Abstract
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the most effective tumor immunotherapy available. Although allo-HSCT provides beneficial graft-versus-tumor effects, acute GVHD (aGVHD) is the primary source of morbidity and mortality after HSCT. Diagnosis of aGVHD is typically based on clinical symptoms in one or more of the main target organs (skin, liver, gastrointestinal tract) and confirmed by biopsy. However, currently available diagnostic and staging tools often fail to identify patients at higher risk of GVHD progression, unresponsiveness to therapy, or death. In addition, there are shortcomings in the prediction of GVHD before clinical signs develop, indicating the urgent need for noninvasive and reliable laboratory tests. Through the continuing evolution of proteomics technologies seen in recent years, plasma biomarkers have been identified and validated as promising diagnostic tools for GVHD and prognostic tools for nonrelapse mortality. These biomarkers may facilitate timely and selective therapeutic intervention but should be more widely validated and incorporated into a new grading system for risk stratification of patients and better-customized treatment. This review identifies biomarkers for detecting GVHD, summarizes current information on aGVHD biomarkers, proposes future prospects for the blinded evaluation of these biomarkers, and discusses the need for biomarkers of chronic GVHD.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a widely used therapy for a variety of malignant and nonmalignant hematologic diseases. In malignant disease, the donor immune system recognizes residual tumor cells as foreign and eradicates them by immunologic means, known as the graft-versus-tumor (GVT) effect. Unfortunately, donor immune cells may also attack normal host tissue, particularly the skin, liver, and gastrointestinal (GI) tract, resulting in acute GVHD (aGVHD). GVHD remains one of the major barriers to a more widespread and successful application of allo-HSCT. Known risk factors that increase the incidence and severity of aGVHD include the use of HLA-mismatched unrelated donors rather than HLA-matched sibling donors.1
A major barrier to GVHD research and treatment is that diagnosis and prognosis rely almost entirely on the presence of clinical symptoms and should be confirmed by biopsy of the involved target organs. No currently used laboratory tests can predict the risk of developing GVHD, responsiveness to treatment, or patient survival. The importance of biomarkers in HSCT settings is crucial because the ability to identify patients at high risk for GVHD early in their transplantation and treatment course has important therapeutic consequences, including more stringent monitoring and/or preventive care. The ability to identify patients who will not respond to traditional treatment and are at particularly high risk for subsequent morbidity and mortality could enable tailored treatment plans, such as additional immunosuppressive treatments that might be more effective if introduced early.
This review provides an update on the discovery and validation of the most clinically relevant biomarkers of aGVHD and insights for specific recommendations on their use in clinical trials.
GVHD biomarker qualification
A good diagnostic GVHD test will be able to distinguish patients with GVHD from those without GVHD (eg, GI GVHD vs infectious colitis, skin GVHD vs drug rash). Furthermore, the biomarker should (1) specifically and sensitively reflect the disease state, (2) be noninvasive (eg, blood or urine test), and (3) be rapid, simple, accurate, inexpensive, and standardized. Ideally, the same test should also be prognostic for outcomes, such as response to treatment, survival, and nonrelapse mortality (NRM) to allow for early risk stratification before initiation of treatment. A biomarker that predicts aGVHD before clinical signs appear will have tremendous impact and allow for preemptive interventions. The ideal biomarker for GVHD should differentiate between GVHD and GVT effects.
Single versus multiple biomarkers
When several biomarkers exist, usually no single biomarker is sufficiently sensitive or specific on its own for either diagnostic or predictive testing. Thus, the simultaneous use of several markers may increase specificity or predictive/diagnostic performance.2 Presumably, a combination of tissue-specific and systemic biomarkers will be more informative for aGVHD diagnosis. However, if a biomarker is not highly correlated to other biomarkers or clinical predictors, one or 2 biomarkers could be sufficient. Proportional odds logistic regression models are used to create a composite panel that will generate a receiver operating characteristic (ROC) curve. The ROC curve is a plot of the true-positive rate (sensitivity = 1 − false negative error rate) versus the false-positive rate (1 − specificity), and is associated with rules that classify a person as “positive” if is marker value is above a threshold c for all possible thresholds.3–6
Types of GVHD biomarkers
miRNAs
MicroRNAs (miRNAs) are 21- to 25-nucleotide transcripts that repress gene function through interaction with target mRNAs. miRNAs control gene activity at multiple levels, specifically transcription, translation, and protein degradation. Circulating miRNAs have recently been reported as promising biomarkers in diverse diseases,7 including GVHD.8
Cellular biomarkers
The functions and numbers of several different immune cell populations are altered in GVHD. Some of these immune cell subsets, particularly regulatory T cells, seem promising as biomarkers of aGVHD9–11 and chronic GVHD (cGVHD).12 Dendritic cells, monocytes, and γ-δ T cells are also promising as markers after HSCT.13,14 In cGVHD, B cells and their modulators (eg, B cell–activating factor) are important biomarkers.15
Proteomic biomarkers
The term “proteomics” indicates PROTEins expressed by a genOME and describes the systematic analysis of protein profiles of a biologic sample. Unlike the genome, the proteome varies with time and is defined as the proteins present in a single sample at a certain time point. Thus, proteins represent ideal biomarkers in the post-transplantation setting and have been widely studied, as detailed in the following sections.
Proteomics strategies in the search for new GVHD biomarkers
The study of blood by proteomics
Ideal clinical tests are based on noninvasive collection, which enables repetitive sample collection from the same patient in a short amount of time. Enormous effort has been made to develop standardized methods for clinical sample collection for proteomic studies.16,17
The levels of individual blood proteins represent a summation of multiple, disparate events that occur in every organ system. Plasma and sera contain proteins shed by the affected tissue as well as proteins that reflect secondary systemic changes. However, plasma and sera are highly complex mixtures containing high levels of many different proteins with a wide dynamic range, spanning 12 orders of magnitude from albumin to the lowest abundance protein. Low-abundance proteins, such as cytokines and their receptors,18,19 are often the most clinically relevant. To detect low-abundance proteins, depletion of predominant proteins and subsequent fractionation of the proteome are required.
Urine samples represent an alternative to plasma/sera samples for biomarker discovery. Urine has 3 main advantages compared with plasma/sera: (1) it can be obtained in large quantities; (2) the protein mixture is far less complex and the variation in protein abundance is low20; and (3) it is more stable.21 However, urine yields better information about diseases in organs that are directly involved in its production and excretion, such as the kidneys.
Tissue proteomics
Although blood biomarkers are ideal for use in a clinical setting, one goal of research into the fundamental biology of GVHD is to identify markers that are target-tissue specific. Thus, the ideal sample for discovery of biologically relevant GVHD proteins may be the target tissue itself. However, finding tissue-specific markers has thus far proven difficult because of the cellular heterogeneity of tissues, and the limited material available in biopsies for tissue proteomics. To date, there is no method capable of amplifying the amount of proteins requiring, at best, pooling of several biopsies.22–24
Proteomics approaches for biomarker discovery
Advances in engineering have increased data throughput, enabling the study of complete sets of molecules (“-omics”) with exponentially increased speed, accuracy, and cost-effectiveness. Thus, analysis of the entire spectrum of molecular and cellular organization is now possible, providing insight into the mechanism of disease, with fewer a priori assumptions. However, arriving at proteins from genes (∼ 20 000) requires 2 more levels of complexity: the transcriptome (∼ 100 000 RNA transcripts) and proteome (∼ 1 000 000 proteins). Here, this review focuses on proteomics for the molecular diagnosis of GVHD after HSCT because proteins are more informative than other cellular metabolites regarding the ongoing pathophysiology of a disease.25 Both mass spectrometry (MS)–based and non-MS–based proteomic approaches have been used to screen potential GVHD biomarkers.
