Abstract
Sulfotransferase catalyzed sulfation regulates the biological activities of various neurotransmitters/hormones and detoxifies xenobiotics. Rat sulfotransferase rSULT1A1 catalyzes the sulfation of neurotransmitters and xenobiotic phenolic compounds. rSULT2A1 catalyzes the sulfation of hydroxysteroids and xenobiotic alcoholic compounds. In this work, Western blot and real-time RT-PCR were used to investigate the effect of methamphetamine on rSULT1A1 and rSULT2A1 protein and mRNA expression in rat cerebellum, frontal cortex, hippocampus, and striatum. After 1-day treatment, significant induction of rSULT1A1 was observed only in the cerebellum; rSULT2A1 was induced significantly in the cerebellum, frontal cortex, and hippocampus. After 7-days of exposure, rSULT1A1 was induced in the cerebellum, frontal cortex, and hippocampus, while rSULT2A1 was induced significantly in all four regions. Western blot results agreed with the real-time RT-PCR results, suggesting that the induction occurred at the gene transcriptional level. Results indicate that rSULT1A1 and rSULT2A1 are expressed in rat frontal cortex, cerebellum, striatum, and hippocampus. rSULT1A1 and rSULT2A1are inducible by methamphetamine in rat brain sections in a time dependable manner. rSULT2A1 is more inducible than rSULT1A1 by methamphetamine in rat brain sections. Induction activity of methamphetamine is in the order of cerebellum > frontal cortex, hippocampus > striatum. These results suggest that the physiological functions of rSULT1A1 and rSULT2A1 in different brain regions can be affected by methamphetamine.
Keywords: sulfotransferase, methamphetamine, gene regulation, rat brain sections, rSULT1A1, rSULT2A1
1. INTRODUCTION
Cytosolic sulfotransferases (SULTs) catalyze the sulfation of a wide range of hydroxyl-containing compounds (Bojarova and Williams, 2008, Chapman et al., 2004, Gamage et al., 2006, Hempel et al., 2007, Kauffman, 2004, Rath et al., 2004, Runge-Morris and Kocarek, 2005b, Wang and James, 2006). Various bio-signaling molecules including neurotransmitters, hydroxysteroids, and glucocorticoids, are regulated by sulfation. Sulfation usually leads to the inactivation of biological signaling molecules, as the sulfated forms are usually unable to bind to receptors. Sulfation of xenobiotics usually leads to detoxification, as sulfated xenobiotics are readily excreted. Although SULTs are most highly expressed in the liver, they are present in many tissues, including the small intestine, brain, adrenal gland, kidney, skin, blood platelets and nucleated blood cells (Alnouti and Klaassen, 2006, Nowell and Falany, 2006, Richard et al., 2001). SULTs are divided into two major subfamilies, phenol SULTs (SULT1) and hydroxysteroid SULTs (SULT2) (Blanchard et al., 2004). Several of the SULTs have been identified in human brain tissues, and the phenol SULTs have been detected both in neurons and glial cells in several different brain regions, such as the cerebellum, hippocampus, stratium, and so on (Alnouti and Klaassen, 2006, Richard, Hume, 2001, Salman et al., 2009, Vietri et al., 2003, Whittemore et al., 1986, Yasuda et al., 2007, Zou et al., 1990). rSULT1A1 shows a very broad distribution and broad substrate specificity (Duffel, 1994, Riches et al., 2007). It is one of the major SULT1 enzymes. It is responsible for the sulfation of neurotransmitters and xenobiotic phenolic compounds. Hydroxysteroid SULTs (SULT2, including SULT2A and SULT2B subfamilies) have also been identified in the brain, catalyzing the sulfation of hydroxyl-containing neurosteroids such as androsterone and allopregnanolone and xenobiotic alcohol compounds (Shimizu et al., 2003, Shimizu and Tamura, 2002). SULT2A1 is one of the major SULT2 enzymes (Fang et al., 2007, Liu et al., 2006). It is also widely distributed and responsible for the sulfation both xenobiotics and hydroxysteroids. rSULT1A1 and rSULT2A1, two of the major SULTs, representing the two major subfamilies, are selected for the studies.
Methamphetamine (METH) is one of the most abused psychostimulants in the world (Brackins et al., 2011). METH administration has been shown to produce long-term decreases in numerous measures of both dopaminergic and serotonergic function such as enzyme activity, monoamine content and monoamine transmitters in experimental animals as well as in human (Gold et al., 2009, Guilarte et al., 2003, Kish, 2008, Kita et al., 2003b, Krasnova and Cadet, 2009, Quinton and Yamamoto, 2006). The pharmacological and toxic effect of METH is also dependent on treatment methods. Acute administration of METH produces locomotor activation (Gaytan et al., 1998), and repeated administration produces a progressive sensitization of this behavioral activation until stereotyped behavior comes to predominate the behavioral repertoire (Robinson and Berridge, 1993). Chronic or intermittent METH abuse may create temporary or permanent disturbances in the dopaminergic systems of the brain that may predispose individuals to Parkinsonism. METH toxicity is frequently reported as a potential model of drug induced Parkinsonism (Gerlach and Riederer, 1996, Guilarte, 2001, Kita et al., 2003a, Tolwani et al., 1999).
