Abstract
Decaprenylphosphoryl-β-D-ribose 2’-epimerase (DprE1) is an essential enzyme in the biosynthesis of cell wall components and a target for development of anti-tuberculosis drugs. We determined the crystal structure of a truncated form of DprE1 from Mycobacterium smegmatis in two crystal forms to up to 2.35 Å resolution. The structure extends from residue 75 to the C-terminus and shares homology with FAD-dependent oxidoreductases of the vanillyl-alcohol oxidase family including the DprE1 homologue from M. tuberculosis. The M. smegmatis DprE1 structure reported here provides further insights into the active site geometry of this tuberculosis drug target.
Keywords: X-ray crystallography, tuberculosis, benzothiazinone, FAD, oxidoreductase
Introduction
Tuberculosis is a major global health threat that is exacerbated by the emergence of multiple-drug resistant, extensively drug resistant, and even totally drug resistant strains of the causative pathogen Mycobacterium tuberculosis. Recent drug development efforts have identified different classes of nitroaromatic compounds that target the enzyme decaprenylphosphoryl-β-D-ribose 2’-epimerase (DprE1), which is crucial for mycobacterial cell wall biosynthesis1. DprE1 catalyzes the FAD-dependent oxidation of decaprenylphosphoryl-β-D-ribose (DPR) to decaprenylphosphoryl-2-keto-D-erythro-pentofuranose (DPX). The NADH-dependent enzyme DprE2 then converts DPX to decaprenylphosphoryl-β-D-arabinose (DPA), completing this two-step epimerization reaction (Fig. 1a)2.
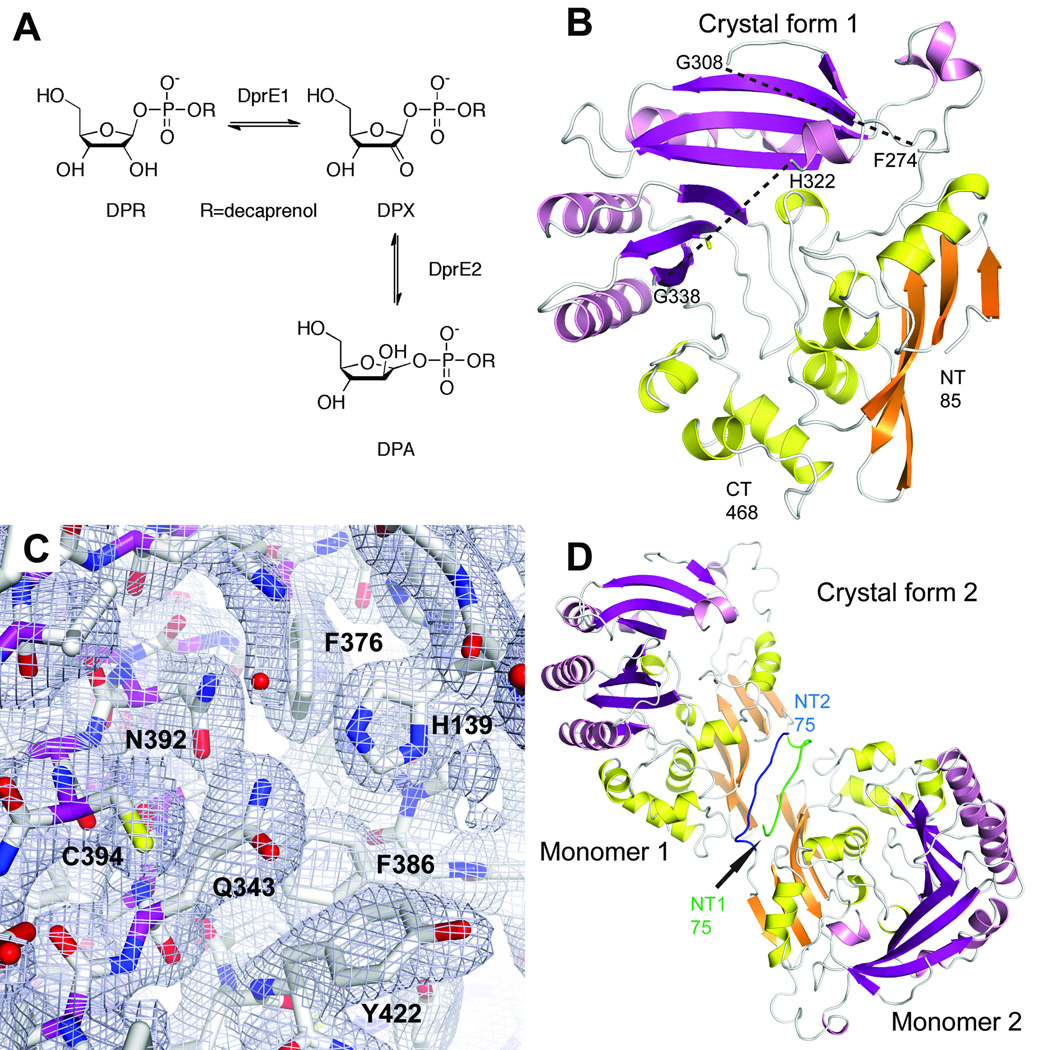
Figure 1. Overall structure of DprE1.
a. Oxidation of decaprenylphosphoryl-β-D-ribose (DPR) to decaprenylphosphoryl-2-keto-D-erythro-pentofuranose (DPX) catalyzed by DprE1 and further reduction of DPX to decaprenylphosphoryl-β-D-arabinose (DPA) by DprE2.