Array-based approaches
Immunoassays are analytical tests that harness the unique properties of antibodies and are therefore extremely sensitive. The unique characteristics of antibodies are derived from 2 important properties: (1) their exceptional binding specificity that enables the measurement of picomolar (10−12) protein amounts in blood samples; and (2) the binding strength between an antibody and its target that makes tests accurate and precise, even at low concentrations.26 To screen for aGVHD biomarkers, microarrays dotted with hundreds of antibodies measure hundreds of proteins in complex biologic matrices.2 The advantages of immunoassays are that they are: (1) suited for the characterization of complex protein mixtures, such as human plasma, (2) quantitative, (3) able to detect low-abundance proteins, such as cytokines, and (4) high-throughput. The disadvantages are limitations to the number of antibodies included in each array (ie, introduction of bias) and high cross-reactivity between antibodies and nontarget proteins.27
The advent of MS for protein analysis
Most nonantibody proteomic strategies are based on MS, which is a powerful tool for characterizing and assessing qualitative and quantitative changes in complex protein mixtures.28 Two types of MS techniques have been used in clinical proteomics: (1) pattern profiling and (2) detailed protein characterization. Pattern profiling compares polypeptide spectra obtained by matrix-assisted laser desorption/ionization time-of-flight MS to show which patients have a particular disease without identifying individual profile components. A variant of this is surface-enhanced laser desorption/ionization MS.29 Because factors influencing the final oligopeptide profiles of body fluid samples are so complex, MS profiling has not yet met the standards required for clinical practice. This technique has been used to screen aGVHD biomarker candidates in both serum30 and saliva.31
Other approaches rely on protein separation using gel-based techniques followed by MS.32–35 However, gel-free separation methods, such as liquid chromatography (LC)36–39 and capillary electrophoresis,40 provide better separation because they overcome several limitations of gel separation, such as lengthy analysis time; poor separation of proteins with low molecular weight, or extreme isoelectric point. MS is the final step in the analytical procedure and reliably identifies proteins and determines their isoforms and post-translational modifications. MS also allows unambiguous quantification, particularly when tandem MS (MS/MS) is used.36,37,41,42 New instrumentation, such as the ultra-high resolution linear ion trap Orbitrap mass spectrometer also facilitates top-down LC-MS/MS.43
These approaches are not suitable for validation purposes because of the time required for analysis, but they remain the most efficient methods for biomarker discovery in clinical research.
Informatics to mine disease proteomes and prioritize candidate biomarkers
Massive amounts of complex and heterogeneous proteomic data are generated from a single experiment. Many proteins are identified, but prioritization is uncertain, with no mechanism for attrition and few available high-quality reagents to validate these discoveries. Proteomics workflows that have led to biomarker discovery include the one detailed here.38,39 First, the acquired spectra are matched to a sequence database to identify proteins, with a false discovery rate < 5%.44,45 False discovery rates are now recognized as markers of significance in -omics studies.46,47 To sequentially refine the list of candidate proteins, proteins can be selected based on relationship to the tissue of origin using the Human Protein Atlas website, which aims to annotate human proteins using antibodies to systematically analyze the cellular distribution of proteins in normal and pathologic tissues.48 Another refinement can be obtained using pathway analysis tools. Physician scientists are usually well versed in the complex pathophysiology of their disease of interest and might be more successful at mining their dataset based on information from published literature than pathway analysis, which depends on the validity of the annotations of the different molecules and their biologic processes. Then, candidate proteins would require further validation by other biotechnologic techniques, ideally those that allow for high-throughput as detailed in the next paragraph. The steps of these workflows are summarized in Figure 1.
Figure 1.
Proteomics biomarkers pipeline.
Proteomics approach for high-throughput validation of GVHD biomarkers
Although proteomics holds great promise for biomarker development, gaps still remain between biomarker discovery and biomarker validation. Most noteworthy is the paucity of affinity-capture reagents, such as high-quality antibodies, which leads to a bias in the prioritization of candidate markers. Furthermore, the number of samples required for validation increases as the biomarker advances through each test phase, augmenting the need for high-throughput assays. The most applicable approach for quantitating individual proteins for validation is the ELISA, which is highly specific and uses 2 antibodies specific for the candidate protein. This procedure is also relatively simple and highly reproducible among performers and laboratories, limiting inter- and intra-assay variability. Finally, because of the urgent need for GVHD blood tests, testing proteins primarily with available antibodies is an excellent strategy. The main disadvantage of validation by ELISA is the large volume of plasma required. Thus, multiplexing technologies are preferred if cross-reactivity does not occur.27 Recently, selected reaction monitoring-MS, which can measure the absolute quantification of multiple native peptides, and by extension, their parent proteins, has emerged as a potentially useful technique to screen and quantify targeted multiplexed proteins in patient plasma samples with high sensitivity, absolute specificity, and high throughput.49–51
Identified and validated GVHD biomarkers
GVHD is not only a systemic immunologic disorder; it also affects specific organ systems, including the skin and GI tract. Noteworthy systemic and target-specific aGVHD biomarkers have been reviewed along with their diagnostic and prognostic values and whether they were identified using aGVHD pathology or a proteomics discovery approach (Table 1).
Table 1.
