and high mass may suggest a multimer product of kb-NB142-70.
MS14 is kb-NB165-09 and was observed in urine and plasma after administration of kb-NB165-09.
MS15 has an m/z ratio of 312 and was observed as a metabolic product of kb-NB165-09. kb-NB142-70 did not yield this metabolite, nor an analogue 14 mass units lower. The mass is in line with that of a tri-oxygenated metabolic product of kb-NB165-09.
Based on the masses observed, we propose the kb-NB142-70and kb-NB165-09 metabolic scheme as depicted in Fig. 4. The introduction of the methoxy group appears to divert metabolism from glucuronidation to primarily oxidation.
Treatment of urine containing kb-NB142-70 or kb-NB165-09 with β-glucuronidase
One metabolite was readily detected in the HPLC-UV assay of plasma and urine of mice treated with kb-NB142-70 or kb-NB165-09. Based on the parent structures, we speculated that it represented an O-glucuronide conjugate of kb-NB142-70 or kb-NB165-09. Therefore, an aliquot of 0-6 h urine from mice treated with kb-NB142-70 or kb-NB165-09 was incubated at 37 °C for 24 h with 10,000 units of β-glucuronidase and compared with an aliquot of the same sample incubated in the absence of β-glucuronidase. Incubation with β-glucuronidase resulted in disappearance of the metabolite and an increase of ~410% in kb-NB142-70, suggesting that the metabolite was indeed the O-glucuronide conjugate of kb-NB142-70 (Supplemental Fig 2). When urine from mice dosed with kb-NB165-09 was incubated with β-glucuronidase there was neither a decrease in the metabolite peak nor an increase in the parental peak indicating that the metabolite was not a glucoronide conjugate.
Discussion
The PKD family is emerging as a novel target for small molecule inhibitors with potential as therapeutics for multiple cancers including pancreas, skin and prostate [41]. The PKD family of serine-threonine kinases consists of three members, PKD1 (mouse PKD or human PKCμ), PKD2 and PKD3 (human PKCν), that mediate important signaling pathways regulating protein trafficking, epigenetic gene expression, oxidative stress signaling, cell proliferation and tumor cell invasion. Aberrant PKD activity and expression have been demonstrated in tumor cell lines and tissues from the pancreas [42], skin [1,43] and prostate [28], with the role in pancreatic cancer especially compelling. In PANC-1 cells treated with neurotensin, a G-protein coupled receptor agonist, PKD has been shown to stimulated Hsp27 Ser-82 phosphorylation separate of the p38/MAPK pathway and overexpression of PKD1 enhanced DNA synthesis induced by neurotensin. It is known that Hsp27 is markedly increased in many cancer cells and its phosphorylation contributes to the malignant properties, including increased tumorgenicity and treatment resistance [23, 44-45]. CRT0066101, a compound that targets the PKD pathway, was shown to be effective in vitro and slowed the growth of a PANC-1 pancreatic cancer xenograft when dosed po at 80 mg/kg qd × 21 [31]. In prostate cancer cell lines, PKD1 has been shown to protect LNCaP prostate cancer cells from phorbol ester-induced apoptosis by promoting ERK1/2 and NF-κB activities [45], while PKD3 plays an important role in the growth and survival of prostate cancer cells [28]. PKD 1 appears to interact in a transcriptional complex with the androgen receptor to support androgen-mediated cell proliferation in both androgen dependent and androgen-independent prostate cancer cell lines [27]. Although some controversies exist with regard to the role PKD1 in certain tumor cells, the PKD inhibitors that have been reported so far exhibit potent anti-tumor activities in diverse tumor cell lines. In prostate cancer cells, Sharlow et al. demonstrated that a small molecule inhibitor of PKD, CID755673, inhibited prostate cancer cell proliferation, migration and invasion, and appeared to be a highly selective non-ATP-competitive inhibitor of PKD1[33]. More recently, LaValle et al. demonstrated the analogues, kb-NB142-70 and kb-NB165-09, were more potent than CID755673 in inhibiting PKD1, PKD2 and PKD3 using radiometric kinase assays. The EC50s of kb-NB142-70 and kb-NB165-09 against PC-3 human androgen-independent prostate cancer cells were 8 and 49 μM, respectively [34].