Similar to cytochrome P450s (CYP), most SULTs are regulated by hormones that remain under the control of the central nervous system (CNS). METH treatment is known to cause the change of dopamine in the CNS. Dopamine is one of the most important endogenous neurotransmitters as well as an important endogenous substrate of SULTs (Lu et al., 2005, Yasuda, Liu, 2007, Yasuda et al., 2009). It has been demonstrated that the expression of CYPs can be regulated by dopamine and other psychostimulants through the dopamine receptor-linked signaling pathway by changing hormone levels in vivo (Konstandi et al., 2008, Wójcikowski, 2004, Wójcikowski et al., 2007, 2008). Psychostimulant regulation of SULTs is not well studied. To the best of our knowledge, there are only two reports on METH regulation of SULTs. One study used a microarray method to screen a series of various candidate genes after a single-dose METH treatment (4.0 mg/kg) in rats (Niculescu et al., 2000). This report showed that the treatment induced rat brain rSULT1A1 mRNA 4.3-fold in the amygdale. Later, our results indicated that METH induced rSULT1A1, rSULT2A1 and rSULT1E1 protein expression both in the rat liver and brain (Zhou et al., 2010). In this study, the effect of short- and long-term treatment of male rats with METH on rSULT1A1 and rSULT2A1 expressions in the cerebellum, frontal cortex, hippocampus, and striatum was studied. Identifying regulation of rSULTs by METH in brain sections will assist in understanding the functions of SULTs in the brain as well as the toxic effects of MEHT on the brain.
2. MATERIALS AND METHODS
2.1 Materials
Methamphetamine was purchased from Sigma-Aldrich (St. Louis, MO). SDS-polyacrylamide gel electrophoresis reagents were obtained from Bio-Rad (Hercules, CA). Western blot chemiluminescence reagent kits (Super Signal West Pico Stable Peroxide and Super Signal West Pico Luminol/Enhancer solutions) were purchased from Pierce Chemical (Rockford, IL). PVDF membranes used for Western blotting analyses were purchased from Millipore Corporation (Bedford, MA). TRI REAGENT for total RNA extraction was purchased from MRC (Cincinnati, OH). M-MLV Reverse Transcriptase was obtained from Promega (Madison, WI). qPCR Master Mix Plus with SYBR® Green I dNTP was purchased from Eurogentec (San Diego, CA). Rabbit anti-rat SULT1A1 was provided by Dr. David Ringer (American Cancer Society). Rabbit anti-STa (rSULT2A1) was provided by Dr. Michael W. Duffel (Division of Medicinal and Natural Products Chemistry, College of Pharmacy, The University of Iowa, Iowa city, IA). Protein assay reagent was purchased from Bio-Rad (Hercules, CA). All other reagents and chemicals were of the highest analytical grade available.
2.2 Animals and Drug Treatment
Male Sprague-Dawley rats (Harlan, Indianapolis, IN), 10 to 11 weeks old and 200-300g body weight were used in this investigation. Rats were housed in a temperature- and humidity-controlled room and supplied with rodent chow and water for at least 1 week before use. Rats with each treatment were divided into 4 groups with four in each group. METH was dissolved in sterilized saline and administrated by intraperitoneal injection at 1, 5 and 20 mg/kg/day (single dose administration) for a 1 day treatment, and 0, 0.2, 1, 5 mg/kg/day for a 7 day treatment, respectively. The corresponding group of control rats received only sterilized saline. Animals didn’t show obvious signs/symptoms of METH intoxication or toxicity under these treatment conditions. The rats were sacrificed 24 hours after the final drug treatment. The cerebellum, frontal cortex, hippocampus, and striatum of the rat brain were collected, washed with sterilized ice-cold NaCl (0.9%, w/v) solution, and snap-frozen. Samples were stored at −80°C until use.
2.3 Cytosol Preparation
The cerebellum, frontal cortex, hippocampus and striatum of the rat brain were homogenized in 50 mM Tris buffer containing 0.25 M sucrose, 3 mM β-mercaptoethanol and 0.02% (v/v) Tween-20, pH 7.4. All homogenates were centrifuged at 100,000 g for 1 h at 4°C. Cytosol aliquots were collected and preserved at −80°C for Western blot analysis.
2.4 Western Blot Analysis
Cytosol protein from the frontal cortex, cerebellum, hippocampus and striatum of the rat brain (40 μg) was used in a 12% polyacrylamide gel in an electrophoresis system (Novex, San Diego, CA). After running at 200 V, the protein bands were transferred overnight at 35 V onto a PVDF membrane. The membranes were blocked for 1 hour by 5% (w/v) nonfat dry milk in Tris-buffered saline (TBS). For these cytosol proteins from different brain regions, membranes were incubated with either rabbit anti-rat AST-IV (rSULT1A1), or rabbit anti-rat STa (rSULT2A1) (1:1000) overnight in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% (v/v) Tween 20) containing 5% (w/v) nonfat dry milk on a shaker at 4°C. After incubation, all membranes were washed with TBST for 3 × 10 min and incubated in secondary antibody (Horseradish peroxidase-conjugated Immuno-Pure goat anti-rabbit IgG (H+L) at 1:8000 dilutions in the same buffer for at least 1 hour. The membranes were washed with TBST for 3 × 10min. The fluorescent bands were developed with 3 ml of substrate containing the same volume of each Super Signal West Pico Luminol Enhancer solution and Super Signal West Pico Stable Peroxidase solution at room temperature for 5 min. The fluorescence image was obtained using a VersaDoc IMAGING SYSTEM 5000MP (BIO-RAD, Hercules, CA). The densitometric quantification of protein bands was obtained using Quantity One 4.6.5 software of VersaDoc imaging system.
