Abstract
Secondary structure-forming DNA motifs have achieved infamy because of their association with a variety of human diseases and cancer. The 3rd FASEB summer conference on dynamic DNA structures focused on the mechanisms responsible for the instabilities inherent to repetitive DNA and presented many exciting and novel aspects related to the metabolism of secondary structures. In addition, the meeting encompassed talks and posters on the dynamic structures that are generated during DNA metabolism including nicked DNA, Holliday junctions and RNA:DNA hybrids. New approaches for analysis and sequencing technologies put forth secondary structures and other DNA intermediates as vital regulators of a variety of cellular processes that contribute to evolution, polymorphisms and diseases.
Keywords: Triplex, G-quartet, hairpin, cruciform, Holliday junction, RNA:DNA hybrid, replication transcription collision
1.0. Introduction
This year is the 50th anniversary of the discovery of the G4 quartet. The 2012 FASEB Summer Research Conference on Dynamic DNA Structures in Biology commemorated this event by dedicating a large fraction of the meeting to the subject. Talks ranged from Martin Gellert’s (NIH) first hand account of how studies of a jelly-like substance formed by guanylic acid led to the identification of G-quadruplexes in 1962 [1] to more recent work demonstrating the functional significance of such structures in vivo. In addition, this meeting, organized by Nancy Maizels (University of Washington) and Sergei Mirkin (Tufts University) at Saxtons River, VT, June 17–22, covered a wide variety of other alternate DNA structures including triplexes, hairpins, cruciforms, RNA:DNA hybrids and Holliday junctions and their roles in the metabolism of prokaryotic and eukaryotic cells. Through a series of talks and poster sessions, the meeting emphasized that unusual DNA secondary structures are widespread in all living organisms where they have profound effects on replication, transcription and genome stability. Some of these effects are positive, affecting normal development and the generation of genetic diversity, whilst other effects are negative and result in a variety of genetic disorders and cancer in humans (Figure 1).
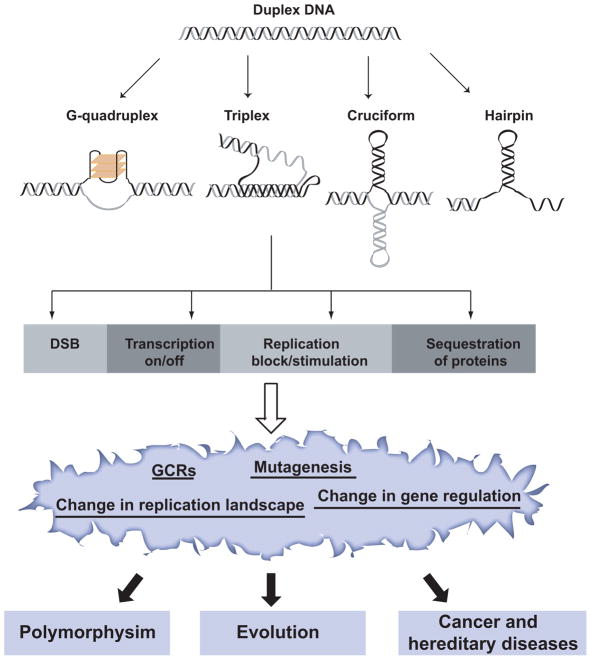
Figure 1.
The role of secondary structure-forming repeats in genome function and integrity. As was evident from the meeting, the formation of secondary structures or the presence of the sequence motif per se can lead to double strand breaks (G-quartets, triplexes, cruciforms and hairpins); induction (G-quartets and GAA/TTC tracts) or inhibition of transcription (G-quartets and triplexes); initiation (triplexes) or stalling of replication (G-quartets, triplexes, cruciforms and hairpins); and sequestration of cellular proteins (G-quartets, triplexes and r(CUG) hairpins). These processes likely contribute to the generation of polymorphisms, to genome evolution and to a variety of diseases via gross-chromosomal rearrangements, mutagenesis, dysregulation of gene expression and alteration of the replication landscape.
While much has been learnt about the behavior of many of these structures, our understanding of their incidence in the genome is still incomplete. As was highlighted in the keynote address by Jeffrey Strathern (NCI), the inability of some of these DNA motifs to be propagated in E. coli (e.g. long palindromes) and to be amplified or sequenced has resulted in their underrepresentation in whole-genome sequencing analyses of complex genomes including humans. However, as was accentuated in the keynote address and other talks in the meeting, the field is entering an exciting new era where more accurate sequencing technologies and bioinformatics tools for analysis of these sequences are emerging.
Below we briefly describe these topics as well as other new findings discussed at this meeting that contribute to our understanding of the dynamic nature of DNA.
