Abstract
Background
Plasmodium falciparum the main causative agent of malaria is an important public health vector. With the use of PCR, its genetic diversity has been extensively studied with dearth information from Nigeria.
Methods
In this study, 100 P. falciparum strains merozoite surface protein 1(msp-1), merozoite surface protein 2 (msp-2) and Glutamate rich protein (Glurp) from Ogun State General Hospitals were characterized. The genetic diversity of P. falciparum isolates was analyzed by restriction fragment length polymorphism following gel electrophoresis of DNA products from nested polymerase chain reactions (PCR) of their respective allelic families KI, MAD 20, RO33 (MSP-1);FC27, 3D7 (MSP-2) and Glutamate rich protein respectively.
Results
Majority of the patients showed monoclonal infections while multiplicity of the infection for msp-1 and msp-2 were 1.1 and 1.2 respectively. The estimated number of genotypes was 8 msp-1 (4 KI; 3 MAD; 1 RO33) and 6 msp-2 (3 FC27; 3 3D7). 80% of the isolates coded for Glurp with allelic size ranged between 700 and 900 bp.
Conclusion
The allelic distributions however were similar to those previously reported in other endemic malaria countries. Future studies will be designed to include other malaria endemic regions of Nigeria such as the oil exploration regions.
Keywords: Genetic diversity, Plasmodium, Merozoite surface protein, monoclonal infection
Introduction
Malaria remains an important public health concern in countries where transmission occurs regularly; 90% of the global burden of malaria occurs in Africa, south of Sahara1,2. Each year an estimated 300 to 650 million clinical cases of malaria occur, making it one of the most common infectious diseases worldwide3. Young children, pregnant women, and non-immune visitors to malarious areas are at greatest risk of severe or fatal illness4. Information on the nature and extent of genetic diversity within P falciparum is essential in understanding the mechanism underlying the pathology of malaria, the acquisition of immunity, the spread of drug resistance, the condition of transmission and the development of vaccines against the parasite5,6,7. However, extensive genetic diversity in natural parasite populations is a major obstacle for the development of an effective vaccine against the human malaria parasite, since antigenic diversity limits the efficacy of acquired protective immunity to malaria4. Molecular techniques have facilitated the studies on genetic diversity of Plasmodium species particularly from field isolates collected directly from patients. Merozoite surface Proteins 1 and 2 (MSP1 and MSP2) are among the membrane proteins characterized to date that are present on the surface of P falciparum merozoites8,9. They have been shown harboring antigenic and specific antibodies directed against MSP-2 epitopes that inhibit merozoite invasion of erythrocytes during in vitro experiments10. Restriction fragment length polymorphism and polymerase chain reaction (PCR) genotyping using the three gene loci (MSP1, MSP2 and Glurp) have been used to distinguish treatment failures from newly acquired infections11.
It has been established that the msp-1 gene is divided into 17 blocks, based on analysis of sequence diversity: seven highly variable blocks are interspersed with five conserved and five semi-conserved regions. Block 2 of the msp-1 gene appears to be subjected to rapid intragenic recombination, and so, is highly polymorphic4. The msp-2 gene, also known as merozoite surface antigen (msa-2) gene, codes for a merozoite surface polymorphic glycoprotein that has been widely studied as one of the major vaccine candidates. The sequencing of DNA has shown that a single copy of msp-2 gene has conserved N- and C-terminal domains (blocks 1 and 5), two non-repetitive variable regions (blocks 2 and 4), and a polymorphic central region (block 3) containing variable numbers of tandem repeats, which also vary in sequence and length 12,13,14. Genes in which polymorphism has arisen through intragenic recombination in repetitive segments are characterized by repeat motifs with length variability differing between strains.
Two highly polymorphic and widely used markers are msp-1 and msp-2 and the large allelic polymorphism has been reported in the block 2 of the msp-1 gene and the central repetitive domain (block3) of the msp-2 gene. Families differing in nucleotide sequences and in number of repetitive sequences (length variation) were used for genotyping purposes. In msp-1 (block 2) three distinct allelic families have been described: K1, MAD20 and RO33 while msp-2 has two distinct families, 3D7 and FC27.