2.5 Quantitative Real-Time PCR
Total RNA was extracted from different regions of the brain using TRI REAGENT from MRC according to supplier’s guidelines. The concentration and purity of the extracted RNA were checked spectrophotometrically by measuring 260/280 absorption ratios. M-MLV Reverse Transcriptase (Promega) with 3 μg of total RNA was used to synthesize cDNA, and 1 μl of reverse-transcribed product served as the template in polymerase chain reactions. Real-time PCR was performed using the qPCR Master Mix Plus with SYBR® Green I kit (EUROGENTEC) following the manufacturer’s instructions. Primers were designed with Primer Express as follows: rActin-F320:5′-AGGCCCCTCTGAACCCTAAG-3′, rActin-R435: 5′-AGAGGCATACAGGGACAACACA-3′, GI NM_031144; rSULT1A1-F530: 5′-AGCTGAGACACACTCACCCTGTT-3′, rSULT1A1-R651: 5′-ATCCACAGTCTCCTCGGGTAGA-3′, GI. L19998; rSULT2A1-F496: 5′-ATCCGTGCCTGGCTGTCTAT-3′, rSULT2A1-R642: 5′-GAGGACCAAATCCAGCTCATCT-3′. GI M33329; Real-time PCR was performed on a 7500 Fast Real-Time PCR System. (Applied Biosystems, Foster City, CA). Initially, regular PCR products were purified with GENECLEAN Turbo (Qbiogene, Carlsbad, CA) for constructing standard curves (10-108 copies). A standard curve was plotted with the threshold cycle (CT) vs. the logarithmic value of the gene copy number. The gene copy number of unknown samples was generated directly from the standard curve by the software Sequence Detector 1.7. At least two duplications were run for each standard or unknown sample. All gene copy numbers were normalized to rat β-Actin mRNA.
2.6 Data Analysis
One-way ANOVA followed by the Dunnett’s test was used to calculate the statistical significance of the difference between the control group means and methamphetamine treatment group means. In all cases, *, P<0.05 was considered significant; **, P<0.01 was considered very significant. Data presented in the figures are means ± SD (standard deviation) of the data collected separately from four individual animals and each experiment was repeated twice.
3. RESULTS
3.1 Regulation of rSULT1A1 in rat cerebellum, frontal cortex, hippocampus, and striatum after 1-day and 7-days treatment with METH
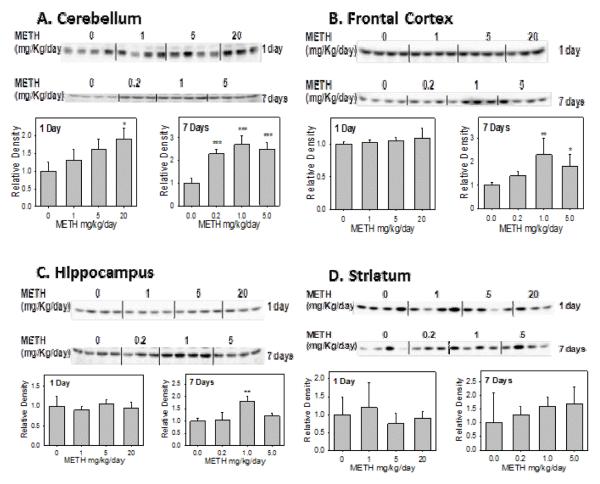
The Western blot and the densitometric analysis data showed that only in the cerebellum, METH significantly induced the rSULT1A1 protein level up to 1.7-fold for the high dose-treated group after 1 day treatment (Figure 1A). In the frontal cortex (Figure 1B), hippocampus (Figure 1C) and striatum (Figure 1D) of the rat brain, rSULT1A1 protein expression was not significantly changed after 1, 5, 20 mg/kg METH treatment for one day. On the other hand, rSULT1A1 protein expression in the rat cerebellum increased by 2.3-, 2.7-, and 1.4-fold respectively, in response to the treatment with 0.2, 1, 5 mg/kg/day METH for 7 days (Figure 1A). Western blot data also showed that the rSULT1A1 protein level in the frontal cortex and hippocampus increased by 2.1- and 1.7-fold respectively, at the treatment of 1 mg/kg/day for 7 days (Figures 1B and 1C). METH didn’t change the rSULT1A1 protein level significantly in rat striatum for the 7 day treatment (Figure 1D). These results indicated that rSULT1A1 regulation by METH in the rat brain was time-dependent and brain region-dependent.
Figure 1.
Representative Western blot and densitometric analysis of rat SULT1A1 in male rat cerebellum (A), frontal cortex (B), hippocampus (C), and striatum (D) after 1 day and 7 days treatment with varying doses of METH.
Values were divided by the smallest densitometric value of the blot. The division factors are plotted and expressed as relative densities. Four animals were used in each group of the treatment and each experiment was repeated twice. *p<0.05; ** p <0.01; and*** p<0.001.
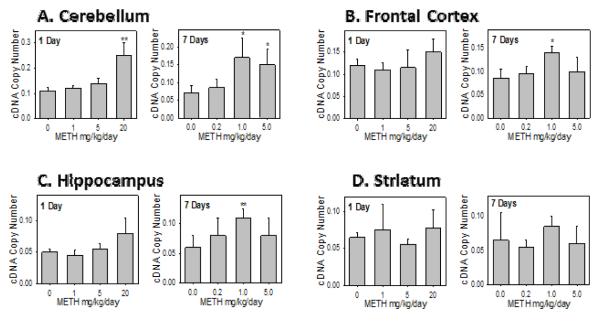
Real-time PCR data demonstrated that the effect of 1day and 7 day METH treatment on the mRNA expressions of the rSULT1A1 (Figure 2) was generally in good agreement with that of protein expression (Figure 1). This applies to all three dose treatments in all four parts of the rat brain. This suggests that the regulation of METH on rSULT1A1 expression is on the gene transcription level.