2.0. G-quartets– the birthday boy
G-rich DNA molecules can form inter- or intramolecular hydrogen bonds to form square planar arrays of 4 guanines known as G-quartets. A series of G-quartets results in a quadruplex structure frequently referred to as a G-quadruplex, G-tetraplex or G4-DNA. The guanines in the quadruplex are held by non-Watson-Crick hydrogen bonds, termed Hoogsteen base-pairs. The topology of the quadruplex varies depending on the orientation of the DNA strands involved, the length of the G-rich region and its nucleotide composition. The consensus sequence commonly used in genome-wide bioinformatic studies to identify G-quartets is d(G3+N1–7)4 [2] However, as was pointed out by Jean-Louis Mergny (IECB), experimental studies demonstrate that this pattern is not perfect and yields many false-positives and false-negatives. For instance, it does not predict human mini-satellite CEB25 to be a G4-forming motif although structural analysis proves it to be so [3]. Mergny’s laboratory has addressed this conundrum by looking for G clusters in the genome which have pronounced GC asymmetry. The new and improved algorithm for detection of G-quadruplex forming motifs also analyzes large genomic regions to take into account the observation that G4-DNA might contain interstitial loops and imperfections that still allow formation of stable secondary-structure.
Kyle Miller (Currently at University of Texas at Austin) presented a study carried out in the labs of Shankar Balasubramanian and Steve Jackson (University of Cambridge). He used pyridostatin (PDS), a small molecule that interacts with G-quadruplexes, to identify G4-forming motifs in the human genome. The loci for PDS binding colocalized with binding sites for Pif1, a known G4-DNA-unwinding helicase. The PDS-mediated stabilization of the G-quadruplex causes double strand breaks (DSBs), allowing the G-quadruplex forming region to be identified by using chromatin immunoprecipitation of γH2AX followed by next-generation sequencing. Twenty-five cancer related genes, including several known oncogenes, were identified including SRC. Interestingly, oncogene expression was down-regulated upon treatment with PDS, suggesting the potential for the use of small molecules that interact with secondary structures as therapeutic agents. The phenomenon of oncogene down-regulation by G-quadruplex stabilizing agents is likely due to the fact that G4-DNA can effectively block the transcription machinery. In general, G4-DNA has been shown to be enriched in gene regulatory regions with preponderance for promoter regions in both prokaryotes and eukaryotes (Reviewed in [2, [4]).
A very clear demonstration of the role of G4 DNA was provided by Laty A. Cahoon (Northwestern University, H. Steven Seifert’s laboratory) who showed that transcription across a G-rich region upstream of the pilE locus of Neisseria gonorrhoeae leads to G4-DNA formation. This pilE G4 DNA structure results in nicks that promote recombination and thus pilin antigenic variation that enables this microbe to evade the host’s immune response.
A G-quadruplex can also present an obstacle for the DNA replication machinery and trigger genomic instability. Alain Nicolas and Aurele Piazza, a graduate student from his laboratory (Curie Institute) studied metabolism of the G-quadruplex-forming human minisatellites CEB1 and CEB25 in the yeast Saccharomyces cerevisiae. Interestingly, these 2 minisatellites behave differently in vivo. Unlike CEB25, CEB1 was found to be extremely prone to size variations when the G-rich strand is a template for leading strand synthesis in the absence of the Pif1 helicase. In addition, fragility at CEB1 could be exacerbated by the bisquinolinium G4 ligands Phen-DC3 and Phen-DC6. On the other hand, CEB25 demonstrated higher level of gross-chromosomal rearrangements (GCR) that presumably result from the fortuitous presence of Cdc13-binding sites within the CEB25 motif. Mutations of the G4-forming sequences of both mini-satellites revealed that structural parameters of G4 determine their efficacy as replication impediments and destabilizing elements.
The CGG repeats responsible for Fragile X Syndrome (FXS) forms G4-DNA structure along with a hairpin containing a mixture of Watson-Crick G•C and Hoogsteen G•G base pairs. These repeats belong to the class of nucleotide repeats that are associated with a growing number of neurological diseases known collectively as the Repeat Expansion Diseases. Expanded CGG repeats can stall DNA replication, down-regulate FMR1 transcription due to repeat-induced gene silencing, and cause chromosome fragility [5, 6]. The mechanism of repeat expansion is still largely unknown and a significant part of this meeting was devoted to related discussions. Using a unique approach that makes it possible to visualize DNA replication by fluorescence microscopy in single DNA molecules, Jeannine Gerhardt (Albert Einstein College of Medicine, Carl Schildkraut’s laboratory) showed that while there are two origins of replication active in human embryonic stem cells (hESCs) from unaffected embryos, only one of these is used in hESCs derived from FXS embryos. She suggested that this could account for the repeat expansion that gives rise to FXS, an event that is thought to be confined to the oocyte or very early embryo.