Candidate aGVHD biomarkers with diagnostic and prognostic significance identified based on GVHD pathology and proteomics discovery
| Protein | Study | No. of patients in the study | Target organ | Diagnosis end point | Association direction* | Prognostic end point | Predictive end point |
|---|---|---|---|---|---|---|---|
| Based on GVHD pathology | |||||||
| Albumin | Rezvani 201176 | 401 | Systemic/GI tract/renal | ND | ↓ | 2 y NRM, 6 mo OS in RIC patients | aGVHD 3-4 in RIC patients |
| CRP | Schots 200264 | 96 | Systemic | All TRC | ↑ | Day 100 NRM | TRC |
| Pihush 200666 | 350 | Systemic | All TRC | ↑ | ND | TRC | |
| CXCL10 | Piper 200770 | 34 | Systemic | aGVHD 0-4 | ↑ | ND | aGVHD 0-4 by day 100 |
| HGF | Okamoto 200171 | 38 | Systemic | aGVHD 0-4 | ↑ | ND | ND |
| Harris 201278 | 826 + 128† | GI tract | aGVHD 0-4 | ↑ | 6 mo NRM | ND | |
| IL-2Rα | Miyamoto 199654 | 30 | Systemic | aGVHD 0-4 | ↑ | ND | ND |
| Foley 199853 | 36 | Systemic | aGVHD 0-4, sepsis, VOD | ↑ | ND | ND | |
| Shaiegan 200656 | 39 | Systemic | aGVHD 2-3 | ↔ | ND | ND | |
| August 201162 | 62 | Systemic | ND | ↑ | ND | Prospective study aGVHD 3-4 by day 60 | |
| IL-6 | Malone 200765 | 147 | Systemic | aGVHD 0-4 by day 100 | ↑ | ND | ND |
| Schots 200361 | 84 | Systemic | All TRC | ↑ | 1 y NRM | ND | |
| Pihush 200666 | 350 | Systemic | All TRC | ↑ | ND | TRC | |
| IL-8 | Uguccioni 199367 | 30 | Systemic | aGVHD 0-4, rejection, liver fibrosis in thalassemic patients | ↑ | ND | ND |
| Schots 200361 | 84 | Systemic | All TRC | ↑ | 1 y NRM | TRC | |
| IL-10 | Liem 199891 | 46 | Systemic | aGVHD 2-4, cGVHD, infection, relapse | ↑ | ND | ND |
| IL-12 | Mohty 200568 | 113 | Systemic | aGVHD 2-4 in RIC patients | ↑ | ND | ND |
| IL-18 | Fujimori 200090 | 37 | Systemic | aGVHD 0-4 | ↑ | ND | ND |
| Shaiegan 200656 | 39 | Systemic | aGVHD 2-3 | ↑ | ND | ND | |
| KRT18 | Luft 200772 | 55 | GI tract | GI GVHD vs non-GVHD-enteritis | ↑ | Response to GVHD treatment | ND |
| Luft 201175 | 48 | Systemic | Refractory GVHD | ↑ | ND | ND | |
| Harris 201278 | 826 + 128† | GI tract | GI GVHD | ↑ | 6 mo NRM | ND | |
| REG3α | Harris 201278 | 826 + 128† | GI tract | GI GVHD | ↑ | 6 mo NRM, response to GVHD treatment | ND |
| Syndecan-1 | Seidel 200355 | 60 | Systemic | aGVHD 0-4 | ↑ | ND | ND |
| TNFα | Holler 199060 | 56 | Systemic | All TRC | ↑ | ND | ND |
| Abdallah 199758 | 80 | Systemic | All TRC, aGVHD 0-1 vs 2-4 | ↔ | ND | ND | |
| Schots 200361 | 84 | Systemic | All TRC | ↑ | 1 y NRM | ND | |
| TNFR1 | Choi 200859 | 438 | Systemic | ND | ↑ | ND | aGVHD 2-4, 1 year NRM |
| Kitko 200859 | 82 (children) | Systemic | ND | ↑ | ND | aGVHD 2-4, 1 year NRM | |
| August 201162 | 62 | Systemic | ND | ↑ | ND | Prospective study aGVHD 3-4 by day 60 | |
| Tryptophan metabolites | Landfried 201174 | 51 | Systemic | ND | ↑ | 6 mo NRM | ND |
| Based on proteomics discovery | |||||||
| Peptides set | Kaiser 200440 | 35 | Systemic | aGVHD 0-4, sepsis | Presence vs Absence | ND | ND |
| Srinivasan 200630 | 34 | Systemic | aGVHD | Presence vs Absence | ND | ND | |
| Imanguli 200731 | 41 | Systemic | 1 month post-HSCT vs pre-HSCT | ↑ | ND | ND | |
| Weissinger 200777 | 33 + 141† | Systemic | aGVHD 2-4 | Presence vs Absence | ND | ND | |
| Proteins set | Wang 200534 | 9 | Systemic | aGVHD vs pre-HSCT | Presence vs Absence | ND | ND |
| Albumin | Kaiser 200440 | 35 | Systemic | aGVHD 0-4, sepsis | Presence vs Absence | ND | ND |
| Imanguli 200731 | 41 | Systemic | 1 month post-HSCT vs pre-HSCT | ↑ | ND | ND | |
| CCL8 | Hori 200869 | 14 | Systemic | Murine model discovery, Validation human serum: aGVHD 0-4 | Presence vs Absence | ND | ND |
| Collagen α-1 | Weissinger 200777 | 33 + 141† | Systemic | aGVHD 2-4 | ↓ while collagen chain III was ↑ | ND | ND |
| Elafin | Paczesny 201084 | 20 + 492† | Skin | Skin aGVHD 0-4 vs non-GVHD rashes | ↑ | NRM, OS | ND |
| HGF | Paczesny 2009 (in a panel)2 | 42 + 282† + 142† | Systemic/GI tract | aGVHD 0-4 vs no GVHD | ↑ | NRM, OS | ND |
| IL-2Rα | Paczesny 2009 (in a panel)2 | 42 + 282† + 142† | Systemic | aGVHD 0-4 vs no GVHD | ↑ | NRM, OS | ND |
| IL-8 | Paczesny 20092 | 42 + 282† + 142† | Systemic | aGVHD 0-4 vs no GVHD | ↑ | NRM, OS | ND |
| Lactoferrin | Imanguli 200731 | 41 | Systemic | 1 month post-HSCT vs pre-HSCT | ↑ | ND | ND |
| Leukotriene A4 hydrolase | Kaiser 200440 | 35 | Systemic | aGVHD 0-4, sepsis | ↑ | ND | ND |
| REG3α | Ferrara 201138 | 20 + 871† + 143† | GI tract | GI-GVHD vs non-GVHD enteritis | ↑ | NRM, OS | ND |
| SLPI | Imanguli 200731 | 41 | Systemic | 1 month post-HSCT vs pre-HSCT | ↑ | ND | ND |
| TNFR1 | Paczesny 20092 | 42 + 282† + 142† | Systemic | aGVHD 0-4 vs no GVHD | ↑ | NRM, OS | ND |
ND indicates not done; OS, overall survival; RIC, reduced intensity conditioning; TRC, transplantation-related complications, major complications, including aGVHD, microangiopathic hemolytic anemia, VOD, pneumonitis, and infection; and VOD, veno-occlusive disease.
↑, plasma protein increased; ↓, plasma protein decreased; and ↔, plasma protein unchanged.
Validation on independent sets of patients.