Figure 2.
Rat SULT1A1 mRNA expression in male rat cerebellum (A), frontal cortex (B), hippocampus (C), and striatum (D) after 1 day and 7 days treatment with varying doses of METH.
Relative copy number of rat SULT1A1 mRNA were standardized by using rat β-actin mRNA. Induction fold was calculated by dividing the copy number of rat SULT mRNA in METH-treated rats by the copy number of corresponding rat SULT mRNA in control rats. Four animals were used in each group of the treatment and each experiment was repeated twice. *p<0.05; and ** p <0.01.
3.2 Regulation of rSULT2A1 in rat cerebellum, frontal cortex, hippocampus, and striatum after 1-day and 7-days treatment with METH
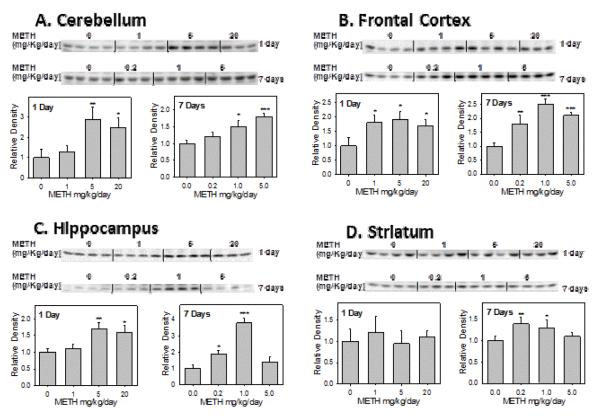
The Western blot and the densitometric analysis results (Figure 3) suggested that the rSULT2A1 protein in the cerebellum (Figure 3A), frontal cortex (Figure 3B) and hippocampus (Figure 3C) of the rat brain was significantly induced after a 1 day treatment of METH. In the cerebellum, the rSULT2A1 protein level increased by 2.8-fold and 2.4-fold in the middle- and high-dose groups respectively (Figure 3A). In the frontal cortex, the rSULT2A1 protein expression increased by about 1.7-, 1.7-, and 1.6-fold, respectively, after 1, 5, 20 mg/kg of METH treatment for 1 day (Figure 3B). In the hippocampus, the METH treatment for 1 day increased rSULT2A1 protein expression by 1.7- and 1.6-fold in the middle- and high-dose groups respectively (Figure 3C). rSULT2A1 was induced the highest by METH in the cerebellum, followed by the frontal cortex, and hippocampus. There was no significant change of rSULT2A1 protein expression in striatum after the 1 day treatment of METH. The cerebellum was the most sensitive section of the brain in which both rSULT2A1 and rSULT1A1were induced after 1 day of treatment.
Figure 3.
Representative Western blot and densitometric analysis of rat SULT2A1 in male rat cerebellum (A), frontal cortex (B), hippocampus (C), and striatum (D) after 1 day and 7 days treatment with varying doses of METH.
Values were divided by the smallest densitometric value of the blot. The division factors are plotted and expressed as relative densities. Four animals were used in each group of the treatment and each experiment was repeated twice. *p<0.05; ** p <0.01; and*** p <0.001.
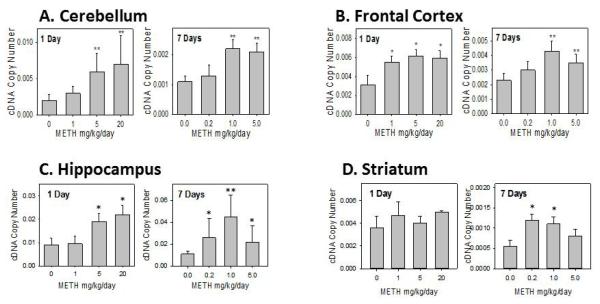
Figure 3 also showed that the rSULT2A1 protein in the cerebellum (Figure 3A), frontal cortex (Figure 3B) and hippocampus (Figure 3C) of the rat brain was significantly induced after the 7 day treatment by METH, similar to that for the 1 day treatment. Western blot results agreed with the real-time PCR data (Figure 4A - 4C). METH increased the rSULT2A1 protein and mRNA expression significantly in cerebellum, frontal cortex, hippocampus as well as striatum (Figure 3D and Figure 4D).
Figure 4.
Rat SULT2A1 mRNA expression in male rat cerebellum (A), frontal cortex (B), hippocampus (C), and striatum (D) after 1 day and 7 days treatment with varying doses of METH.
Relative copy number of rat SULT2A1 mRNA were standardized by using rat β-actin mRNA. Induction fold was calculated by dividing the copy number of rat SULT2A1 mRNA in METH-treated rats by the copy number of corresponding rat SULT2A1 mRNA in control rats. Four animals were used in each group of the treatment and each experiment was repeated twice. *p<0.05; and ** p <0.01.
In summary, after a 1-day treatment of METH, significant induction of rSULT1A1 was observed only in cerebellum, while evident induction of rSULT2A1 was found in the cerebellum, frontal cortex, and hippocampus, except striatum. After 7-day of treatment, METH induced rSULT1A1 in the cerebellum, frontal cortex and hippocampus, while it significantly induced rSULT2A1 in all four parts of the rat brain. These data indicated that rSULT2A1 is more inducible than rSULT1A1 in the rat brain sections. rSULT1A1 and rSULT2A1 are more inducible by METH in the cerebellum than in the frontal cortex and hippocampus. rSULT1A1 and rSULT2A1 are much less sensitive to METH in stratum.