Studies on genetic diversity, differentiation of the different strains within a Plasmodium species and presence of multiple parasite strains in individual host have been reported from different regions of the globe5,11,14,15,16,17. However, limited reports are available on the genetic diversity existing among P. falciparum population of Nigeria. In this report, polymorphic markers in P. falciparum isolates were used to examine genetic diversity and complexity of parasite populations in patients with uncomplicated malaria infections in Ogun State, South Western Nigeria.
Methods
Study setting
Ogun State is in the transitional zone between the tropical rain forest and derived savannah zone in the southwest, Nigeria. The area experiences two seasons, the dry season (November to March) and the wet season (April to October). The annual temperature range from 22.8 °C to 34.9 °C and the mean annual rainfall is about 107.3 mm. Malaria is present throughout the year with a marked increase during the raining season19. The State was divided into 4 zones for sample collection. This was based on geopolitical and health zoning. The 4 zones are Sango - Ota (Yewa), Abeokuta (Egba), Ijebu Ode (Ijebu) and Sagamu (Remo).
Sample collection
A total of 4066 subjects comprising of 1839 males and 2227 females presenting with malaria in 4 different health districts of Ogun State were sampled for malaria screening. Safety procedures were adopted in the collection of finger-prick blood samples by swabbing the area to be sampled with 70% alcohol and allowed to dry before collection. 100 subjects were included in this pilot study for molecular characterization of P falciparum. The criteria for inclusion in the study were fever or a history of fever in the 24 – 48 hours preceding presentation; parasite density of more than 2,000 asexual forms per microliter of blood confirmed by microscopy; no history of antimalarial drug ingestion two weeks preceding presentation. The bleeding (venepuncture) was done in the hospitals by clinicians/Medical laboratory scientists and blood was transported to the laboratory in heparin bottles at 4 °C. Peripheral blood was collected on 1.5 × 7.0-cm strips of Whatman (Brentford, United Kingdom) 3 MM filter paper and in heparinised bottles. The strips were then air-dried and kept in plastic bags until use.
Ethical consideration
Scientific and Ethical permit/clearance was obtained from the Nigerian Institute of Medical Research Institutional Review Board (NIMR-IRB) and Covenant University Ethics Committee. The Ogun State Ministry of health (Hospitals Management Board) was also informed before this research was carried out. Written informed consent was obtained from patients prior to recruitment into this study. Consent for the children was provided by the parents/guardians while some of the children participants provided the assents by nodding and thumb printing.
Microscopic examination
Thick and thin films of the blood samples were prepared. Thin films were fixed with methanol and all films were stained with 3% Giemsa stain of pH 7.0 for 30 min as recommended by Cheesbrough20. Blood films were examined microscopically using 100X (oil immersion) objectives as described by Cheesbrough20. The thick films were used to determine the parasite densities while thin films were used to identify the parasite species and infective stages. Parasite density per microliter of blood was estimated from the thick film by taking the number of leucocytes per microliter of blood as 8,000 and expressed as follows:
DNA extraction and Molecular analyses
DNA was extracted from patient blood samples on 3MM Whatman filter paper (Brentford, United Kingdom) using the QiaAmp DNA Blood Minikit Blood and Body Fluid Spin Protocol (Qiagen, Valencia, CA). Molecular Genotyping of isolates using merozoite surface proteins 1 and 2 (msp1 and msp2) and Glutamate rich protein (Glurp) primers and PCR protocols were followed as previously described by Snounou et al9 for family specific allele analysis of msp-1 (block 2), msp-2 (block 3) and Glurp. PCR amplification was performed on thermal cycler (Perkin Elmer 9700/2400, UK) in a final volume of 20 /µl. The PCR products were visualized by UV transillumination at 302 nm on gel documentation system (Syngenta, USA) after electrophoresis on 2% agarose gel (Promega/ Boehringer) using 0.5 × TBE buffer at 80–100 volts. Allele sizes were calculated using Gene tool programme.