Biomarkers identified using the pathology of aGVHD
Systemic biomarkers of aGVHD
Because of the cytokine storm that occurs early after donor graft infusion, cytokines and their receptors have been tested as potential aGVHD biomarkers.52 Soluble IL-2 receptor α chain (sIL-2Rα) concentrations were increased in aGVHD patients in many studies.53–57 In 2 studies, IL-18 concentrations were closely correlated with IL-2Rα concentrations, and IL-2Rα concentrations with GVHD severity.56 However, some studies found that sIL-2Rα concentrations were also increased in patients with other transplantation-related complications, such as veno-occlusive disease and sepsis.53 Similarly, TNF-α and its receptors, particularly TNFR1 concentrations, were higher in aGVHD patients than in patients without GVHD.58–62 The same precautions used to evaluate sIL-2Rα levels in other transplantation-related complications should be considered when evaluating TNF-α and TNFR1. The roles of TNF-α/TNFR1 and IL-2/IL-2Rα in aGVHD pathogenesis are supported by evidence suggesting that antibodies directed against TNFR1 or IL-2Rα are effective therapies for steroid-refractory aGVHD.63 C-reactive protein is a nonspecific inflammatory protein that is elevated in GVHD patients along with IL-6, the main cytokine that induces C-reactive protein release.61,64–66 Uguccioni et al found that IL-8 concentrations correlated with aGVHD.67 However, Schots et al showed that IL-8 was released in all types of transplantation-related complications, rather than specifically in cases of aGVHD.61 Similarly, increases in IL-8 and other cytokines (eg, IL-6, IL-10, and IL-18) were not confirmed in a study of patients who underwent a reduced intensity conditioning regimen; however, this same study found that IL-12 was elevated in association with aGVHD development.68 Chemokines and chemokine receptors that were also implicated in immune cell migration from lymphoid organs to target organs were also elevated in patients with aGVHD.69,70
Hepatocyte growth factor (HGF) is a multifunctional cytokine secreted by mesenchymal cells that acts primarily on cells of epithelial origin. Okamoto et al observed higher serum HGF concentrations in patients who developed severe aGVHD than in patients who did not develop the disease.71 HGF appears to belong to a different category of biomarkers that represent a physiologic response to GVHD damage. In this respect, HGF seems similar to cytokeratin-18 (KRT18) fragments, markers of epithelial apoptosis that are associated with intestinal and hepatic GVHD damage.72 Indoleamine 2,3-dioxygenase, the rate-limiting enzyme in the kynurenine pathway of tryptophan degradation, functions in a potent immunoregulatory loop and is involved in the pathology of GVHD.73 Recently, LC-MS/MS was used to measure major tryptophan metabolites in serial urine specimens from 51 patients. Surviving patients had significantly lower metabolite levels on days 28, 42, and 90 after HSCT than patients dying of GVHD.74
Target-specific biomarkers of aGVHD
As mentioned in the previous paragraph, KRT18 is the only target-specific marker of GVHD that is based on GVHD pathology.72 In addition, few biomarker studies have evaluated their prognostic value for outcomes, such as NRM, primarily because of small sample sizes. One exception is a recent study that measured KRT18 along with markers of endothelial dysfunction and found that patients with steroid-refractory GVHD were not exposed to an overwhelming T-cell attack but to a progressive microangiopathy that led to organ failure.75 Few studies have evaluated the predictive value of biomarkers. Studies measuring the kinetics of cytokine concentrations during the first month after HSCT showed that monitoring IL-2Rα and TNFR1 concentrations during the engraftment period allows for early detection of aGVHD. The mean serum IL-2Rα and TNFR1concentrations appear to increase during the first 2 weeks after transplantation in patients with aGVHD.53,54,56,59–61 Recently, August et al reported that sIL-2R, TNFR1, and CD8 have high predictive values for aGVHD occurrence.62 A recent study by Rezvani et al found that a decrease of 0.5 g/dL in serum albumin from pretransplantation levels predicted the development of grade 3 or 4 aGVHD and overall survival 6 months after initiation of aGVHD treatment in a cohort of 401 patients with aGVHD grades 2-4 after reduced intensity HSCT.76 Because measuring serum albumin concentration is easy and inexpensive, the authors suggest incorporating albumin into the set of other validated biomarkers to improve the prediction of aGVHD severity and mortality.
Biomarkers identified using proteomics discovery
Systemic biomarkers of aGVHD
Four studies have identified the proteomic pattern of aGVHD using MS-based approaches on small sample sizes. One group identified a pattern that was validated in a larger study. Imanguli et al used complementary proteomic techniques to characterize the salivary proteome of 41 patients who underwent allo-HSCT.31 Hori et al used the SELDI technique to screen plasma proteins specific for aGVHD in a mouse model.69 One peak appeared to distinguish GVHD plasma from non-GVHD plasma and was identified as CCL8. Using surface-enhanced laser desorption/ionization time-of-flight MS, Srinivasan et al identified an aGVHD-specific peptide pattern in training samples that was validated in an independent set of aGVHD samples obtained on the day aGVHD symptoms appeared.30 Kaiser et al used capillary electrophoresis-MS to identify peptide patterns in urine samples as early indicators of aGVHD development.40 Two prominent GVHD-indicative polypeptides were identified as a 1.85-kDa peptide from leukotriene A4 hydrolase and a 1.83-kDa peptide from albumin. This aGVHD-specific peptide set pattern was used to screen 63 samples collected from 33 patients after allo-HSCT.77 A subsequent blind evaluation of 599 samples from 141 patients enabled the prediction of aGVHD before the appearance of clinical symptoms with a sensitivity of 83% and specificity of 76%. Using MS/MS, 3 of the 31 peptides that contributed to the aGVHD pattern were identified as fragments from collagen α-1 chain I (down-regulated) and α-1 chain III (up-regulated).
Paczesny et al identified and validated a panel of proteins using an antibody microarray.2 To validate this aGVHD biomarker panel, they randomly divided the samples into a training set (282 patients) and a validation set (142 patients), which is currently one of the most reliable approaches for validation. This approach identified and validated a 4-protein biomarker panel for GVHD diagnosis (IL-2Rα, TNFR1, IL-8, and HGF) with high specificity. Because of the large sample size of this study, Paczesny et al provided the first evidence that these biomarkers are associated with GVHD clinical outcomes and prognosis.2
Target-specific biomarkers of aGVHD
Clinical symptoms of the skin (eg, maculopapular rash) and GI tract (eg, nausea, diarrhea) caused by GVHD can be difficult to distinguish from other causes (eg, infection, drugs). Thus, biomarkers specific for GVHD and target organs may improve diagnosis. Using intact-protein analysis system,36,37 plasma pooled from 10 patients with skin GVHD labeled with the heavy isotope was compared with plasma from 10 patients without GVHD labeled with the light isotope for the analysis of thousands of spectra and hundreds of proteins. Elafin emerged as the lead biomarker candidate for detecting skin GVHD at the time of clinical diagnosis. Paczesny et al demonstrated that plasma elafin concentrations in samples from 492 patients had significant diagnostic and prognostic value, including the prediction of long-term survival.39 These data also provide proof-of-principle demonstration that biomarkers of disease-related tissue-specific changes can be detected in plasma. Using the same proteomics strategy, Ferrara et al discovered 74 proteins with increased ratios (ie, heavy to light) in patients with GI GVHD, and 5 were of GI origin.38 Regenerating islet-derived 3-α (REG3α) was the lead candidate and was validated as a biomarker of lower GI GVHD using ELISA in 2 independent sets including 1014 patients from 3 centers.38 In a follow-up study, REG3α was compared with KRT18 and HGF, which were previously identified as GI GVHD markers, and showed higher diagnostic precision for lower GI GVHD than the other 2 biomarkers.78
There are also limitations in predicting response to GVHD therapy. Recently, Luft et al showed that markers of endothelial dysfunction are elevated in steroid-refractory GVHD.75 Levine et al measured 6 previously validated diagnostic biomarkers from samples prospectively obtained at treatment initiation and on days 14 and 28 in a multicenter, randomized, 4-arm phase 2 clinical trial for newly diagnosed aGVHD.79 At each time point, the panel of 6 biomarkers predicted important clinical outcomes (post-therapy nonresponse at day 28 and mortality at day 180 from onset).79
Decision-making strategies
Because GVHD target-specific markers are primarily indicative of GVHD-damaged tissue, they represent indicators of the disease when clinical signs are present. Therefore, GVHD target-specific markers complementary to biomarkers and infectious agents' analyses are useful in distinguishing skin rashes and diarrhea from other causes at time of diagnosis. If only one target, such as diarrhea, is involved, the most specific GI GVHD biomarker described so far, REG3α, should be tested.38,78 If both diarrhea and rashes are present at the same time, which is suggestive of a more systemic disease, REG3α and elafin should be tested along with systemic biomarkers.2,38,39
GVHD should be analyzed as a single disease process early in the transplantation course because mechanisms involved in alloreactivity start as soon as donor T cells are activated and in contact with host antigen-presenting cells. We expect that biomarkers will be identified that correlate with subclinical disease. A good example of such a marker is sIL-2Rα, which was shown in a prospective study to be elevated at days 7 and 15 after HCST and to predict severe aGVHD with 57% sensitivity and 82% specificity.62
Biomarkers of GVHD have also recently been used to follow the response to GVHD treatment.79 However, the key to developing useful surrogate end points is to identify biomarkers that reflect fundamental aspects of the treatment's effects on disease pathogenesis. A “perfect” surrogate end point, as described by De Gruttola et al, can be presented as T induces S that induces O, where T is the treatment, S is the surrogate end point, and O is a clinical outcome. In this case, the biomarker S measures all the effects of T on O.80 A more likely situation arises when T has a direct effect on O that is not mediated through S.80
Design considerations for future interventional trials with biomarkers
Given the progress in GVHD biomarker identification and validation, clinical trial design will begin incorporating biomarkers. The number of specimens that should be tested depends on the objective of the study and the extent of biomarker variability in the study. The following factors contribute to variability: the prevalence of the disease; the subtypes of disease among the study samples (eg, skin GVHD or GI GVHD); the capacity of the biomarkers to discriminate among the different disease subtypes; the number of biomarkers under study; and the statistical algorithm used to select promising biomarkers. Thus, as suggested by Pepe et al,81 there are no simple methods for recommending samples sizes. They proposed that computer simulations guide the choice of sample sizes with the guidance of investigators on biologically plausible models to generate data.