Data supporting the development of small molecule inhibitors targeting PKD in pancreatic cancer is compelling. The two compounds evaluated here, kb-NB142-70 and kb-NB165-09, are analogues of CID755673 and were more potent than CID755673 in inhibiting PKD1 [34]. Although these compounds were effective against PKD1 in vitro, and had IC50’s in the low to mid μM range against PC-3, PANC-1 and CFPAC-1 cell lines, they were not active at the maximum soluble dose when administered to mice bearing subcutaneous xenografts; and no consistent effects on protein kinase D protein expression were observed by Western blot analysis in the tissue samples of tumor, liver and kidney collected as part of the pharmacokinetic studies. Based on the pharmacokinetic and metabolism data presented in this study, the high plasma protein binding and rapid metabolism of kb-NB142-70 and kb-NB165-09 limits their potential as anticancer therapeutics. Unlike CRT0066101, kb-NB165-09 was not bioavailable when given orally to mice as a single dose. Future studies will be directed at structural modifications to develop PKD inhibitors with better pharmacokinetic and metabolic characteristics. Newer analogs based on the benzothienothiazepinone structures have been synthesized and analogs such as kmg-NB4-23 with a pyrimidine ring appear to have good in vitro activity [35, 36] and should be more metabolically stable. Unfortunately this analog has poor aqueous solubility. Thus, alternative formulation and/or further structural modification will be required to enable the further evaluation of this analog in vivo.
Supplementary Material
Acknowledgments
This study was supported in part by U.S. Public Health Service National Institutes of Health grant CA78039, CA129127 and CA142580. This project used the UPCI Clinical Pharmacology Analytical Facility (CPAF) and was supported in part by award P01CA078039. We acknowledge Diane Mazzei and other members of DLAR for the excellent care of the animals used in these studies. We also acknowledge the UPCI Writing Group for their helpful comments.
References
- 1.Rennecke PA, Rehberger G, Fürstenberger FJ, Johannes M, Stöhr F, Marks, Richter KH. Protein-kinase-Cmu expression correlates with enhanced keratinocyte proliferation in normal and neoplastic mouse epidermis and in cell culture. Int. J. Cancer. 1999;80:98–103. doi: 10.1002/(sici)1097-0215(19990105)80:1<98::aid-ijc19>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 2.Zhukova E, Sinnett-Smith J, Rozengurt E. Protein Kinase D Potentiates DNA Synthesis and Cell Proliferation Induced by Bombesin, Vasopressin, or Phorbol Esters in Swiss 3T3 Cells. J. Biol. Chem. 2001;276:40298–40305. doi: 10.1074/jbc.M106512200. [DOI] [PubMed] [Google Scholar]
- 3.Johannes FJ, Horn J, Link G, Haas E, Siemienski K, Wajant H, Pfizenmaier K. Protein kinase Cmu downregulation of tumor-necrosis-factor-induced apoptosis correlates with enhanced expression of nuclear-factor-kappaB-dependent protective genes. Eur. J. Biochem. 1998;257:47–54. doi: 10.1046/j.1432-1327.1998.2570047.x. [DOI] [PubMed] [Google Scholar]
- 4.Endo K, Oki E, Biedermann V, Kojima H, Yoshida K, Johannes FJ, Kufe D, Datta R. Proteolytic cleavage and activation of protein kinase C [micro] by caspase-3 in the apoptotic response of cells to 1-beta - D-arabinofuranosylcytosine and other genotoxic agents. J. Biol. Chem. 2000;275:18476–18481. doi: 10.1074/jbc.M002266200. [DOI] [PubMed] [Google Scholar]
- 5.Brändlin I, Hübner S, Eiseler T, Martinez-Moya M, Horschinek A, Hausser A, Link G, Rupp S, Storz P, Pfizenmaier K, Johannes FJ. Protein kinase C (PKC)eta-mediated PKC mu activation modulates ERK and JNK signal pathways. J. Biol. Chem. 2002;277:6490–6496. doi: 10.1074/jbc.M106083200. [DOI] [PubMed] [Google Scholar]
- 6.Rey O, Young SH, Cantrell D, Rozengurt E. Rapid Protein Kinase D Translocation in Response to G Protein-coupled Receptor Activation. J. Biol. Chem. 2001;276:32616–32626. doi: 10.1074/jbc.M101649200. [DOI] [PubMed] [Google Scholar]
- 7.Paolucci L, Rozengurt E. Protein Kinase D in Small Cell Lung Cancer Cells: Rapid Activation through Protein Kinase C. Cancer Research. 1999;59:572–577. [PubMed] [Google Scholar]
- 8.Sidorenko SP, Law CL, Klaus SJ, Chandran KA, Takata M, Kurosaki T, Clark EA. Protein kinase C mu (PKC mu) associates with the B cell antigen receptor complex and regulates lymphocyte signaling. Immunity. 1996;5:353–363. doi: 10.1016/s1074-7613(00)80261-7. [DOI] [PubMed] [Google Scholar]