Data interpretation
Based on the visual inspection of amplified fragments separated by agarose gel electrophoresis, alleles were classified according to the number of fragments, size of each fragment, and allelic family to which each fragment belongs. Fragment sizes were estimated by comparing with molecular size standards. Positive controls were set up using characterized P. falciparum isolates. To understand the identity of Nigerian isolates with respect to isolates of other regions, sequence data available in public domains were downloaded for allelic families of msp-1 & 2 and details are given below;
Thailand (K1-M77730, MAD20-M77722, R033-AAA29684, 3D7-U91676),
Vietnam (K1-AF509651, MAD20-AF509653&94, FC27-AF104696, 3D7-AF104693),
Tanzania (K1-AF061134, FC27-AY532386),
Brazil (K1-AF509714, MAD20-AY714585, FC27-DQ115973, 3D7-AF177389),
China (MAD20-AF251345),
Sudan (MAD20-AF034635),
Iran (MAD20-AY138509, R033-AY138507, FC27DQ338451),
Indonesia (K1-AF191061, R033-AAF18431),
Western Africa (R033-PFAMSA1),
Kenya (R033-AAM21583),
Ghana (FC27-AF329577), PNG(FC27-AF329579),
Gambia (FC27-U91668, 3D7-U91665)
Nigeria (3D7-AF148224)
Statistical analysis
Statistical analysis was performed using Stat View 5.0.1 (SAS Institute Inc., Cary, NC). Statistical significance was based on á level of 0.05.