Study using diagnostic biomarkers in place of invasive biopsies
Target-specific diagnostic biomarkers that can differentiate skin GVHD from other rashes and GI GVHD from other forms of enteritis will replace invasive biopsies. Observational trials in which samples and biopsies taken at GVHD onset are prospectively compared should be the first step. The goal is to replace invasive tests (biopsies) that have specificity and sensitivity > 85% with a blood test that should achieve similar specificity and sensitivity. The outcome evaluated is the presence or absence of GVHD, which has an incidence in most centers of ∼ 50%. If we define similar sensitivity and specificity as being within 15 percentage points of 85% (≥ 70%), 50 patients per arm or a total of 100 patients would be needed to conclude that a blood test measuring biomarkers can replace biopsies.
Study using biomarkers that predict unresponsiveness to GVHD therapy
Another potential clinical application of GVHD biomarkers is to stratify patients based on risk at the time of GVHD onset before initiation of therapy. GI GVHD is considered high risk for mortality, but without further risk stratification, the standard of care for all patients with GI GVHD is prompt initiation of systemic steroid treatment, with second-line agents reserved for patients who fail initial therapy. Unfortunately, most patients who require second-line therapy die, highlighting the need to refine risk beyond the current grading system. The University of Michigan BMT group recently developed a risk stratification algorithm for patients with new-onset GI GVHD that incorporates clinical stage, histologic grade, and plasma levels of the newly discovered GI GVHD biomarker, REG3α.38
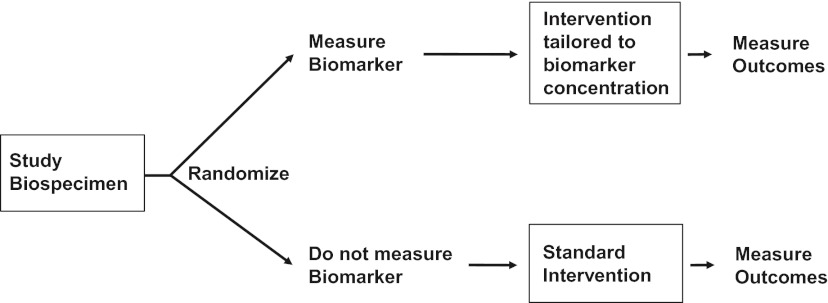
In addition, early identification of patients at high risk for steroid unresponsiveness may allow alternative testing or additional therapies before the development of refractory disease. Martin et al recently highlighted the urgent need to develop well-designed prospective phase 2 treatment trials with a well-established benchmark of success, good statistical plan, and well-defined primary end point for GVHD treatment studies, which still need to be standardized between centers.82 Biomarkers might facilitate decision making if their incorporation can be validated in a prospective randomized trial. To estimate the effectiveness of measuring a biomarker, the trial must be designed such that participants are randomized to different groups: one in which a biomarker is measured and provided to the clinician and another in which it is not (Figure 2). The next step is to include the intervention and control strategies.83 A schema for treating newly diagnosed GVHD using biomarkers is shown in Figure 3. Comparison of box B and box D shows whether additional treatment at onset of GVHD improves response rates and lowers NRM in high-risk patients identified by biomarkers. Comparison of box A and box C shows whether GVHD treatment is less toxic (faster taper and shorter steroids exposition may decrease infection rates) in low-risk patients identified by biomarkers. As previously mentioned, the “best” treatment for treating GVHD is still under debate.82 The expectation is that these markers will also have a specificity and sensitivity similar to diagnostic markers. The outcomes measured in this study are response 28 days after treatment and NRM 180 days after treatment. Approximately 50-70 patients may need to be included in each arm for a total of 200-280 patients in the study.
Figure 2.
Fundamental design for a randomized trial to evaluate biomarker utility.
Figure 3.
Biomarker strategy design for newly diagnosed GVHD.
Study using biomarkers that predict risk for future occurrence of GVHD to preemptively treat GVHD
To determine whether their validated biomarkers can predict GVHD before the appearance of clinical symptoms, Paczesny et al evaluated the 4 most informative biomarkers (ie, IL2R-α, TNFR1, elafin, and REG3α) in day 7 and 14 post-HSCT samples from 513 patients who underwent unrelated HSCT and had not yet developed GVHD. Measurement of this biomarker panel before HSCT predicted grade 2-4 GVHD with a specificity of 75% and sensitivity of 57%.84 Indicators of success must include not only a lower incidence of GVHD but also reductions in infectious complications and relapse. Ultimately, a randomized trial is needed to assess the effectiveness of GVHD preemption. Assuming an early biomarker (eg, at day 14 after HSCT) exists with reasonable specificity and sensitivity, the study design should be defined narrowly, such that a single treatment strategy is clinically reasonable in the absence of the biomarker result. For GVHD, a reasonable treatment strategy would be standard steroid treatment (2 mg/kg per day) or low-dose steroid treatment (1 mg/kg per day), which may produce similar outcomes with less toxicity.85 A schema for a preemptive trial to decrease GVHD incidence using biomarkers is shown in Figure 4. Comparison of box B and box D shows whether preemptive treatment lowers aGVHD in high-risk patients identified by biomarkers. The expectation is that these markers will have a lower specificity and sensitivity than diagnostic markers. It is also likely that at least one or 2 clinical parameters, most likely age and conditioning intensity, will need to be included. The outcomes measured are incidence of GVHD and NRM 180 days after HSCT. Taking an optimistic approach and assuming a simplified algorithm (no more than 4 parameters) with specificity and sensitivity > 80% has been identified, ∼ 80-120 patients may need to be included in each arm for a total of 320-480 patients for the study.
Figure 4.
GVHD preemptive trial design.