b. Cartoon representation of the DprE1 core structure in space group P3221. Secondary structure elements in the FAD-binding domain and in the substrate-binding domain are colored in orange and yellow and magenta and pink, respectively. The approximate positions of two mobile loop regions (between residues 274 and 308, and between residues 322 and 338) are indicated with dashes.
c. Final σA-weighted 2mFo-DFc electron density for the active site region in space group P3221.
d. Dimer formation in space group P21. The additional N-terminal residues 75–84 in the two monomers establish an antiparallel β-strand-like interaction. N-terminal residues from monomers 1 and 2 are colored in green and blue, respectively.
One class of DprE1 inhibitors, benzothiazinones, is active against mycobacteria at low nanomolar concentrations in vitro and in a mouse model1. Biochemical studies showed that the aromatic nitro group of benzothiazinones is enzymatically reduced to a nitroso group by DprE13 and that this reduced derivative then acts as a suicide inhibitor by forming a semimercaptal with a cysteine residue in the DprE1 active site (Cys394 in Mycobacterium smegmatis)4. Resistance to these compounds in M. smegmatis arose from mutation of this cysteine residue1.
Structural information is essential for further development of these inhibitors and for drug development programs against tuberculosis and infectious diseases in general. This is exemplified by the structural genomics approach carried out by the tuberculosis structural genomics consortium5. In this study, we determined the crystal structure of a proteolytic core fragment of DprE1 from M. smegmatis. No closely related structures were available in the Protein Databank, and the structure was solved with the single-wavelength anomalous dispersion method (SAD). The structure confirms the predicted homology with FAD-dependent oxidoreductases belonging to the vanillyl-alcohol oxidoreductase (VAO) family6 and reveals the presence of unique loop regions in the substrate-binding domain that contribute to define access to the enzyme’s active site.