Results
One hundred P. falciparum isolates analyzed during the study have demonstrated highly diverse nature of field isolates in respect of msp-1 (block 2) and msp-2 (central repeat region, block3).
As an estimate of multiclonality of infection, the minimal number of P. falciparum strains in each individual was determined by assessing the number of alleles of the genes encoding merozoite surface protein 1 (MSP-1) and MSP-2 families (table 1). The MSP-1 and MSP-2 alleles were defined by gene family-specific PCR assays and analysis of the resulting length polymorphisms of the PCR products
Table 1.
Genetic diversity of Plasmodium falciparum isolates from Ogun State, South Western Nigeria
| Loci | No. positive by PCR (%) |
No. of distinct alleles |
Sizes of alleles (bp) |
MOI |
| MSP-1 | ||||
| K1 | 68 (68) | 4 | 100–300 | |
| MAD20 | 40 (40) | 3 | 100–300 | 2.1 |
| RO33 | 20 (20) | 1 | 200 | |
| MSP-2 | ||||
| 3D7 | 76 (76) | 3 | 450–600 | |
| FC27 | 56 (56) | 3 | 300–500 | 2.2 |
| GLURP | 80 (80) | 5 | 700–900 | 1.3 |
* bp _ base pairs; MSP _ merozoite surface protein; GLURP _ glutamate-rich protein. MOI = Multiplicity of Infection
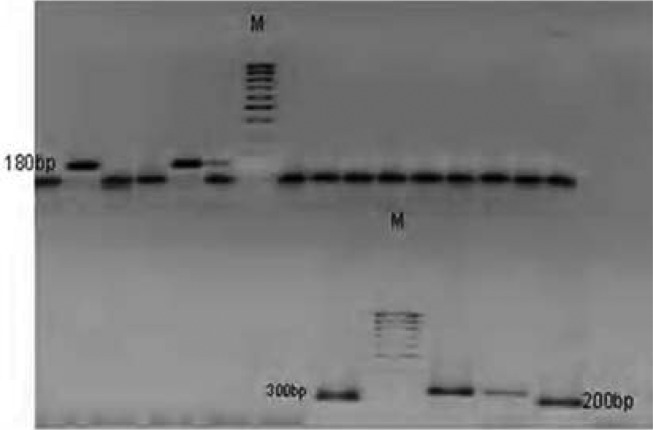
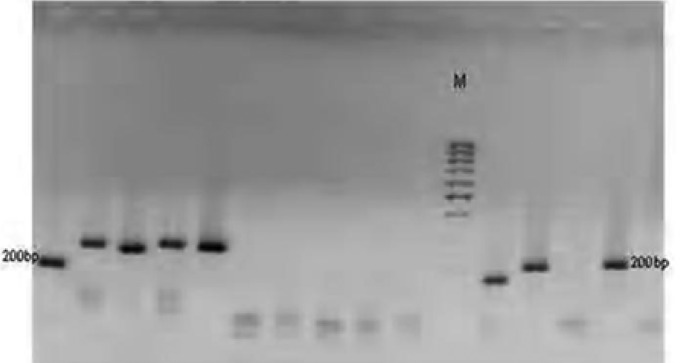