CGG repeats can also stall the progression of the replication fork in yeast, as was highlighted by Catherine Freudenreich (Tufts University). Fork stalling and fragility at the repeats are increased in an srs2 null mutant, implicating the Srs2 helicase in removal of the barrier imposed by these repeats during DNA replication. Sergei Mirkin (Tufts University) described an exciting new system to study expansions of CGG repeats in mammalian cells. The carrier-size repeats were positioned between the FMR1 promoter and the ORF for a thymidine kinase reporter. Expansions in this unique cassette led to the inactivation of this marker making cells ganciclovir-resistant. Moreover, CGG repeats also led to increased levels of mutagenesis in the body of the reporter gene. This data corroborates the observation that secondary structures in the genome can be a source of mutations, as was described in the meeting by the laboratories of Kirill Lobachev and Sergei Mirkin (see below). Anna Aksenova (Mirkin lab) described a yeast screen utilizing a cyclic peptide library to find compounds that promote expansions of quadruplex-forming telomeric repeats. Two out of 50,000 tested cyclic peptides were found to stimulate expansions of interstitial telomeric repeats. This approach might be useful in developing of alternative therapeutics for the premature aging diseases caused by telomere shortening, such as Werner and Bloom syndromes, ataxia telangiectasia, dyskeratosis congenita and others.
Besides helicases, other DNA repair proteins and chromatin remodelers can influence G4 DNA metabolism [2, 8]. The complexity of interactions likely reflects the differences in structural organization of the quadruplexes, cellular processes that are affected by the secondary structure, the location of G4-forming repeats in the genome, the type and age of the cells carrying repeats. There were many talks on the roles played by checkpoint response, mismatch repair (MMR) and the SWI/SNF family proteins in preventing or promoting repeat instability. Karen Usdin (NIH) described work from her lab on a mouse model for CGG/CCG-repeat expansions in the Fragile X-related disorders. This work demonstrates that expansions and deletions occur via different mechanisms and that expansions are seen in both oocytes and neurons. This, along with her lab’s demonstration that oxidative damage exacerbates expansion, supports the idea that expansions in this mouse model arise from aberrant DNA repair rather than problems arising from the ability of CGG/CCG-repeats to block DNA replication. In addition, her group have found some interesting differences between the Fragile X mouse and other mouse models with respect to the effect of the mismatch repair protein Msh2 on repeat instability that may shed light on the role of this protein in repeat expansion.
MutSα has been shown to bind to G4-DNA in the switch regions of immunoglobulin genes and telomeric repeats. MutS binds G4-DNA with high affinity and the reaction cannot be reversed with addition of ATP [7]. Erik Larson (Illinois State University) showed that the specificity of E.coli MutS for G4-DNA is independent of its activity on normal DNA heteroduplexes. A mutation of F36A in MutS that prevents it from binding to DNA heteroduplexes, had no effect on its binding efficiency to G4-DNA. Through analyses of 1000 human genomes he also found that G-rich sequences are associated with small nucleotide polymorphisms (SNPs) and microsatellite instability. He thus suggested that G4-mediated blockage of mismatch repair may contribute to instability at G-rich loci.
David Clynes (University of Oxford, Richard Gibbons’s laboratory) discussed the human protein ATRX, a member of the snf2 family of chromatin remodeling proteins, that along with its interaction partner DAXX, is implicated in the deposition of the histone variant H3.3 at G-rich regions and tandem repeats. Mutations in ATRX lead to alpha-thalassaemia perhaps via G4-dependent down regulation of the a-globin gene. This downregulation may involve inhibition of DNA replication by G4-structures and DSB formation that affects gene expression. The role of ATRX in G4-DNA metabolism was underscored by the finding that this protein is largely deleted in the U2OS cancer cell line where telomere lengths are maintained via telomerase-independent homologous recombination between telomeric repeats (the Alternative Lengthening of Telomeres (ALT) pathway). Re-introduction of ATRX into U2OS cells leads to suppression of the ALT pathway as evidenced by a decrease in the level of extra-chromosomal telomeric circles. A model is proposed where in ALT cells, without ATRX, stalled replication forks at telomeres trigger this alternative HR pathway.
3.0. Triplex forming repeats-the split personality
Triplex DNA (H-DNA) is formed by homopurine-homopyrimidine sequences that have mirror symmetry. In the secondary structure, a third strand is folded into the major groove of the duplex DNA, an interaction that is stabilized by Hoogsteen-base pairing. The best known triplex structure-forming repeat is the GAA/TTC tract, whose expansion down regulates the frataxin (FXN) gene and causes Friedreich ataxia (FRDA) in humans [8].