Future directions
Future directions include a blinded evaluation of these biomarkers with samples collected in a multicenter prospective study that will reduce center effects and facilitate the successful design of subsequent trials. Ideally, a single formula will be developed to predict a patient's risk for aGVHD, allowing for personalized medicine.
Furthermore, biomarkers may represent novel therapeutic targets that could be inhibited by future aGVHD-specific drugs. Because these drugs would target the appropriate effector T cells, they should increase efficacy and lower toxicity.
GVHD was traditionally categorized as acute (ie, arising before day 100 after HSCT) or chronic (ie, occurring after that time).86,87 This definition was updated by the new National Institutes of Health classification, which includes late-onset aGVHD (after day 100) and an overlap syndrome with features of both aGVHD and cGVHD.88,89 So far, development of post-HSCT biomarkers has focused on aGVHD biomarkers. However, future efforts in biomarker discovery and validation will be particularly valuable for cGVHD.
In conclusion, proteomics is a revolutionary field that includes detection technologies for proteins, which are the molecules that best represent the real-time pathophysiology of alloreactivity. In a short time, proteomics has led to the identification of novel mechanisms of allo-HSCT that probably would not have been discovered by traditional hypothesis-driven research. A promising approach is to use protein biomarkers in risk stratification to better use current treatment modalities. Furthermore, the findings presented in this review demonstrate the potential of biomarkers for exploring targeted therapeutics. The principal barrier to be circumvented is the validation of biomarker concentrations in different allo-HSCT settings, such as conditioning intensity or donor sources (particularly cord blood, T cell–depleted graft). Achieving this aim will require a much larger validation study, ideally a multicenter prospective trial. Once an algorithm for each setting is established, personalized medicine will be possible.
Acknowledgments
The author thanks Drs James Ferrara and Samir Hanash for their guidance and mentorship in discovery and validation of GVHD biomarkers and Drs Thomas Braun and John Levine for their counseling and valuable comments on statistical design.
S.P. was supported by the National Institutes of Health (grants RC1HL101102 and P01-CA039542), and is an investigator of the Eric Hartwell Research Fund and the Amy Strelzer Manasevit Research Program.
Authorship
Contribution: S.P. wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
The current affiliation for S.P. is Bone Marrow and Stem Cell Transplantation Program, Indiana University Melvin and Bren Simon Cancer Center, Riley Hospital for Children, and Wells Center for Pediatric Research, Indianapolis, IN.
Correspondence: Sophie Paczesny, Bone Marrow and Stem Cell Transplantation Program, Indiana University Melvin and Bren Simon Cancer Center and Wells Center for Pediatric Research, 1044 W Walnut St, Rm 425, Indianapolis, IN 46202; e-mail: sophpacz@iupui.edu.
References
- 1.Flowers ME, Inamoto Y, Carpenter PA, et al. Comparative analysis of risk factors for acute graft-versus-host disease and for chronic graft-versus-host disease according to National Institutes of Health consensus criteria. Blood. 2011;117(11):3214–3219. doi: 10.1182/blood-2010-08-302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paczesny S, Krijanovski OI, Braun TM, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113(2):273–278. doi: 10.1182/blood-2008-07-167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu W, Pepe MS. Estimating the diagnostic likelihood ratio of a continuous marker. Biostatistics. 2011;12(1):87–101. doi: 10.1093/biostatistics/kxq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker SG. The central role of receiver operating characteristic (ROC) curves in evaluating tests for the early detection of cancer. J Natl Cancer Inst. 2003;95(7):511–515. doi: 10.1093/jnci/95.7.511. [DOI] [PubMed] [Google Scholar]
- 5.Pepe MS, Longton G. Standardizing diagnostic markers to evaluate and compare their performance. Epidemiology. 2005;16(5):598–603. doi: 10.1097/01.ede.0000173041.03470.8b. [DOI] [PubMed] [Google Scholar]
- 6.Schlatzer DM, Dazard JE, Ewing RM, et al. Human biomarker discovery and predictive models for disease progression for idiopathic pneumonia syndrome following allogeneic stem cell transplantation. Mol Cell Proteomics. 2012;11(6):M111015479. doi: 10.1074/mcp.M111.015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 2012;12(3):3359–3369. doi: 10.3390/s120303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranganathan P, Heaphy CE, Costinean S, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119(20):4786–4797. doi: 10.1182/blood-2011-10-387522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magenau JM, Qin X, Tawara I, et al. Frequency of CD4(+)CD25(hi)FOXP3(+) regulatory T cells has diagnostic and prognostic value as a biomarker for acute graft-versus-host-disease. Biol Blood Marrow Transplant. 2010;16(7):907–914. doi: 10.1016/j.bbmt.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teshima T, Maeda Y, Ozaki K. Regulatory T cells and IL-17-producing cells in graft-versus-host disease. Immunotherapy. 2011;3(7):833–852. doi: 10.2217/imt.11.51. [DOI] [PubMed] [Google Scholar]
- 11.Zinocker S, Sviland L, Dressel R, Rolstad B. Kinetics of lymphocyte reconstitution after allogeneic bone marrow transplantation: markers of graft-versus-host disease. J Leukoc Biol. 2011;90(1):177–187. doi: 10.1189/jlb.0211067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koreth J, Matsuoka K, Kim HT, et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N Engl J Med. 2011;365(22):2055–2066. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau J, Sartor M, Bradstock KF, Vuckovic S, Munster DJ, Hart DN. Activated circulating dendritic cells after hematopoietic stem cell transplantation predict acute graft-versus-host disease. Transplantation. 2007;83(7):839–846. doi: 10.1097/01.tp.0000258731.38149.61. [DOI] [PubMed] [Google Scholar]
- 14.Akhtari M, Giver CR, Ali Z, et al. Receiver operating characteristic curve analysis of circulating blood dendritic cell precursors and T cells predicts response to extracorporeal photopheresis in patients with chronic graft-versus-host disease. Transfusion. 2010;50(11):2424–2431. doi: 10.1111/j.1537-2995.2010.02712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine JE, Paczesny S, Sarantopoulos S. Clinical applications for biomarkers of acute and chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2012;18(1 suppl):S116–S124. doi: 10.1016/j.bbmt.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Court M, Selevsek N, Matondo M, et al. Toward a standardized urine proteome analysis methodology. Proteomics. 2011;11(6):1160–1171. doi: 10.1002/pmic.201000566. [DOI] [PubMed] [Google Scholar]
- 17.Rai AJ, Gelfand CA, Haywood BC, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5(13):3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 18.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics. 2003;2(10):1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Thongboonkerd V. Recent progress in urinary proteomics. Proteomics Clin Appl. 2007;1(8):780–791. doi: 10.1002/prca.200700035. [DOI] [PubMed] [Google Scholar]