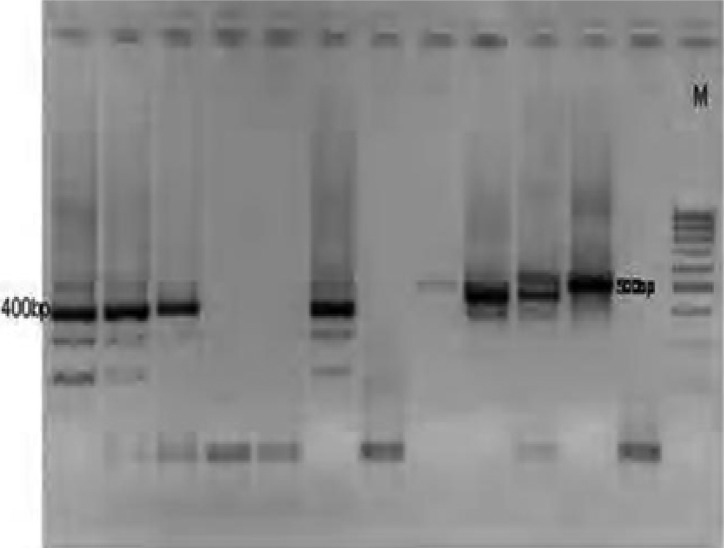
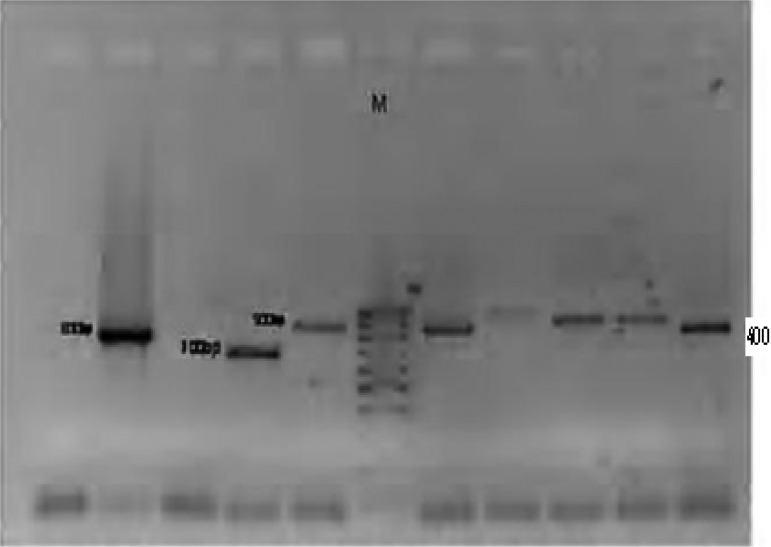
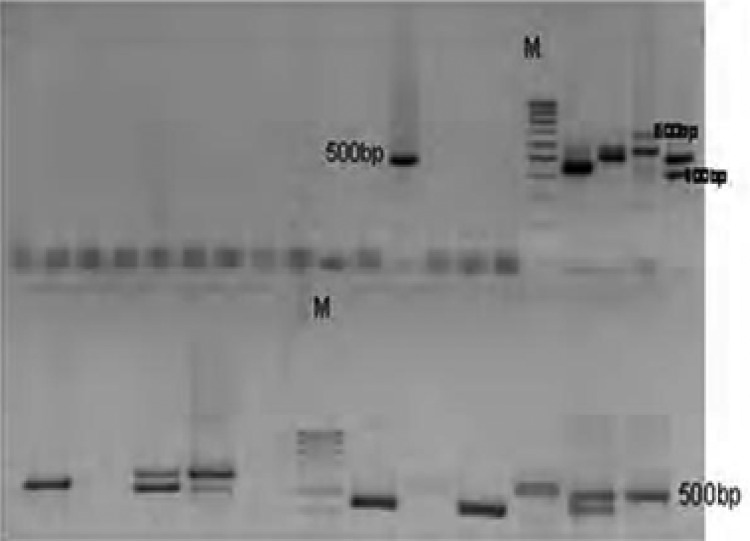
One hundred P. falciparum isolates analyzed during the study have demonstrated highly diverse nature of field isolates in respect of msp-1 (block 2) and msp-2 (central repeat region, block3). All the three reported families of msp-1(K1, MAD20 and RO33) as shown on plates 1–3 and two of msp-2 (FC27 and 3D7), were observed among the isolates (Plates 5 and 6). DNA bands of the P. falciparum MSPP2 3D& are shown in plate 4. Proportion of isolates with K1 family was 68 % with 4 alleles in the range of 100 to 300 bp. Proportion of isolates with MAD20 family was 40 % and a total of 3 alleles were observed within 100 to 300 bp. RO33 proportion was 20 % and the family was observed to be monomorphic with an allele size of 200 bp. Observed proportions, numbers and size range of alleles among the isolates are given in table 1.
Plate 1.
DNA bands of P. falciparum MSP1 MAD20 on gel
Plate 3.
DNA bands of P. falciparum MSP1 RO33 on gel
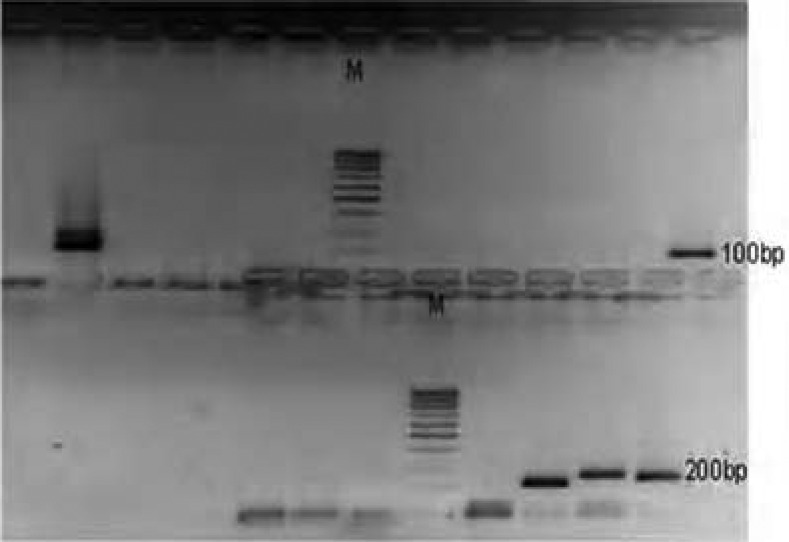
Plate 5.
DNA bands of P. falciparum Merozoite surface protein FC27 on gel
Plate 6.
DNA bands of P. falciparum Glutamate rich protein
Plate 4.
DNA bands of P. falciparum MSP2 3D7 on gel
Plate 2.