Similar to G4-DNA, it has been shown that GAA/TTC repeats can block replication and transcription efficiently. However, this meeting revealed novel, and at first glance paradoxical properties of GAA/TTC repeats to promote replication and transcription. These attributes seem to be important factors that determine the ability of GAA/TTC tracts to expand, to inactivate gene expression, to trigger DSBs and mutations. Cindy Follonier (University of Zurich, Massimo Lopes’ laboratory) presented evidence that these repeats block replication of SV40-based plasmids in human cells, as has previously been seen in yeast. Taking advantage of isolating plasmid intermediates in large quantities and using a combination of 2-dimensional gel electrophoresis analysis and electron microscopy, she also identified frequent fork reversals and homology-driven postreplicative junctions, that are possible mediators of repeat instability. Maria Krasilnikova (Penn State University) also reported a strong replication arrest at the tracts on a plasmid transfected into monkey and human cell lines. However, the arrest only occurs during the first replication cycle when nucleosome occupation on the plasmid is sparse, but not in the following cycles after the DNA becomes chromatinized. One interesting observation was that despite GAA repeat-mediated replication stalling, the repeats also promote origin-independent plasmid replication. Consistently, an enrichment of Orc, DNA primase and Mcm proteins was observed at the repeats. As was highlighted in Kirill Lobachev’s (see below) talk, the association of these proteins with the repeats may protect the cell from repeat instability.
GAA/TTC-induced replication blockage can result in repeat instability that is manifested as fragility, repeat size variations and mutations in yeast. Sergei Mirkin (Tufts University) demonstrated that the size of the Okazaki fragment is an important determinant of repeat instability. He showed that the expansion potential of GAA/TTC repeats is elevated in pol2 and pol3 mutants, while the scale of expansions is not changed. In contrast, in pol1 mutants deficient in Okazaki fragment initiation larger expansions are seen, but the expansion rates do not change significantly.
Marek Napierala’s (University of Texas MD Anderson Cancer Center) studies underlined the role of transcription on the stability of the trinucleotide repeat tracts. He demonstrated that both GAA/TTC and CTG/CAG tracts accumulate R-loops where stable RNA·DNA hybrids lead to formation of long unpaired ssDNA. These known recombinogenic intermediates are key suspects in triggering repeat-mediated instability.
Jintang Du (The Scripps Research Institute, Joel Gottesfeld’s laboratory) presented data showing that induced pluripotent stem cells derived from FRDA patients are also extremely prone to expansion indicating that disease-associated expansion might be aggressive during embryogenesis. Treatment of cells with a B-DNA-stabilizing polyamide, or knockdown of mismatch repair genes, caused a decline in the expansion rate of the repeats, implicating mismatch repair in repeat expansion and suggesting a potential therapeutic target for preventing FRDA.
In an attempt to elucidate the mechanisms underlying instabilities at GAA/TTC tracts, Kirill Lobachev (Georgia Institute of Technology) presented the results of an unbiased genome-wide screen in yeast aimed at identifying mutants prone to DSB formation and expansions. Consistent with the previously mentioned talks, he showed that down-regulation of almost any component of the replication fork results in repeat instability. In addition the study revealed several surprises. First, depletion of the proteins constituting the Orc complex or telomere maintenance complex (Cdc13-Ten1-Stn1) led to increased fragility and expansion rates, indicating that these complexes can bind to and influence the stability of the repeats (see data from Krasilnikova’s lab above). Second, it was found that GAA/TTC tracts can also serve as non-canonical promoter elements and transcription initiation mutants dramatically amplify fragility and expansions at the repeats. These observations taken together with Napierala’s studies (see above) imply impaired transcription, probably initiated within the repeats, can result in accumulation of R-loops at GAA/TTC tracts culminating in instabilities in non-dividing cells. On the other hand, defects in the DNA replication pathway stimulate repeat-mediated breakage in actively dividing cells.
4.0. Hairpins and Cruciforms – Intrastrand acrobatics prone to instability
Palindromic DNA sequences or inverted repeats are sequences with internal symmetry such that they can switch between inter-strand and intra-strand base pairing. Consequently, these repeats can form hairpins when only one DNA strand is involved, a cruciform structure when both strands are involved, or slipped DNA when both strands form hairpins that are offset from one another. Several talks addressed the following questions: how to identify loci containing palindromes, what nucleases lead to DSB formation, when does this occur and what are the consequences of the breakage. Alison Rattray (NCI) expanded on the problems of identifying palindromes in complex genomes raised during the keynote address by Jeffrey Strathern (See above). She presented novel methodology to determine the palindromic content of the cancer cell line MCF7. This approach is based on the fact that closely spaced inverted repeats anneal faster than normal duplex DNA after denaturation. Treatment with S1 nuclease preferentially removes the denatured ssDNA and enriches templates with hairpins for PCR and sequencing. Using this methodology more than 100 novel palindromes were identified in the cancer cell-line MCF-7 that were missed using classical sequencing methods. Their group is optimizing conditions for sequencing palindromic regions and their results are eagerly anticipated at the next meeting.