4. DISCUSSION
Rat rSULT1A1 and rSULT2A1 expression level in brain sections is very important for the homeostasis of endogenous signaling molecules and the detoxification of xenobiotics in CNS. It is important to understand how rSULT1A1 and rSULT2A1 are regulated by drugs such as METH. Our results showed that both rat rSULT1A1 and rSULT2A1 were expressed in four regions of the rat brain: frontal cortex, cerebellum, hippocampus and striatum. Real-time PCR results indicate that the relative cDNA copy number of rSULT1A1 mRNA standardized by rat β-actin is much greater than that of rSULT2A1 mRNA, this is consistent with the previous human data (Salman, Kadlubar, 2009, Zou, Pentney, 1990). Among the four different regions of brain, there is no significant difference of expression of both rSULT1A1 and rSULT2A1 in the control group without METH treatment. This work is the first report to investigate rSULT1A1 and rSULT2A1 expression distribution in rat brain sections and their induction by xenobiotics in rat brain sections.
Rat rSULT1A1 gene transcription has been reported to occur through a glucocorticoid receptor-mediated mechanism. A low dose of a synthetic glucocorticoid can have physiologically significant effects attributable to the increased expression of rSULT1A1 (Runge-Morris and Kocarek, 2005b). METH has been reported to differentially regulate the glucocorticoid receptor in the rat brain (Kabbaj et al., 2003). This may partially explain the mechanism for METH regulation of rSULT1A1. Studies indicated that METH can differentially regulate the activity or expression of some important functional protein such as dopamine transporter in the brain after acute and chronic treatment (Fleckenstein et al., 1997, Isao and Akiyama, 2004, Iwazaki et al., 2006, Kokoshka et al., 1998). This may partially explain the differential METH regulation activities toward rSULT1A1 and rSULT2A1 in different rat brain sections. A varying pattern of regulation between brain regions is likely due to the different physiological functions of SULTs as well as different action mechanism of METH in different regions.
Endogenous hormones including glucocorticoids and cytokines are under CNS control. METH is a CNS stimulant which increases synaptic levels of the monoamines dopamine, serotonin and norepinephrine (Sofuoglu and Sewell, 2009). METH can induce extracellular dopamine in the prefrontal cortex (Ago et al., 2008). As the structural analogue of METH, dopamine increased the activities of CYP2B, CYP2C11 and CYP3A significantly and caused a substantial increase in the growth hormone level and a significant decrease in the T3 level in rat blood plasma (Wójcikowski, Gołembiowska, 2008) (Brackins, Brahm, 2011, Wojcikowski and Daniel, 2009). Another analogue of METH, amphetamine, significantly elevated CYP3A activity and produced a substantial increase in corticosterone concentration (Wójcikowski, Gołembiowska, 2008). The induction of rSULTs by METH may be related to the dopamine signaling pathway. METH may have variable effects on dopamine signaling pathway in different brain regions. Different rat brain regions have different sensitivity to the treatment with METH. The difference may be due to varying dopamine responses in different brain sections. Dopamine responses may change hormone level changes or nuclear receptor expression level changes, therefore change the expression of SULTs. This work showed that rSULT2A1 is more inducible than rSULT1A1 in different regions of brain. This may be due to a great dependence of rSULT1A1 regulation predominantly on glucocorticoid (Runge-Morris and Kocarek, 2005b), while the regulation of rSULT2A1 is shared by different hormones (e.g. GH, thyroid hormones, glucocorticoids) (Duanmu et al., 2002, Fang et al., 2005 -a, Fang et al., 2005 -b, Liu and Klaassen, 1996, Runge-Morris and Kocarek, 2005a).
The METH effect on rSULTs can have physiological and toxicological significance, since SULTs are important for drug detoxification and the regulation of endogenous biosignaling molecules including neurotransmitters and hormones. METH toxicity is frequently reported as a potential model of drug induced Parkinsonism (Gerlach and Riederer, 1996, Guilarte, 2001, Kita, Wagner, 2003a, Tolwani, Jakowec, 1999). It will be important to future investigate the regulation mechanisms of SULTs by METH in different brain regions. Monoamines including dopamine are sulfated in vivo (Akimov et al., 2009, Schwaninger et al., 2011, Yoshinari et al., 2011). Dopamine exists in vivo mainly as sulfated form (Goldstein et al., 2003, Itaaho et al., 2007). Neuronal toxicity of dopamine is mainly derived from its instability; it is easily oxidized by oxygen and producing reactive oxygen species (ROS) to cause damage. Sulfated dopamine is much more chemically stable. The sulfation can decrease the oxidative stress caused by dopamine. rSULT1A1 catalyzes the sulfation of dopamine. In human, hSULT1A1 and hSULT1A3 are responsible for the sulfation of dopamine. Gene induction of these SULTs may have an effect on dopamine sulfation and dopamine stability in vivo. It will be interesting to investigate the potential roles of SULT induction in the development of certain diseases such as Parkinson’s disease etc.
Highlights.
rSULT1A1 and rSULT2A1 are expressed in rat frontal cortex, cerebellum, striatum, and hippocampus.
rSULT1A1 and rSULT2A1are inducible by METH in rat brain sections in a time dependable manner.
rSULT2A1 is more inducible than rSULT1A1 by METH in rat brain sections.
Induction activity of METH is in the order of cerebellum > frontal cortex, hippocampus > striatum.