DNA bands of P. falciparum MSP1K1 on gel
In msp-2, the reported families FC27 and 3D7 were observed among the isolates (Table 2). Proportion of FC27 family was 76 % and that of 3D7 was 56 %. Proportional prevalence of FC27 and 3D7 families was significantly different (chi square = 16.5, P = 0.002). Three alleles of FC27 in the range of 300–500 bp and 3 alleles of 3D7 in the range of 450–600 bp were observed. Proportion of multiclonal isolates (multiple infection) is given in table 2 and multiplicity of the infection (MOI) was estimated by dividing the total number of fragments detected in the individual system by the number of samples positive in the particular system (either msp-1 or msp-2). Eighty percent of the isolates harboured the glutamate rich protein with allelic size ranging from 700–900 bp
Table 2.
Zone wise genetic diversity of P. falciparum from Ogun State, South western Nigeria
| Zone | No of Isolates | MSP 1 | MSP 2 | ||||
| K1 (%) | MAD20 (%) | RO33 (%) | 3D7 (%) | FC27 (%) | Glurp (%) | ||
| Ijebu | 20 | 14 (20.6) | 9 (22.5) | 6 (30) | 16 (21.1) | 13 (23.2) | 16 (20) |
| Yewa | 43 | 31 (45.6) | 21 (52.5) | 8 (40) | 38 (50) | 18 (32.1) | 33 (41.3) |
| Egba | 25 | 15 (22.1) | 7 (17.5) | 4 (20) | 12 (15.8) | 18 (32.1) | 21(26.3) |
| Remo | 12 | 8 (11.8) | 3 (7.5) | 2 (10) | 10 (13.2) | 7 (12.5) | 10 (12.5) |
| Total | 100 | 68 (100) | 40 (100) | 20 (100) | 76 (100) | 56 (100) | 80 (100) |
Discussion
In this study, polymorphic markers in P. falciparum isolates were used to examine genetic diversity and complexity of parasite populations in patients with uncomplicated malaria infections in Ogun State, South Western Nigeria. It bridges an important gap in the understanding of the molecular characteristics of the parasite population in Ogun State. As limited reports are available on the genetic diversity among the P falciparum population of Nigeria, this pilot study evaluated the extent of genetic diversity in the field isolates of P. falciparum in South Western Nigeria.
The Merozoite Surface Protein1 (MSP-1) and Merozoite Surface Protein2 (MSP-2) are highly polymorphic markers and the large allelic polymorphism has been reported in the block 2 of the msp-1 gene and the central repetitive domain (block3) of the msp-2 gene. Families differing in nucleotide sequences and in number of repetitive sequences (length variation) were used for genotyping purposes.
The population structure of P. falciparum isolates analyzed with the polymorphic markers (MSP-1), MSP-2, and Glutamate Rich Protein (GLURP) in this study showed extensive diversity in parasite populations in the four zones in Ogun State, South Western Nigeria. The MSP-1, MSP-2, and GLURP markers showed 8, 6, and 5 allelic families respectively (Table 1). This diversity of the P. falciparum population in South Western Nigeria is reflected in the complexity of parasite populations in the samples. A catalog of genetically distinct parasite populations co-infecting those infected with malaria, based on PCR amplification of the Glutamate rich protein (Glurp), Merozoite Surface Protein 1 (MSP-1), and Merozoite surface Protein 2 (MSP-2) markers showed that multiplicity of infection was common. Multiclonality of infections has been shown to be a common feature in most malaria-endemic areas20,21,22.23. Epidemiologic data from some study sites in Africa suggest that the multiplicity of P. falciparum infection may be directly related to the intensity of transmission21,22,23.
The multiplicity of infection observed in this study is high, compared to the ones observed in similar studies from other regions 8,9,17,18,20,21. This suggests that the intensity of transmission of malaria in this region is high. This multiplicity of infections may also have important implications for the epidemiology of drug-resistant P. falciparum malaria and the outcome of treatment in patients. The initial presence of several parasite populations with different drug response profiles would result in elimination of drug sensitive populations and selection of resistant parasites.