Hidehito Inagaki (Fujita Health University) transfected human cells with plasmids containing preformed cruciform structures and found DSB products containing hairpin-capped termini. In an attempt to identify the nucleases that create these breaks and taking into account that cruciform structures resemble recombination intermediates, he knocked-down the expression of known Holliday junction resolvases. While MUS81 and SLX4 were found to have no effect, GEN1 knock-down led to a decline in hairpin-capped DSBs. These molecules were identified to be substrates for another nuclease, Artemis, which opens the hairpin to form DSB ends. He speculated that this process might be responsible for the palindromic AT-rich repeat-mediated translocation seen in patients with Emanuel syndrome. It is interesting to note that in yeast Mus81 has been shown to attack secondary structures formed by palindromes placed in plasmids. Why MUS81 and its yeast homolog behave differently may perhaps be accounted for by the differences in the mechanisms of breakage in yeast and humans. However, as was brought up during the discussions, these studies are plasmid based and it would be important to establish what nuclease is responsible for the breakage when palindromes are present in a chromosomal context.
Secondary structures formed by inverted repeats are associated with blocks to replication fork progression in yeast suggesting that breaks at inverted repeats might occur during S-phase. Rachel Parent (Eastern Michigan University, Anne Casper’s laboratory) showed that DNA breaks at the inverted Ty1 elements that comprise the yeast fragile site FS2 are formed during DNA replication. Using a galactose-inducible Pol1 strain they showed that breaks at this locus occur in S and G2 phases of the cell cycle in cells under replication stress. Sgs1 was not required for break formation but plays a crucial role in DSB-repair at this locus. Thomas Petes (Duke University) determined that breaks at inverted Ty1 elements occur in the G1 phase of the cell cycle in wild-type diploid yeast. His system relies on the detection of loss of heterozygosity of small nucleotide polymorphisms (SNPs) after break-induced cross-overs are generated between homologous chromosomes. On the other hand, in cells with decreased Pol1 levels, the breakage-associated crossovers occur predominantly in the G2 phase. Natalie Saini (Georgia Institute of Technology, Kirill Lobachev’s laboratory) highlighted the fact that breaks at inverted repeats might occur in either the late-S phase or the G2 phase of the cell cycle and cause mutagenesis in the flanking genes. At these stages DSBs can be repaired using the sister chromatid. Extensive resection generating long single-stranded DNA in combination with recruitment of Polζ for repair synthesis leads to mutagenesis up to 8 kb away in the regions flanking the repeats. Remarkably, repair of the break involving error-prone synthesis restores inverted repeats, making them a long-term source of mutations.
Susan Lovett (Brandeis University) described the interesting case of mutagenesis at inverted repeats in E. coli. She showed that diverged inverted repeats could undergo a template switch in which one arm of the repeat serves as a template for synthesis of the second arm. This phenomenon results in mutations within the repeats. The template switching was dependent on DnaE (Polymerase III) and mutations in dnaE which impair this process led to a reduction in mutation frequencies. The chain-terminating nucleosides azidothymidine and didehydrodideoxythymidine dramatically augmented mutagenesis pointing towards replication stalling in the vicinity of inverted repeats as the trigger for template-switch mutagenesis.
DNA palindromes are also implicated in gene amplification events. These events can either be beneficial and play an important role in evolution, or lead to carcinogenesis as was emphasized in Hisashi Tanaka’s (Cleveland Clinic) talk. His study on copy number alterations in the ERBB2 oncogene locus in breast tumors revealed that a 400 kb region comprised of a number of direct and inverted repeats located 1.5 Mb telomeric of ERBB2 was the initial breakpoint for gene amplification and that in tumors such unstable breakpoints are the major source of recurrent amplification.