- 9.Rozengurt E, Sinnett-Smith J, Van Lint J, Valverde AM. Protein kinase D: a novel target for diacylglycerol and phorbol esters. Mutat Res. 1995;326:545–551. doi: 10.1016/0027-5107(95)00141-7. [DOI] [PubMed] [Google Scholar]
- 10.Haworth RS, Sinnett-Smith J, Rozengurt E, Avkiran M. Protein kinase D inhibits plasma membrane Na+/H+exchanger activity. Am. J. Physiol. 1999;277:C1202–C1209. doi: 10.1152/ajpcell.1999.277.6.C1202. [DOI] [PubMed] [Google Scholar]
- 11.Rykx A, Kimpe LD, Mikhalap S, Vantus T, Seufferlein T, Vandenheede JR, Lint JV. Protein kinase D: a family affair. FEBS Lett. 2003;546:81–86. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- 12.Eiseler T, Döppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11(1):R13. doi: 10.1186/bcr2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papazyan R, Rozengurt E, Rey O. The C-terminal tail of protein kinase D2 and protein kinase D3 regulates their intracellular distribution. Biochem Biophys Res Commun. 2006;342(3):685–689. doi: 10.1016/j.bbrc.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Lu G, Chen J, Espinoza LA, Garfield S, Toshiyuki S, Akiko H, Huppler A, Wang QJ. Protein kinase D 3 is localized in vesicular structures and interacts with vesicle-associated membrane protein 2. Cell Signal. 2007;19(4):867–79. doi: 10.1016/j.cellsig.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Marklund U, Lightfoot K, Cantrell D. Intracellular location and cell context-dependent function of protein kinase D. Immunity. 2003;19(4):491–501. doi: 10.1016/s1074-7613(03)00260-7. [DOI] [PubMed] [Google Scholar]
- 16.Hassan S, Biswas MH, Zhang C, Du C, Balaji KC. Heat shock protein 27 mediates repression of androgen receptor function by protein kinase D1 in prostate cancer cells. Oncogene. 2009;28(49):4386–96. doi: 10.1038/onc.2009.291. [DOI] [PubMed] [Google Scholar]
- 17.Matthews SA, Navarro MN, Sinclair LV, Emslie E, Feijoo-Carnero C, Cantrell DA. Unique functions for protein kinase D1 and protein kinase D2 in mammalian cells. Biochem J. 2010;432(1):153–63. doi: 10.1042/BJ20101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American Cancer Society . Cancer Facts and Figures 2009. American Cancer Society; Atlanta, Ga: [Last accessed May 7, 2009]. 2009. Also available online. [Google Scholar]
- 19.Amadesi S, Grant AD, Cottrell GS, Vaksman N, Poole DP, Rozengurt E, Bunnett NW. Protein kinase D isoforms are expressed in rat and mouse primary sensory neurons and are activated by agonists of protease-activated receptor 2. J Comp Neurol. 2009;516(2):141–56. doi: 10.1002/cne.22104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen LA, Li J, Silva SR, Jackson LN, Zhou Y, Watanabe H, Ives KL, Hellmich MR, Evers BM. PKD3 is the predominant protein kinase D isoform in mouse exocrine pancreas and promotes hormone-induced amylase secretion. J Biol Chem. 2009;284(4):2459–71. doi: 10.1074/jbc.M801697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rey O, Yuan J, Rozengurt E. Intracellular redistribution of protein kinase D2 in response to G-protein-coupled receptor agonists. Biochem Biophys Res Commun. 2003a;302:817–824. doi: 10.1016/s0006-291x(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 22.Rey O, Yuan J, Young SH, Rozengurt E. Protein kinase C nu/protein kinase D3 nuclear localization, catalytic activation, and intracellular redistribution in response to G protein-coupled receptor agonists. J Biol Chem. 2003b;278:23773–23785. doi: 10.1074/jbc.M300226200. [DOI] [PubMed] [Google Scholar]
- 23.Yuan J, Rozengurt E. PKD, PKD2, and p38 MAPK mediate Hsp27 serine-82 phosphorylation induced by neurotensin in pancreatic cancer PANC-1 cells. J Cell Biochem. 2008;103(2):648–62. doi: 10.1002/jcb.21439. [DOI] [PubMed] [Google Scholar]
- 24.Seufferlein T. Novel protein kinases in pancreatic cell growth and cancer. Int J Gastrointest Cancer. 2002;31(1-3):15–21. doi: 10.1385/IJGC:31:1-3:15. [DOI] [PubMed] [Google Scholar]
- 25.Guha S, Rey O, Rozengurt E. Neurotension Induces Protein Kinase C-dependent Protein Kinase D Activation and DNA Synthesis in Human Pancreatic Carcinoma Cell Line PANC-1. Cancer Res. 2002;62:1632–1640. [PubMed] [Google Scholar]
- 26.Du C, Jaggi M, Zhang C, Balaji KC. Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res. 2009;69(3):1117–24. doi: 10.1158/0008-5472.CAN-07-6270. [DOI] [PubMed] [Google Scholar]