The findings in this study are in agreement with previous studies involving asymptomatic carriers and symptomatic patients in holoendemic areas, both in the complexity of population structure and multiplicity of parasites in human hosts5,7,8,9. All the MSP 1 and MSP2 families were represented in each of the four zones in Ogun State, South Western Nigeria where samples were collected. Genotyping of these samples showed that the allelic families of MSP-1 and MSP-2 were often represented in parasite DNA derived from a single patient, indicating a polyclonal infection. Specifically, three allelic families of MSP-1 and two of MSP-2 were assessed respectively. Alleles were classified according to the size of PCR fragments. Prevalence of identical allelic composition of alleles in the four study areas of the Ogun State, Nigeria suggests that there is a considerable amount of gene flow between the P. falciparum populations of different zones.
Conclusion
The present study shows that field isolates of Ogun State, South Western Nigeria are highly diverse in respect of msp-1 (block 2) and msp-2 (central repeat region, block 3) and glutamate rich protein. This pilot study has shown the diversity and complexity of P. falciparum populations in South Western, Nigeria. The MSP-2 marker appears to be the most reliable genetic marker to evaluate diversity and complexity of P. falciparum infections8,23. This approach in addition to other molecular methods may be useful as part of the surveillance program for monitoring drug-resistant malaria infections.
Conflict of interest
None declared.
Acknowledgements
This work was supported by Third World organization for women in Science (TWOWS), National Institute of Malaria Research, Delhi, India and Covenant University, Nigeria. We are also grateful to Board of NIMR for their support.
References
- 1.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman F, Batchelor A. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol Microbiol. 2004;52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 2.Takala SL, Escalante AA, Branch OH, Kariuki S, Biswas S, Chaiyaroj SC, Lal AA. Genetic diversity in the Block 2 region of the merozoite surface protein 1 (MSP-of Plasmodium falciparum: additional complexity and selection and convergence in fragment size polymorphism. Infect Genet Evol. 2006;6:417–424. doi: 10.1016/j.meegid.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hay S, Guerra C, Tatem A, Noor A, Snow R. “The global distribution and population at risk of malaria: past, present, and future”. Lancet. 2004;4(6):327–336. doi: 10.1016/S1473-3099(04)01043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourdy M, Willcox L, Ginsburg H, Rasoanaivo P, Graz B, Deharo E. Ethnopharmacology and malaria: New hypothetical leads or old efficient antimalarials? International Journal for Parasitology. 2008;38:33–41. doi: 10.1016/j.ijpara.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Basco LK, Tahar R, Escalante A. Molecular epidemiology of malaria in Cameroon. Xviii. Polymorphisms of the Plasmodium falciparum Merozoite Surface Antigen-2 gene in isolates from symptomatic patients. Am J Trop Med Hyg. 2004;70(3):238–244. [PubMed] [Google Scholar]
- 6.Yah CS, Fatumo S. In-silico studies of multi drug resistance [MDR] genetic markers of Plasmodium species. J Comput Biol Bioinform Res. 2010;2(1):005–009. [Google Scholar]
- 7.Basco LK, Ringwald P. Molecular epidemiology of malaria in Yaounde, Cameroon. VII. Analysis of recrudescence and reinfection in patients with uncomplicated falciparum malaria. Am J Trop Med Hyg. 2000;63:215–221. [PubMed] [Google Scholar]