Materials and Methods
Protein expression, purification and crystallization
The full-length decaprenylphosphoryl-β-D-ribose 2’-epimerase from Mycobacterium smegmatis strain ATCC 607 was cloned into the pET28a vector (Novagen) and over-expressed in E. coli strain BL21Star (Invitrogen) at 293 K. Bacterial cells were lysed by ultrasonification on ice in a buffer containing 20 mM Tris pH 7.5, 200 mM NaCl, 5 mM β-mercaptoethanol, 0.1 % Triton-X100, and 5 % glycerol. Soluble N-terminally hexahistidine-tagged DprE1 was bound to nickel-agarose affinity resin (Qiagen), washed with a buffer containing 20 mM Tris (pH 7.5), 200 mM NaCl, and 10 mM imidazole, and eluted with a buffer containing 20 mM Tris (pH 7.5), 250 mM NaCl, and 150 mM imidazole. The eluted protein was concentrated and diluted with a buffer containing 50mM Tris (pH 7.5) and 150 mM NaCl and digested with thrombin for 12 – 15 hours at 277 K. The protein was further purified with anion exchange chromatography, using a linear gradient of 10 mM to 1 M NaCl concentration, and size exclusion chromatography at pH 7.5 and 200 mM NaCl. Purified cut DprE1 was concentrated to 25 mg/ml without buffer exchange. SDS polyacrylamide gel electrophoresis of purified protein and of re-dissolved protein crystals showed one major band at an approximate molecular weight of 44 kDa, indicating partial proteolysis of the full-length protein after thrombin digest. For the production of selenomethionyl protein, the expression construct was transformed into B834(DE3) cells (Novagen). Crystals were obtained with the sitting drop vapor diffusion method at 277 K. For native protein in crystal form 1, 1 µl of protein was mixed with 1 µl of a solution containing 15 – 20 % isopropanol, 0.2 M sodium citrate pH 6.8, and 0.5 % N,N-dimethyldodecylamine-N-oxide. Selenomethionyl-labeled protein crystals in the same crystal form were obtained from a solution containing 15 – 20 % 1, 4-butane diol, 0.2M NaCl, 0.1 M Tris pH 7.5, and 0.5 % N,N-dimethyldodecylamine-N-oxide. To obtain crystals in form 2, 1 µl protein (10 mg/ml) was mixed with 1 µl of a solution containing 30% butanol, 0.2 M NaCl, 0.1 M Tris pH 7.5, and 0.5 % N,N-dimethyldodecylamine-N-oxide. Crystals were flash-frozen in liquid nitrogen.
Data collection, structure determination and refinement
Diffraction data were collected at the National Synchrotron Light Source in Brookhaven. Data for crystal form 1 were collected at the X4C beamline on a MAR CCD detector at a wavelength of 0.979 Å at 100 K. Data for crystal form 2 were collected at the X4A beamline on an ADSC Quantum Q4 detector at a wavelength of 0.979 Å at 100 K. For structure solution, a selenomethionyl single wavelength anomalous dispersion data set was collected to 2.6 Å at the X29 beamline7 on an ADSC Quantum Q315 detector at a wavelength of 0.9789 Å at 100 K. Diffraction images for both crystal forms were processed and scaled with XDS8, the anomalous data set was processed and scaled with the HKL2000 package9. Data processing statistics are summarized in Table 1. The locations of nine selenium atoms were determined with the Phenix program package10. An initial model from Phenix was manually rebuilt with Coot11 and further refined with Phenix. The structure in form 2 was solved by molecular replacement with the program Phaser12 with the refined structure in form 1 as a search model. Phenix was used for final crystallographic refinement. The final model in crystal form 1 contains one monomer with residues 85 – 274, 308 – 322, and 338 – 468. The final model in crystal form 2 contains two monomers with residues 75 – 274, 308 – 322, and 338 – 468. The Ramachandran statistics calculated with Procheck13 are (most favored/additionally allowed/generously allowed/disallowed) 90.8 / 9.2 / 0.0 / 0.0 % for crystal form 1 and 90.7 / 9.3 / 0.0 / 0.0 % for crystal form 2. Structure refinement statistics are summarized in Table 1. Figures were produced with Pymol (www.pymol.org). Coordinates and structure factors have been deposited at the Protein Data Bank with accession codes 4G3T and 4G3U.
Table 1.