ACKNOWLEDGMENT
The authors are grateful to Dr. David Ringer (American Cancer Society) for the generous gift of rabbit anti-rat SULT1A1 antibody; and to Dr. Michael Duffel (The University of Iowa) for the generous gift of rabbit anti-rat SULT2A1 antibody. This work was supported in part by NIH grant GM078606 (G.C.), American Cancer Society grant RSG-07-028-01-CNE (G.C.), USDA grant 2006-35200-17137 (G.C.), Oklahoma Center for the Advancement of Science and Technology (OCAST) grant HR05-015 (G.C.), and National Natural Science Foundation of China (NSFC) grant 81072699 (T.Z.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ago Y, Arikawa S, Yata M, Yano K, Abe M, Takuma K, et al. Role of prefrontal dopaminergic neurotransmission in glucocorticoid receptor-mediated modulation of methamphetamine-induced hyperactivity. Synapse. 2008;63:7–14. doi: 10.1002/syn.20575. [DOI] [PubMed] [Google Scholar]
- Akimov MG, Nazimov IV, Gretskaya NM, Zinchenko GN, Bezuglov VV. Sulfation of N-acyl dopamines in rat tissues. Biochemistry. 2009;74:681–5. doi: 10.1134/s0006297909060133. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue Distribution and Ontogeny of Sulfotransferase Enzymes in Mice. Toxicological Sciences. 2006;93:242–55. doi: 10.1093/toxsci/kfl050. [DOI] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MWH. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- Bojarova P, Williams SJ. Sulfotransferases, sulfatases and formylglycine-generating enzymes: a sulfation fascination. Current Opinion in Chemical Biology. 2008;12:573–81. doi: 10.1016/j.cbpa.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Brackins T, Brahm NC, Kissack JC. Treatments for methamphetamine abuse: a literature review for the clinician. J Pharm Pract. 2011;24:541–50. doi: 10.1177/0897190011426557. [DOI] [PubMed] [Google Scholar]
- Chapman E, Best MD, Hanson SR, Wong C-H. Sulfotransferases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angewandte Chemie, International Edition. 2004;43:3526–48. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- Duanmu Z, Locke D, Smigelski J, Wu W, Dahn M, Falany C, et al. Effects of dexamethasone on aryl (SULT1A1)- and hydroxysteroid (SULT2A1)-sulfotransferase gene expression in primary cultured human hepatocytes. Drug Metab Dispos. 2002;30:997–1004. doi: 10.1124/dmd.30.9.997. [DOI] [PubMed] [Google Scholar]
- Duffel MW. Molecular specificity of aryl sulfotransferase IV (tyrosine-ester sulfotransferase) for xenobiotic substrates and inhibitors. Chem-Biol Interact. 1994;92:3–14. doi: 10.1016/0009-2797(94)90048-5. [DOI] [PubMed] [Google Scholar]
- Fang H, Abdolalipour M, Duanmu Z, Smigelski J, Weckle A, Kocarek T, et al. Regulation of glucocorticoid-inducible hydroxysteroid sulfotransferase (SULT2A-40/41) gene transcription in primary cultured rat hepatocytes: role of CCAAT/enhancer-binding protein liver-enriched transcription factors. Drug Metab Dispos. 2005-a;33:147–56. doi: 10.1124/dmd.104.000281. [DOI] [PubMed] [Google Scholar]
- Fang H, Strom S, Cai H, Falany C, Kocarek T, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol Pharmacol. 2005-b;67:1257–67. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, et al. Positive and Negative Regulation of Human Hepatic Hydroxysteroid Sulfotransferase (SULT2A1) Gene Transcription by Rifampicin: Roles of Hepatocyte Nuclear Factor 4{alpha} and Pregnane X Receptor. J Pharmacol Exp Ther. 2007 doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- Fleckenstein AE, Metzger RR, Wilkins DG, Gibb JW, Hanson GR. Rapid and reversible effects of methamphetamine on dopamine transporters. Journal of Pharmacology and Experimental Therapeutics. 1997;282:834–8. [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, et al. Human Sulfotransferases and their Role in Chemical Metabolism. Toxicological Sciences. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- Gaytan O, Swann A, Dafny N. Time-dependent differences in the rat’s motor response to amphetamine. Pharmacology, biochemistry, and behavior. 1998;59:459–67. doi: 10.1016/s0091-3057(97)00438-3. [DOI] [PubMed] [Google Scholar]
- Gerlach M, Riederer P. Animal models of Parkinson’s disease: an empirical comparison with the phenomenology of the disease in man. J Neural Transm. 1996;103:987–1041. doi: 10.1007/BF01291788. [DOI] [PubMed] [Google Scholar]
- Gold MS, Kobeissy FH, Wang KK, Merlo LJ, Bruijnzeel AW, Krasnova IN, et al. Methamphetamine- and trauma-induced brain injuries: comparative cellular and molecular neurobiological substrates. Biol Psychiatry. 2009;66:118–27. doi: 10.1016/j.biopsych.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DS, Eisenhofer G, Kopin IJ. Sources and significance of plasma levels of catechols and their metabolites in humans. J Pharmacol Exp Ther. 2003;305:800–11. doi: 10.1124/jpet.103.049270. [DOI] [PubMed] [Google Scholar]
- Guilarte T. Is methamphetamine abuse a risk factor in parkinsonism? Neurotoxicology. 2001;22:725–31. doi: 10.1016/s0161-813x(01)00046-8. [DOI] [PubMed] [Google Scholar]