- 8.Happi CT, Gbotosho GO, Sowunmi A, Falade CO, Akinboye BO, Gerena L, Kyle KE, Milhous W, Wirth DF, Oduola AMJ. Molecular Analysis of Plasmodium falciparum recrudescent malaria infections in children treated with chloroquine in Nigeria. Am J Trop Med Hyg. 70(1):20–26. [PubMed] [Google Scholar]
- 9.Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown K, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 10.Fatumo SA, Yah CS. In-silico detection of chokepoint enzymes in four Plasmodium species. J Pharm and Allied Sci. 2009;6(3):632–640. [Google Scholar]
- 11.Smythe JA, Coppel RL, Brown GV, Ramasamy R, Kemp DJ, Anders RF. Identification of two integral membrane proteins of Plasmodium falciparum. Proc Natl Mol Biochem Parasitol. 1988;34:79–86. doi: 10.1073/pnas.85.14.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenton B, Clark JT, Wilson CF, McBride JS, Walliker D. Polymorphism of a 35–48 kDa Plasmodium falciparum merozoite surface antigen. Acad Sci. 1989;85:5195–5199. doi: 10.1016/0166-6851(89)90022-4. [DOI] [PubMed] [Google Scholar]
- 13.Berzins K, Anders RF. The malaria antigens. In: Wahlgren M, Perlmann P, editors. Malaria: Molecular and Clinical Aspects. Amsterdam: Harwood Academic Publisher; 1999. pp. 181–216. [Google Scholar]
- 14.Ohrt C, Mirabelli-Primdahl L, Karnasuta C, Chantakulkij S, Kain KC. Distinguishing Plasmodium falciparum treatment failures from reinfections by restrictions fragment length polymorphism and polymerase chain reaction genotyping. Am J Trop Med Hyg. 1997;57:430–437. doi: 10.4269/ajtmh.1997.57.430. [DOI] [PubMed] [Google Scholar]
- 15.Basco LK, Tahar R, Escalante A. Molecular epidemiology of malaria in Cameroon. XVIII. Polymorphisms of the Plasmodium falciparum merozoite surface antigen-2 gene in isolates from symptomatic patients. Am J Trop Med Hyg. 2004;70(3):238–244. [PubMed] [Google Scholar]
- 16.Gertrude N K. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997–2007. J Vector Borne Dis. 2009;46:1–12. [PubMed] [Google Scholar]
- 17.Felger I, Irion A, Steiger S, Beck HP. Genotypes of merozoite surface protein 2 of Plasmodium falciparum in Tanzania. Trans R Soc Trop Med Hyg. 1999;93(Suppl 1):3–9. doi: 10.1016/s0035-9203(99)90320-6. [DOI] [PubMed] [Google Scholar]
- 18.Eisen D, Billman-Jacobe H, Marshall VF, Fryauff D, Coppel RL. Temporal variation of the merozoite surface protein-2 gene of Plasmodium falciparum. Infect Immun. 1998;66:239–246. doi: 10.1128/iai.66.1.239-246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojo DA. Malaria in Endemic Area of Ogun State, Nigeria: Investigation of Perceptions and practices among the residents and health providers. Pakistan Journal of Social Sciences. 2005;3(4):564–571. [Google Scholar]
- 20.Cheesbrough M. District Laboratory Practice Manual in Tropical Countries Part 2. Cambridge University Press; 2000. pp. 178–179. [Google Scholar]
- 21.Happi TC, Thomas SM, Gbotosho GO, Falade CO, Akinboye DO, Gerena L, Hudson T, Sowunmi A, Kyle DE, Milhous W, Wirth DF, Oduola AM. Point mutations in the pfcrt and pfmdr-1 genes of Plasmodium falciparum and clinical response to chloroquine, among malaria patients from Nigeria. Ann Trop Med Parasitol. 2003;97:439–451. doi: 10.1179/000349803235002489. [DOI] [PubMed] [Google Scholar]
- 22.Babiker HA, Lines J, Hill WG, Walliker D. Population structure of Plasmodium falciparum in villages with different malaria endemicity in East Africa. Am J Trop Med Hyg. 1997;6:141–147. doi: 10.4269/ajtmh.1997.56.141. [DOI] [PubMed] [Google Scholar]
- 23.Ghanchi NK, Mårtensson A, Ursing J, Jafri S, Bereczky S, Hussain R, Be MA. Genetic diversity among Plasmodium falciparum field isolates in Pakistan measured with PCR genotyping of the merozoite surface protein 1 and 2. Malaria Journal. 2010;9:1. doi: 10.1186/1475-2875-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]