Besides palindromic DNA, a subset of long repetitive micro- and minisatellites that have internal inverted symmetry can form hairpins. These motifs are highly unstable as they can induce DSBs and are prone to repeat size variations. For example, the CTG/CAG repeats can form intra-strand hairpins containing a mixture of C-G base pairs and either T-T or A-A mispairs [9]. These repeats have been implicated in many neurological diseases including myotonic dystrophy, Kennedy disease, spinocerebellar ataxia and Huntington disease. Formation of hairpins at CTG/CAG repeats is proposed to cause replication blockage and DSBs that might culminate into repeat size variations and GCRs. Catherine Freudenreich (Tufts University) demonstrated that the helicase Srs2 is required for replication across long CTG/CAG or CGG/CCG repeats. Mutations in the ATPase and the PCNA binding domains of Srs2 caused an increase in replication blockage accompanied with increased chromosome fragility at the repeats. However, a deletion of the Rad51 binding domain in Srs2 had no effect on fork stalling and fragility, implicating a role for Srs2 in unwinding hairpin structures encountered during replication. In contrast, Rad51 binding and displacement were important for preventing repeat instability, indicating that uncontrolled recombination is a pathway to repeat expansions. Michael Leffak’s (Wright State University) presented work that added credence to the concept that hairpins at the tracts stall replication leading to repeat instabilities. He showed that both CTG and CAG strands are capable of adopting hairpin-structures in human cells, as evidenced by cleavage using structure- and sequence-specific zinc finger nucleases. Formation of hairpins was associated with fork stalling and repeat size variations. The replication block and repeat expansion or contraction frequencies were increased during prolonged growth in culture and on treatment with agents that inhibit either the leading or lagging strand synthesis. Transfection with oligonucleotides capable of binding the repeats and abolishing structure formation reduced fork stalling and repeat size variations.
Guy-Franck Richard (Pasteur Institute) raised the possibility that replication stalling and the instability of CTG/CAG repeats might be unrelated events. He showed using 2-D gels that replication progression is blocked when either strand is the template for lagging strand synthesis. However, when the CTG-strand was the template for lagging strand synthesis repeat contraction occurred at a higher frequency than when the CAG-strand was the template. Interestingly, replication stalling was dependent on Msh2, suggesting that binding of mismatch repair machinery to the hairpins can create a strong barrier to fork progression.
Similar to G4 and triplex-forming motifs, the stability of hairpin-forming trinucleotide repeats is modulated by DNA damage response and transcription. Oxidative DNA base damage at CTG/CAG repeats yields expansions and contractions via involvement of the base excision repair (BER) machinery as was underscored in the talk by Yuan Liu (Florida International University). Treatment of human cells with hydrogen peroxide, bromate and chromate led to increased expansions and contractions at CTG/CAG repeats. Using an in vitro BER system, she further elucidated that the instability varies with the site of the oxidative damage, with damages near the 5′ end of CTG repeats giving rise predominantly to expansions, while damage in the middle of the repeat generated contractions.
Robert Lahue (National University of Ireland) discovered that a specific subset of histone acetyle transferases (HATs) and deacetylase complexes (HDACs) have a pronounced influence on the stability of CTG/CAG repeats in both yeast and human cells. He found that several HDACs promote expansions of CTG/CAG repeats and consistently HATs are crucial for their stability. He postulated that HATs and HDACs alter the acetylation status and therefore the activity of different proteins that in turn influence CTG/CAG expansion levels. One culprit responsible for expansions and the target for deacetylation by the HDACs Rpd3 and Hda1 was found to be the Sae2 protein. In the model to explain the data, Lahue suggested that in the absence of the deacetylation, Sae2 is degraded, the hairpin is not processed and expansion frequencies decline. Intriguingly, the disruption of HDACs had a stronger effect than eliminating Sae2 implicating that there may be other important factors in expansions under the control of acetylation that are still under wraps. One of the possible candidates is chromatin.
Charles Thornton (University of Rochester) showed that transcription through CTG repeats in myotonic dystrophy poses a dual threat to the genome. In patients, bidirectional transcription across the repeat tract led to large-scale expansions and instability in somatic cells. The RNA molecules that contain expanded CUG tracts are extremely toxic to the cell. CUG hairpins in RNA transcripts sequester the splicing factor Muscleblind-like 1 protein, and at the same time cause an upregulation of CUG-binding protein 1. Dysregulation of these protein levels leads to aberrant splicing of various genes including Cav1.1, a calcium ion channel. Abnormal splicing of this calcium channel may exacerbate the myopathy characteristic of this disorder. Targeting of the CUG RNA with a specific locked nucleic acid oligonucleotide, and the subsequent degradation of the RNA by RNaseH leads to reversal of the RNA toxicity-associated phenotypes in mice. These studies pave the road towards drugs that can provide a treatment for myotonic dystrophy patients.
4.0. Branches, forks and head-on collisions: Hazards on the road to replication, recombination, repair and transcription
The genome is employed in a variety of often simultaneous metabolic operations including replication, transcription and recombination which besides aiding formation of secondary structures as described above, can yield other transient deviations from the canonical B-DNA conformation. Various talks at this meeting discussed how defects in such operations can lead to the accumulation of intermediates that result in genetic instability. Another interesting related topic that attracted attention was the effect of RNA on DNA’s integrity (Figure 2).