- 27.Mak P, Jaggi M, Syed V, Chauhan SC, Hassan S, Biswas H, Balaji KC. Protein kinase D1 (PKD1) influences androgen receptor (AR) function in prostate cancer cells. Biochem Biophys Res Commun. 2008;373(4):618–23. doi: 10.1016/j.bbrc.2008.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68(10):3844–53. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 29.Sturany S, Lint JV, Gilchrist A, Vandenheede JR, Adler G, Seufferlein T. Mechanism of Activation of Protein Kinase D2(PKD2) by the CCKB/Gastrin Receptor. J. Biol. Chem. 2002;277:29431–29436. doi: 10.1074/jbc.M200934200. [DOI] [PubMed] [Google Scholar]
- 30.Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, Balaji KC. E-Cadherin Phosphorylation by Protein Kinase D1/Protein Kinase C{mu} is Associated with Altered Cellular Aggregation and Motility in Prostate Cancer. Cancer Res. 2005;65:483–492. [PubMed] [Google Scholar]
- 31.Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, Kelland L, Jamieson S, Sutherland R, Raynham T, Charles M, Bagherzadeh A, Foxton C, Boakes A, Farooq M, Maru D, Diagaradjane P, Matsuo Y, Sinnett-Smith J, Gelovani J, Krishnan S, Aggarwal BB, Rozengurt E, Ireson CR, Guha S. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol. Cancer Ther. 2010;9:1136–1145. doi: 10.1158/1535-7163.MCT-09-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart JR, Christman KL, O’Brian CA. Effects of resveratrol on the autophosphorylation of phorbol ester-responsive protein kinases: inhibition of protein kinase D but not protein kinase C isozyme autophosphorylation. Biochem Pharmacol. 2000;60(9):1355–9. doi: 10.1016/s0006-2952(00)00450-0. [DOI] [PubMed] [Google Scholar]
- 33.Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, Bravo-Altamirano K, Wipf P, Lazo JS, Wang QJ. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem. 2008;283(48):33516–26. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaValle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, Wipf P, Wang QJ. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5. doi: 10.1186/1472-6769-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bravo-Altamirano K, George KM, Frantz MC, Lavalle CR, Tandon M, Leimgruber S, Sharlow ER, Lazo JS, Wang QJ, Wipf P. Synthesis and structure-activity relationships of benzothienothiazepinone inhibitors of protein kinase D. ACS Med. Chem. Lett. 2011;2(2):154–159. doi: 10.1021/ml100230n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George KM, Frantz M-CL, Bravo-Altamirano K, Lavalle CR, Tandon M, Leimgruber S, Sharlow ER, Lazo JS, Wang QJ, Wipf P. Design, synthesis, and biological evaluation of PKD inhibitors. Pharmaceutics. 2011;3(2):186–228. doi: 10.3390/pharmaceutics3020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Argenio DZ, Schumitzky A. ADAPT, Biomedical Simulation Resource, USC. bmsr.usc.edu/Software/ADAPT/ADAPT.html.
- 38.Yeh KC, Kwan KC. A comparison of numerical integrating algorithms by trapezoidal, Lagrange, and spline approximation. J Pharmacokinet Biopharm. 1978;6:79–98. doi: 10.1007/BF01066064. [DOI] [PubMed] [Google Scholar]
- 39.Rocci ML, Jusko WJ. LAGRAN program for area and moments in pharmacokinetic analysis. Comput. Programs Biomed. 1983;16:203–216. doi: 10.1016/0010-468x(83)90082-x. [DOI] [PubMed] [Google Scholar]
- 40.Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP. Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health. 1997;13:407–84. doi: 10.1177/074823379701300401. [DOI] [PubMed] [Google Scholar]
- 41.LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. Protein kinase D as a potential new target for cancer therapy. Biochim Biophys Acta. 2010;1806:183–92. doi: 10.1016/j.bbcan.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochi N, Tanasanvimon S, Matsuo Y, Tong Z, Sung B, Aggarwal BB, Sinnett-Smith J, Rozengurt E, Guha S. Protein kinase D1 promotes anchorage-independent growth, invasion, and angiogenesis by human pancreatic cancer cells. J Cell Physiol. 2011;226:1074–81. doi: 10.1002/jcp.22421. [DOI] [PubMed] [Google Scholar]
- 43.Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br J Dermatol. 2006;154:586–93. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
- 44.Kisfalvi K, Hurd C, Guha S, Rozengurt E. Induced overexpression of protein kinase D1 stimulates mitogenic signaling in human pancreatic carcinoma PANC-1 cells. J Cell Physiol. 2010;223:309–16. doi: 10.1002/jcp.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Giridhar KV, Zhang L, Xu S, Wang QJ. A protein kinase C/protein kinase D pathway protects LNCaP prostate cancer cells from phorbol ester-induced apoptosis by promoting ERK1/2 and NF-{kappa}B activities. Carcinogenesis. 2011;32:1198–206. doi: 10.1093/carcin/bgr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.