Data collection and refinement statistics
| Crystal Form 1 | Crystal Form 2 |
|
|---|---|---|
| Data collection1 | ||
| Space group | P3221 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 64.2, 64.2, 275.5 | 105.2, 65.8, 108.5 |
| α, β, γ (°) | 90, 90, 120 | 90, 109.0, 90 |
| Resolution (Å)2 | 40 – 2.35 (2.49 – 2.35) | 35 – 2.69 (2.85 – 2.69) |
| Rmeas | 0.130 (1.285) | 0.135 (0.744) |
| I / σI | 10.7 (2.0) | 10.8 (2.3) |
| Completeness (%) | 99.7 (99.5) | 92.7 (92.6) |
| Redundancy | 9.1 (9.1) | 3.2 (3.1) |
| Refinement | ||
| Resolution (Å)2 | 40 – 2.35 (2.44 – 2.35) | 35 – 2.69 (2.76 – 2.69) |
| No. reflections | 28,621 (2,823) | 36,552 (1,838) |
| Rwork / Rfree | 0.227/0.256 (0.350/0.356) | 0.220/0.254 (0.321/0.366) |
| No. atoms | ||
| Protein | 2,618 | 5,370 |
| Water | 77 | 287 |
| B-factors | ||
| Protein | 69.8 | 47.15 |
| Water | 59.6 | 38.74 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (°) | 0.72 | 0.72 |
One crystal was used for each data set.
Values in parentheses are for highest-resolution shell.
Results and Discussion
The gene encoding the full-length decaprenylphosphoryl-β-D-ribose 2’-epimerase (Genbank MSMEG_6382, 468 amino acids) from Mycobacterium smegmatis (strain ATCC 607) was subcloned from genomic DNA, and the recombinant N-terminally histidine-tagged protein was expressed in E. coli and purified using standard protocols. The recombinant protein was slightly yellow colored after elution from a Ni-NTA affinity column but gradually lost this color throughout further purification. The histidine-tagged protein failed to crystallize in an extensive crystallization screen. Therefore, the N-terminal histidine tag was removed by cleavage with thrombin. The cleaved protein was smaller than expected with an estimated molecular weight of 44 kDa based on size-exclusion chromatography and SDS gel electrophoresis, indicating proteolytic cleavage by thrombin between residues 62 and 63. This truncated protein form crystallized in several conditions and diffracting crystals were obtained in space groups P3221 and P21. Single-wavelength anomalous dispersion diffraction data to 2.6 Å resolution were used to solve the structure in space group P3221. The initial model (one monomer) was rebuilt and refined against a native data set to 2.35 Å resolution. This model was used to locate two monomers in space group P21 by molecular replacement. The final model in space group P3221 consists of residues 85 to 274, 308 to 322, and 338 to 468. No density was observed for the two loop regions with residues 275 – 307 and 323 – 337. Additional electron density enabled us to build the N-terminal residues 75 to 84 in the second crystal form in space group P21. The DprE1 structure remains essentially identical between the two monomers in space group P21 (0.12 Å rmsd for 346 Cα atoms) and between the two crystal forms (0.36 Å for 336 Cα atoms).
The overall structure of DprE1 belongs to the family of vanillyl-alcohol oxidases and consists of two domains with an α+β fold (Fig. 1b, c). The FAD-binding domain consists of a larger N-terminal part extending from the N-terminus (based on structural homology discussed below) to residue 200 and a smaller C-terminal part between residues 422 and 468. The substrate-binding domain consists of residues 201 to 421. Analysis of packing interactions in the P21 space group reveals that the additionally ordered N-terminal residues 75 to 84 assume an extended conformation and interact symmetrically between two DprE1 monomers in a cross-over antiparallel β-strand contact (Fig. 1d). This interaction partially mimics the formation of an N-terminal β-sheet that is predicted for these residues in the full-length structure based on homologous structures.