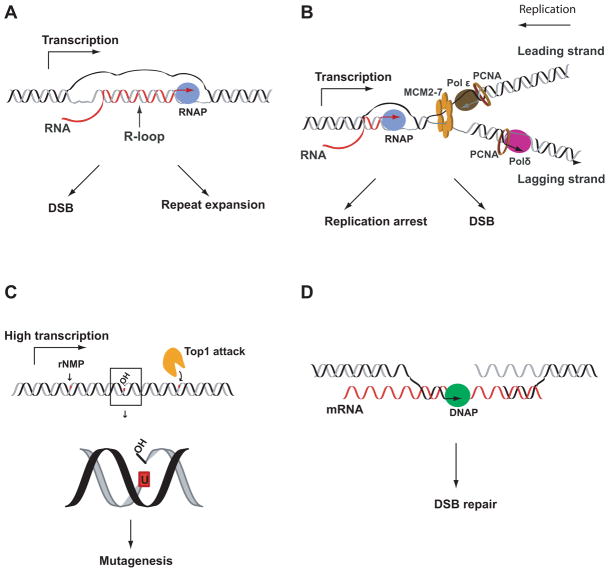
Figure 2.
The impact of the interplay between RNA and DNA on genome stability. (A) R-loop could be formed due to persistent interaction of nascent pre-mRNA and the repetitive DNA template during transcription, leading to repeat expansion and chromosomal fragility, as indicated in Marek Napierala’s and Kirill Lobachev’s studies. (B) Collision between transcription and replication machineries could be hazardous and result in replication arrest as shown by Jue D. Wang and Houra Merrikh in Bacillus subtilis, or induce DSBs as demonstrated by Evgeny Nudler in E. coli. (C) rNMPs mis-incorporated into DNA are targeted by topoisomerase I during transcription, causing mutagenesis in yeast S. cerevisiae (Sue Jinks-Robertson’s study). (D) mRNA serves as a template for DSB repair, as shown by Francesca Storici in yeast.
Holliday junction is one of the transient intermediates that is formed during recombination processes and whose resolution often leads to crossing over. Lorraine Symington’s (Columbia University) talk focused on discerning the roles of the endonucleases in cross-over generation during DSB repair. She found that during mitotic homologous recombination, Mus81 endonuclease is crucial for resolution of the intermediates. In mus81yen1 double mutants there is a sharp decline in crossovers relative to those seen in mus81 mutants, indicating that in the absence of Mus81, another endonuclease, Yen1 takes over. On the other hand crossovers resulting from recombination between homeologous, but not homologous, molecules required the action of the Rad1-Rad10 clippase. Removal of non-homologous tails during the formation of intermediates by the Rad1-Rad10 complex appears to be a prerequisite for channeling homeologous exchange into the crossing-over pathway. Rebecca Billmire (University of Pennsylvania, Brad Johnson’s laboratory) further highlighted the complexity of the pathways that govern the metabolism of branched recombination and replication intermediates. She investigated the role of the components of the Sgs1/Top3/Rmi1 complex in dissolution of Holliday junctions and X-shaped intermediates from template switch during replication (Rec-X). She demonstrated that Δsgs1 Δtop3 strains are extremely sensitive to MMS and this phenomenon can be partially reversed in Δsgs1 strains with Top3 overexpression. These data indicate that Top3 can work independently of Sgs1/Rmi1. Furthermore, she found that Top3 activity is sufficient for decatenation of Rec-X structures but not for dissolution of Holliday junctions.
David Leach (University of Edinburgh) presented data on the accumulation of repair intermediates in E.coli mutants defective in branch migration. He took advantage of the fact that in bacteria long hairpins formed by palindromes on the lagging strand are attacked by the SbcCD nuclease. Using a regulatable SbcCD nuclease, a DSB can be introduced on the lagging strand in a controlled manner allowing for repair using the leading strand as a template. He ascertained that mutations in ruvAB helicase led to accumulation of Y-shaped and branched intermediates while in mutants for the recG helicase, only the Y-shaped structures were persistent. In double mutants for ruvAB and recG severe DNA loss occurred. To account for these data, a model was proposed wherein branch migration is required to stabilize joint molecules formed during recombination.