- Guilarte T, Nihei M, McGlothan J, Howard A. Methamphetamine-induced deficits of brain monoaminergic neuronal markers: distal axotomy or neuronal plasticity. Neuroscience. 2003;122:499–513. doi: 10.1016/s0306-4522(03)00476-7. [DOI] [PubMed] [Google Scholar]
- Hempel N, Gamage N, Martin JL, McManus ME. Human cytosolic sulfotransferase SULT1A1. International Journal of Biochemistry & Cell Biology. 2007;39:685–9. doi: 10.1016/j.biocel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Isao T, Akiyama K. Effect of acute and chronic treatment with methamphetamine on mRNA expression of synaptotagmin IV and 25 KDa-synaptic-associated protein in the rat brain. Psychiatry and Clinical Neurosciences. 2004;58:410–9. doi: 10.1111/j.1440-1819.2004.01276.x. [DOI] [PubMed] [Google Scholar]
- Itaaho K, Alakurtti S, Yli-Kauhaluoma J, Taskinen J, Coughtrie MW, Kostiainen R. Regioselective sulfonation of dopamine by SULT1A3 in vitro provides a molecular explanation for the preponderance of dopamine-3-O-sulfate in human blood circulation. Biochem Pharmacol. 2007;74:504–10. doi: 10.1016/j.bcp.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Iwazaki T, McGregor IS, Matsumoto I. Protein expression profile in the striatum of acute methamphetamine - treated rats. Brain Research. 2006;1097:19–25. doi: 10.1016/j.brainres.2006.04.052. [DOI] [PubMed] [Google Scholar]
- Kabbaj M, Yoshida S, Numachi Y, Matsuoka H, Devine DP, Sato M. Methamphetamine differentially regulates hippocampal glucocorticoid and mineralocorticoid receptor mRNAs in Fischer and Lewis rats. Molecular Brain Research. 2003;117:8–14. doi: 10.1016/s0169-328x(03)00257-2. [DOI] [PubMed] [Google Scholar]
- Kauffman FC. Sulfonation in Pharmacology and Toxicology. Drug Metabolism Reviews. 2004;36:823–43. doi: 10.1081/dmr-200033496. [DOI] [PubMed] [Google Scholar]
- Kish S. Pharmacologic mechanisms of crystal meth. CMAJ. 2008;178:1679–82. doi: 10.1503/cmaj.071675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita T, Wagner G, Nakashima T. Current research on methamphetamine-induced neurotoxicity: animal models of monoamine disruption. J Pharmacol Sci. 2003a;92:178–95. doi: 10.1254/jphs.92.178. 2003 Jul;92(3): [DOI] [PubMed] [Google Scholar]
- Kita T, Wagner G, Nakashima T. Current research on methamphetamine-induced neurotoxicity: animal models of monoamine disruption. J Pharmacol Sci. 2003b;92:178–95. doi: 10.1254/jphs.92.178. [DOI] [PubMed] [Google Scholar]
- Kokoshka JM, Vaughan RA, Hanson GR, Fleckenstein AE. Nature of methamphetamine -induced rapid and reversible changes in dopamine transporters. European Journal of Pharmacology. 1998;361:269–75. doi: 10.1016/s0014-2999(98)00741-9. [DOI] [PubMed] [Google Scholar]
- Konstandi M, Harkitis P, Kostakis D, Marselos M, Johnson E, Lang M. D2-receptor-linked signaling pathways regulate the expression of hepatic CYP2E1. Life Sci. 2008;82:1–10. doi: 10.1016/j.lfs.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Krasnova I, Cadet J. Methamphetamine toxicity and messengers of death. Brain Res Rev. 2009;60:379–407. doi: 10.1016/j.brainresrev.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Klaassen CD. Regulation of hepatic sulfotransferases by steroidal chemicals in rats. Drug metabolism and disposition. 1996;24:854–8. [PubMed] [Google Scholar]
- Liu Y, Apak TI, Lehmler HJ, Robertson LW, Duffel MW. Hydroxylated polychlorinated biphenyls are substrates and inhibitors of human hydroxysteroid sulfotransferase SULT2A1. Chem Res Toxicol. 2006;19:1420–5. doi: 10.1021/tx060160+. [DOI] [PubMed] [Google Scholar]
- Lu J-H, Li H-T, Liu M-C, Zhang J-P, Li M, An X-M, et al. Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3′-phosphoadenosine 5′-phosphate. Biophysical Research Communications. 2005;335:417–23. doi: 10.1016/j.bbrc.2005.07.091. [DOI] [PubMed] [Google Scholar]
- Niculescu ABr, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiological genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–8. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- Quinton MS, Yamamoto BK. Causes and consequences of methamphetamine and MDMA toxicity. The AAPS journal. 2006;8:E337–47. doi: 10.1007/BF02854904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath VL, Verdugo D, Hemmerich S. Sulfotransferase structural biology and inhibitor discovery. Drug Discovery Today. 2004;9:1003–11. doi: 10.1016/S1359-6446(04)03273-8. [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, Coughtrie MWH. Sulfation of thyroid hormone and dopamine during human development : ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. Journal of Clinical Endocrinology and Metabolism. 2001;86:2734–42. doi: 10.1210/jcem.86.6.7569. [DOI] [PubMed] [Google Scholar]