- 21.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol. 2004;15(1):219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 22.Tangrea MA, Wallis BS, Gillespie JW, Gannot G, Emmert-Buck MR, Chuaqui RF. Novel proteomic approaches for tissue analysis. Expert Rev Proteomics. 2004;1(2):185–192. doi: 10.1586/14789450.1.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Hwang SI, Thumar J, Lundgren DH, et al. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26(1):65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 24.Kondo T. Tissue proteomics for cancer biomarker development: laser microdissection and 2D-DIGE. BMB Rep. 2008;41(9):626–634. doi: 10.5483/bmbrep.2008.41.9.626. [DOI] [PubMed] [Google Scholar]
- 25.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21(3):255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 26.Wild D, editor. The Immunoassay Handbook. 3rd Ed. Oxford, United Kingdom: Elsevier; 2005. [Google Scholar]
- 27.Schweitzer B, Roberts S, Grimwade B, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20(4):359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boja E, Hiltke T, Rivers R, et al. Evolution of clinical proteomics and its role in medicine. J Proteome Res. 2011;10(1):66–84. doi: 10.1021/pr100532g. [DOI] [PubMed] [Google Scholar]
- 29.Rodland KD. Proteomics and cancer diagnosis: the potential of mass spectrometry. Clin Biochem. 2004;37(7):579–583. doi: 10.1016/j.clinbiochem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan R, Daniels J, Fusaro V, et al. Accurate diagnosis of acute graft-versus-host disease using serum proteomic pattern analysis. Exp Hematol. 2006;34(6):796–801. doi: 10.1016/j.exphem.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Imanguli MM, Atkinson JC, Harvey KE, et al. Changes in salivary proteome following allogeneic hematopoietic stem cell transplantation. Exp Hematol. 2007;35(2):184–192. doi: 10.1016/j.exphem.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 33.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Clouthier SG, Galchev V, et al. Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol Cell Proteomics. 2005;4(5):618–625. doi: 10.1074/mcp.M400126-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321(5895):1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faca V, Coram M, Phanstiel D, et al. Quantitative analysis of acrylamide labeled serum proteins by LC-MS/MS. J Proteome Res. 2006;5(8):2009–2018. doi: 10.1021/pr060102+. [DOI] [PubMed] [Google Scholar]
- 37.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 38.Ferrara JL, Harris AC, Greenson JK, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118(25):6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paczesny S, Braun TM, Levine JE, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Sci Transl Med. 2010;2(13):13ra12. doi: 10.1126/scitranslmed.3000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser T, Kamal H, Rank A, et al. Proteomics applied to the clinical follow-up of patients after allogeneic hematopoietic stem cell transplantation. Blood. 2004;104(2):340–349. doi: 10.1182/blood-2004-02-0518. [DOI] [PubMed] [Google Scholar]
- 41.Brewis IA, Brennan P. Proteomics technologies for the global identification and quantification of proteins. Adv Protein Chem Struct Biol. 2010;80:1–44. doi: 10.1016/B978-0-12-381264-3.00001-1. [DOI] [PubMed] [Google Scholar]
- 42.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44(12):1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 43.Michalski A, Damoc E, Lange O, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11(3):O111013698. doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22(22):2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 45.Rauch A, Bellew M, Eng J, et al. Computational Proteomics Analysis System (CPAS): an extensible, open-source analytic system for evaluating and publishing proteomic data and high throughput biological experiments. J Proteome Res. 2006;5(1):112–121. doi: 10.1021/pr0503533. [DOI] [PubMed] [Google Scholar]
- 46.Pawitan Y, Michiels S, Koscielny S, Gusnanto A, Ploner A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics. 2005;21(13):3017–3024. doi: 10.1093/bioinformatics/bti448. [DOI] [PubMed] [Google Scholar]
- 47.Butte A. The use and analysis of microarray data. Nat Rev Drug Discov. 2002;1(12):951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 48.Berglund L, Bjorling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7(10):2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Addona TA, Abbatiello SE, Schilling B, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27(7):633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(13):1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Huttenhain R, Soste M, Selevsek N, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2012;4(142):142ra194. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toubai T, Tanaka J, Paczesny S, Shono Y, Reddy P, Imamura M. Role of cytokines in the pathophysiology of acute graft-versus-host disease (GVHD): are serum/plasma cytokines potential biomarkers for diagnosis of acute GVHD following allogeneic hematopoietic cell transplantation (Allo-HCT)? Curr Stem Cell Res Ther. 2012;7(3):229–239. doi: 10.2174/157488812799859856. [DOI] [PubMed] [Google Scholar]
- 53.Foley R, Couban S, Walker I, et al. Monitoring soluble interleukin-2 receptor levels in related and unrelated donor allogenic bone marrow transplantation. Bone Marrow Transplant. 1998;21(8):769–773. doi: 10.1038/sj.bmt.1701163. [DOI] [PubMed] [Google Scholar]
- 54.Miyamoto T, Akashi K, Hayashi S, et al. Serum concentration of the soluble interleukin-2 receptor for monitoring acute graft-versus-host disease. Bone Marrow Transplant. 1996;17(2):185–190. [PubMed] [Google Scholar]
- 55.Seidel C, Ringdén O, Remberger M. Increased levels of syndecan-1 in serum during acute graft-versus-host disease. Transplantation. 2003;76(2):423–426. doi: 10.1097/01.TP.0000074316.76104.A5. [DOI] [PubMed] [Google Scholar]
- 56.Shaiegan M, Iravani M, Babaee G, Ghavamzadeh A. Effect of IL-18 and sIL2R on aGVHD occurrence after hematopoietic stem cell transplantation in some Iranian patients. Transpl Immunol. 2006;15(3):223–227. doi: 10.1016/j.trim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Visentainer JEL, Lieber SR, Persoli LBL, et al. Serum cytokine levels and acute graft-versus-host disease after HLA-identical hematopoietic stem cell transplantation. Exp Hematol. 2003;31(11):1044–1050. doi: 10.1016/j.exphem.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 58.Abdallah AN, Boiron JM, Attia Y, Cassaigne A, Reiffers J, Iron A. Plasma cytokines in graft vs host disease and complications following bone marrow transplantation. Hematol Cell Ther. 1997;39(1):27–32. doi: 10.1007/s00282-997-0027-2. [DOI] [PubMed] [Google Scholar]
- 59.Choi SW, Kitko CL, Braun T, et al. Change in plasma tumor necrosis factor receptor 1 levels in the first week after myeloablative allogeneic transplantation correlates with severity and incidence of GVHD and survival. Blood. 2008;112(4):1539–1542. doi: 10.1182/blood-2008-02-138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holler E, Kolb HJ, Moller A, et al. Increased serum levels of tumor necrosis factor alpha precede major complications of bone marrow transplantation. Blood. 1990;75(4):1011–1016. [PubMed] [Google Scholar]
- 61.Schots R, Kaufman L, Van Riet I, et al. Proinflammatory cytokines and their role in the development of major transplant-related complications in the early phase after allogeneic bone marrow transplantation. Leukemia. 2003;17(6):1150–1156. doi: 10.1038/sj.leu.2402946. [DOI] [PubMed] [Google Scholar]