Problems during DNA synthesis can result in the accumulation of branched molecules in the form of stalled replication forks and generation of damaged DNA that stimulates genetic instability. Using single molecule microscopy Giuseppe Lia (University of Paris XI, Bénédicte Michel’s laboratory) found that within 20 minutes of replication blockage in E. coli, the replication machinery falls off leaving the replicative polymerase, Pol III, behind. RecA binding then promotes dissociation of 2 out of 3 subunits of the Pol III holoenzyme suggesting a novel role of RecA in replication fork restart.
Andrei Kuzminov (University of Illinois at Urbana-Champaign) reported that in E. coli a deficiency in DNA ligase induces formation of nicked nascent DNA that triggers fork stalling, DSBs and cell death. He presented evidence that in ligase-deficient conditions, extensive resection at DSBs in both sister chromatids generates large double stranded gaps behind replication forks. Consistent with this model, inactivation of linear DNA degradation in the presence of functional DSB repair, eliminates the lethality of ligase-deficient replication. These observations suggest that bacterial ligase inhibitors may be good targets for the development of novel antibiotics.
Genetic instabilities can also arise upon collisions between replication and transcription machineries. In Bacillus subtilis ~75% of the genes are co-oriented with DNA replication in order to avoid head-on collisions [10]. Head-on transcription-replication collisions are extremely deleterious and can disrupt replication progression as was emphasized in the talk by Jue D. Wang (Baylor College of Medicine). She showed that reversing the orientation of transcription so as to increase collision frequencies resulted in an increase in the doubling time of Bacillus subtilis due to slowed replication. Impaired replication was further established as the source of increased mutations in the genome, lending a strong bias towards co-orientation of essential genes with the direction of the replication fork. Houra Merrikh (University of Washington) demonstrated that co-directional transcription can also pose a conflict for DNA replication as seen for the rDNA operons in the Bacillus subtilis genome. Her work demonstrates that replication restart proteins are recruited to places where the replication and transcription machineries collide, most likely to help resume synthesis.
Evgeny Nudler (New York University) highlighted another problem arising from replication-transcription collisions. He showed that co-directional transcriptional collisions with the replication machinery are a source of DSBs due to polymerase backtracking, and formation of recombinogenic R-loops. Active translation of the mRNA being transcribed prevented polymerase backtracking and reduced breakage.
Sue Jinks-Robertson (DukeUniversity) showed that in yeast active transcription also drives instability that is manifested as mutations with specific signatures. She identified hotspots consisting of tandem 2–5 bp repeats that underwent Top1-dependent deletions upon induction of transcription. The deduced mechanisms for generation of mutations at these hotspots are Top1-mediated DNA cleavage during the removal of transcription generated supercoiling and Top1 incision at incorporated ribonucleotide monophosphates (rNMPs) in DNA. Interestingly, an additional type of hotspot was discovered that seems to be generated by rNMPs near quasi-palindromes.
Francesca Storici (Georgia Institute of Technology) presented an exciting discovery that accentuates the interplay between DNA and RNA. She found that in yeast cellular mRNA can serve as a template for DSB repair either acting in cis or trans, although repair in cis was more frequent than in trans. This repair process is under the control of RNase H1 and RNase H2, raising the possibility that RNA-dependent repair may be a robust way to repair actively transcribed genes, particularly in the absence of RNase H.
6.0. Concluding remarks
Although G4 quartets were described half a century ago, it is only recently that we have begun to appreciate the dynamic role that they and other non-B DNA structures play in the evolution and function of the genomes in which they are found. This conference provided an excellent forum for discussion on the progress that has been made in this area and provided many fascinating ideas for new avenues of research that it is hoped will make the next meeting in the series, planned for June of 2014, very interesting indeed.
Acknowledgments
We are grateful to all the participants at the meeting who agreed to present unpublished results in this review. On behalf of the organizers, we would also like to thank ABCAM, American Society for Biochemistry and Molecular Biology, Annual Reviews, Elsevier, New England Biolabs, Inc., Public Library of Science and Tufts University, School of Arts and Sciences for their generous support for the meeting. The studies in KL laboratory were supported by the grants MCB-0818122 from NSF and R01GM082950 from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8.0. References
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–8. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat Struct Mol Biol. 2006;13:1055–9. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 3.Amrane S, Adrian M, Heddi B, Serero A, Nicolas A, Mergny JL, Phan AT. Formation of pearl-necklace monomorphic G-quadruplexes in the human CEB25 minisatellite. J Am Chem Soc. 2012;134:5807–16. doi: 10.1021/ja208993r. [DOI] [PubMed] [Google Scholar]
- 4.Huppert JL. Structure, location and interactions of G-quadruplexes. FEBS J. 2010;277:3452–8. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 5.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 6.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 7.Larson ED, Duquette ML, Cummings WJ, Streiff RJ, Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr Biol. 2005;15:470–4. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 8.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–7. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 9.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–54. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–56. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]