- Riches Z, Bloomer JC, Coughtrie MW. Comparison of 2-aminophenol and 4-nitrophenol as in vitro probe substrates for the major human hepatic sulfotransferase, SULT1A1, demonstrates improved selectivity with 2-aminophenol. Biochem Pharmacol. 2007;74:352–8. doi: 10.1016/j.bcp.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. The neural basis of drug craving: an incentive-sensitization theory of addiction. Brain Research Reviews. 1993;18:247–91. doi: 10.1016/0165-0173(93)90013-p. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek T. Regulation of sulfotransferases by xenobiotic receptors. Curr Drug Metab. 2005a;6:299–307. doi: 10.2174/1389200054633871. [DOI] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. Regulation of sulfotransferases by xenobiotic receptors. Current Drug Metabolism. 2005b;6:299–307. doi: 10.2174/1389200054633871. [DOI] [PubMed] [Google Scholar]
- Salman ED, Kadlubar SA, Falany CN. Expression and localization of cytosolic sulfotransferase (SULT) 1A1 and SULT1A3 in normal human brain. Drug Metabolism and Disposition. 2009;37:706–9. doi: 10.1124/dmd.108.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaninger AE, Meyer MR, Zapp J, Maurer HH. Sulfation of the 3,4-methylenedioxymethamphetamine (MDMA) metabolites 3,4-dihydroxymethamphetamine (DHMA) and 4-hydroxy-3-methoxymethamphetamine (HMMA) and their capability to inhibit human sulfotransferases. Toxicol Lett. 2011;202:120–8. doi: 10.1016/j.toxlet.2011.01.026. [DOI] [PubMed] [Google Scholar]
- Shimizu C, Fuda H, Yanai H, Strott CA. Conservation of the hydroxysteroid sulfotransferase SULT2B1 gene structure in the mouse: Pre- and postnatal expression, kinetic analysis of isoforms, and comparison with prototypical SULT2A1. Endocrinology. 2003;144:1186–93. doi: 10.1210/en.2002-221011. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Tamura H. Identification and localization of two hydroxysteroid sulfotransferases in the human brain. Journal of Health Science. 2002;48:467–72. [Google Scholar]
- Sofuoglu M, Sewell R. Norepinephrine and stimulant addiction. Addict Biol. 2009;14:119–29. doi: 10.1111/j.1369-1600.2008.00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolwani R, Jakowec M, Petzinger G, Green S, Waggie K. Experimental models of Parkinson’s disease: insights from many models. Lab Anim Sci. 1999;49:363–71. [PubMed] [Google Scholar]
- Vietri M, Vaglini F, Cantini R, Pacifici G. Quercetin inhibits the sulfation of R(−)-apomorphine in human brain. International Journal of Clinical Pharmacology and Therapeutics. 2003;41:30–5. [PubMed] [Google Scholar]
- Wang L-Q, James MO. Inhibition of sulfotransferases by xenobiotics. Current Drug Metabolism. 2006;7:83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- Whittemore RM, Pearce LB, Roth JA. Purification and kinetic characterization of a phenol-sulfating form of phenol sulfotransferase from human brain. Archives of Biochemistry and Biophysics. 1986;249:464–71. doi: 10.1016/0003-9861(86)90023-8. [DOI] [PubMed] [Google Scholar]
- Wójcikowski J. Potential role of the brain dopaminergic system in the regulation of cytochrome P-450 expression. Pol J Pharmacol. 2004;56:701–8. [PubMed] [Google Scholar]
- Wojcikowski J, Daniel WA. The brain dopaminergic system as an important center regulating liver cytochrome P450 in the rat. Expert Opin Drug Metab Toxicol. 2009;5:631–45. doi: 10.1517/17425250902973703. [DOI] [PubMed] [Google Scholar]
- Wójcikowski J, Gołembiowska K, Daniel W. The regulation of liver cytochrome p450 by the brain dopaminergic system. Curr Drug Metab. 2007;8:631–8. doi: 10.2174/138920007781368872. [DOI] [PubMed] [Google Scholar]
- Wójcikowski J, Gołembiowska K, Daniel W. Regulation of liver cytochrome P450 by activation of brain dopaminergic system: physiological and pharmacological implications. Biochem Pharmacol. 2008;76:258–67. doi: 10.1016/j.bcp.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu M-Y, Suiko M, Sakakibara Y, Liu M-C. Hydroxylated serotonin and dopamine as substrates and inhibitors for human cytosolic SULT1A3. Journal of Neurochemistry. 2007;103:2679–89. doi: 10.1111/j.1471-4159.2007.04948.x. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Yasuda T, Hui Y, Liu M-Y, Suiko M, Sakakibara Y, et al. Concerted action of the cytosolic sulfotransferase, SULT1A3, and catechol-O-methyltransferase in the metabolism of dopamine in SK-N-MC human neuroblastoma cells. Neuroscience Research. 2009;64:273–9. doi: 10.1016/j.neures.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Yoshinari K, Sakamoto M, Senggunprai L, Yamazoe Y. Clioquinol is sulfated by human jejunum cytosol and SULT1A3, a human-specific dopamine sulfotransferase. Toxicol Lett. 2011;206:229–33. doi: 10.1016/j.toxlet.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Zhou T, Huang C, Chen Y, Shanbhag P, Chen G. Methamphetamine Regulation of Sulfotransferases in Rat Liver and Brain. American Journal of Pharmacology and Toxicology. 2010;5:125–32. [Google Scholar]
- Zou J, Pentney R, Roth JA. Immunohistochemical detection of phenol sulfotransferase-containing neurons in human brain. Journal of Neurochemistry. 1990;55:1154–8. doi: 10.1111/j.1471-4159.1990.tb03119.x. [DOI] [PubMed] [Google Scholar]