- 62.August KJ, Chiang KY, Bostick RM, et al. Biomarkers of immune activation to screen for severe, acute GVHD. Bone Marrow Transplant. 2011;46(4):601–604. doi: 10.1038/bmt.2010.165. [DOI] [PubMed] [Google Scholar]
- 63.Paczesny S, Choi SW, Ferrara JL. Acute graft-versus-host disease: new treatment strategies. Curr Opin Hematol. 2009;16(6):427–436. doi: 10.1097/MOH.0b013e3283319a6f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schots R, Van Riet I, Othman TB, et al. An early increase in serum levels of C-reactive protein is an independent risk factor for the occurrence of major complications and 100-day transplant-related mortality after allogeneic bone marrow transplantation. Bone Marrow Transplant. 2002;30(7):441–446. doi: 10.1038/sj.bmt.1703672. [DOI] [PubMed] [Google Scholar]
- 65.Malone FR, Leisenring WM, Storer BE, et al. Prolonged anorexia and elevated plasma cytokine levels following myeloablative allogeneic hematopoietic cell transplant. Bone Marrow Transplant. 2007;40(8):765–772. doi: 10.1038/sj.bmt.1705816. [DOI] [PubMed] [Google Scholar]
- 66.Pihusch M, Pihusch R, Fraunberger P, et al. Evaluation of C-reactive protein, interleukin-6, and procalcitonin levels in allogeneic hematopoietic stem cell recipients. Eur J Haematol. 2006;76(2):93–101. doi: 10.1111/j.0902-4441.2005.00568.x. [DOI] [PubMed] [Google Scholar]
- 67.Uguccioni M, Meliconi R, Nesci S, et al. Elevated interleukin-8 serum concentrations in beta-thalassemia and graft-versus-host disease. Blood. 1993;81(9):2252–2256. [PubMed] [Google Scholar]
- 68.Mohty M, Blaise D, Faucher C, et al. Inflammatory cytokines and acute graft-versus-host disease after reduced-intensity conditioning allogeneic stem cell transplantation. Blood. 2005;106(13):4407–4411. doi: 10.1182/blood-2005-07-2919. [DOI] [PubMed] [Google Scholar]
- 69.Hori T, Naishiro Y, Sohma H, et al. CCL8 is a potential molecular candidate for the diagnosis of graft-versus-host disease. Blood. 2008;111(8):4403–4412. doi: 10.1182/blood-2007-06-097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piper KP, Horlock C, Curnow SJ, et al. CXCL10-CXCR3 interactions play an important role in the pathogenesis of acute graft-versus-host disease in the skin following allogeneic stem-cell transplantation. Blood. 2007;110(12):3827–3832. doi: 10.1182/blood-2006-12-061408. [DOI] [PubMed] [Google Scholar]
- 71.Okamoto T, Takatsuka H, Fujimori Y, Wada H, Iwasaki T, Kakishita E. Increased hepatocyte growth factor in serum in acute graft-versus-host disease. Bone Marrow Transplant. 2001;28(2):197–200. doi: 10.1038/sj.bmt.1703095. [DOI] [PubMed] [Google Scholar]
- 72.Luft T, Conzelmann M, Benner A, et al. Serum cytokeratin-18 fragments as quantitative markers of epithelial apoptosis in liver and intestinal graft-versus-host disease. Blood. 2007;110(13):4535–4542. doi: 10.1182/blood-2006-10-049817. [DOI] [PubMed] [Google Scholar]
- 73.Reddy P, Sun Y, Toubai T, et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest. 2008;118(7):2562–2573. doi: 10.1172/JCI34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landfried K, Zhu W, Waldhier MC, et al. Tryptophan catabolism is associated with acute GVHD after human allogeneic stem cell transplantation and indicates activation of indoleamine 2,3-dioxygenase. Blood. 2011;118(26):6971–6974. doi: 10.1182/blood-2011-06-357814. [DOI] [PubMed] [Google Scholar]
- 75.Luft T, Dietrich S, Falk C, et al. Steroid-refractory GVHD: T-cell attack within a vulnerable endothelial system. Blood. 2011;118(6):1685–1692. doi: 10.1182/blood-2011-02-334821. [DOI] [PubMed] [Google Scholar]
- 76.Rezvani AR, Storer BE, Storb RF, et al. Decreased serum albumin as a biomarker for severe acute graft-versus-host disease after reduced-intensity allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2011;17(11):1594–1601. doi: 10.1016/j.bbmt.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weissinger EM, Schiffer E, Hertenstein B, et al. Proteomic patterns predict acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Blood. 2007;109(12):5511–5519. doi: 10.1182/blood-2007-01-069757. [DOI] [PubMed] [Google Scholar]
- 78.Harris AC, Ferrara JL, Braun TM, et al. Plasma biomarkers of lower gastrointestinal and liver acute GVHD. Blood. 2012;119(12):2960–2963. doi: 10.1182/blood-2011-10-387357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levine JE, Logan BR, Wu J, et al. Acute graft-versus-host disease biomarkers measured during therapy can predict treatment outcomes: a Blood and Marrow Transplant Clinical Trials Network study. Blood. 2012;119(16):3854–3860. doi: 10.1182/blood-2012-01-403063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Gruttola VG, Clax P, DeMets DL, et al. Considerations in the evaluation of surrogate endpoints in clinical trials: summary of a National Institutes of Health workshop. Control Clin Trials. 2001;22(5):485–502. doi: 10.1016/s0197-2456(01)00153-2. [DOI] [PubMed] [Google Scholar]
- 81.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 82.Martin PJ, Rizzo JD, Wingard JR, et al. First- and second-line systemic treatment of acute graft-versus-host disease: recommendations of the American Society of Blood and Marrow Transplantation. Biol Blood Marrow Transplant. 2012;18(8):1150–1163. doi: 10.1016/j.bbmt.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McShane LM, Hunsberger S, Adjei AA. Effective incorporation of biomarkers into phase II trials. Clin Cancer Res. 2009;15(6):1898–1905. doi: 10.1158/1078-0432.CCR-08-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paczesny S, Braun T, Lugt MV, et al. Three biomarker panel at day 7 and 14 can predict development of grade II-IV acute graft-versus-host disease [abstract]. Blood (ASH Annual Meeting Abstracts) 2010;116(21):675. [Google Scholar]
- 85.Mielcarek M, Storer BE, Boeckh M, et al. Initial therapy of acute graft-versus-host disease with low-dose prednisone does not compromise patient outcomes. Blood. 2009;113(13):2888–2894. doi: 10.1182/blood-2008-07-168401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin P, Schoch G, Fisher L, et al. A retrospective analysis of therapy for acute graft-versus-host disease: initial treatment. Blood. 1990;76(8):1464–1472. [PubMed] [Google Scholar]
- 87.Sullivan KM, Mori M, Sanders J, et al. Late complications of allogeneic and autologous marrow transplantation. Bone Marrow Transplant. 1992;10(Suppl 1):127–134. [PubMed] [Google Scholar]
- 88.Filipovich AH, Weisdorf D, Pavletic S, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transplant. 2005;11(12):945–956. doi: 10.1016/j.bbmt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 89.Griffith LM, Pavletic SZ, Lee SJ, Martin PJ, Schultz KR, Vogelsang GB. Chronic graft-versus-host disease: implementation of the National Institutes of Health Consensus Criteria for Clinical Trials. Biol Blood Marrow Transplant. 2008;14(4):379–384. doi: 10.1016/j.bbmt.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liem LM, van Lopik T, van Houwelingen HC, Aarden L, Goulmy E. Soluble Fas levels in sera of bone marrow transplantation recipients are increased during acute graft-versus-host disease but not during infections. Blood. 1998;91(4):1464–1468. [PubMed] [Google Scholar]
- 91.Fujimori Y, Takatsuka H, Takemoto Y, et al. Elevated interleukin (IL)-18 levels during acute graft-versus-host disease after allogeneic bone marrow transplantation. Br J Haematol. 2000;109(3):652–657. doi: 10.1046/j.1365-2141.2000.02095.x. [DOI] [PubMed] [Google Scholar]