Sequence analysis and a structural homology search with the program Dali14 identify two flavin-dependent VAO family members as closest related structures. The alditol oxidase from Streptomyces coelicolor shares 18% sequence identity with DprE1 (Fig. S1) and superimposes with a root-mean-square deviation (rmsd) of 2.3 Å for 254 Cα atoms (Fig. 2a). The cytokinin dehydrogenase from Zea mays shares 16% sequence identity and superimposes with an rmsd of 2.4 Å for 228 Cα atoms (Fig. 2b). The comparison with these related structures confirms the overall homology of the core structural elements including β-sheets and the large flanking helices in both domains. In both superpositions, the structural homology is substantially higher in the cofactor-binding domain (Fig. 2c, d). Significant differences can be seen in the conformation of loops connecting core structural elements in the substrate-binding domain. Both structures provide only weak guidance for homology-based modeling of the flexible loop regions in DprE1 as the length of the loops as well as their conformation and interaction with core structural elements are unique in each structure.
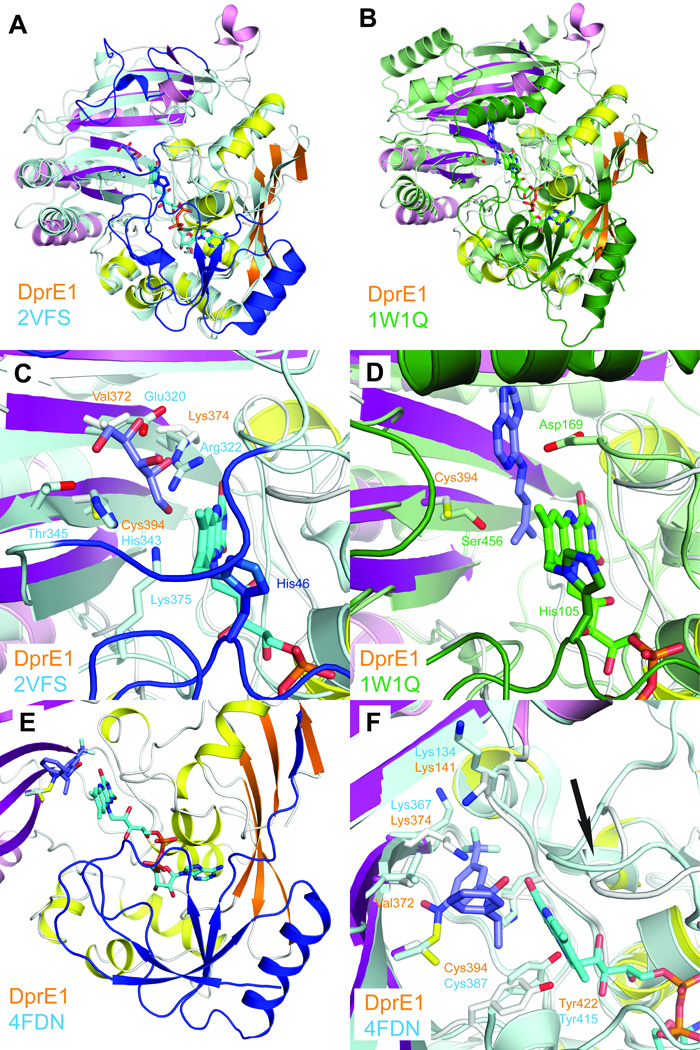
Figure 2. Comparison of DprE1 with structurally related enzymes.
a. Least-squares superposition of DprE1 with alditol oxidase (colored in cyan and blue, PDB entry 2VFS). N-terminal residues absent in the DprE1 structure and residues approximately equivalent to the flexible loop regions are colored in blue.
b. Least-squares superposition of DprE1 with cytokinin dehydrogenase (colored in green, PDB entry 1W1Q). N-terminal residues absent in the DprE1 structure and residues approximately equivalent to the flexible loop regions are colored in dark green.
c. Close-up view comparing the active site region of DprE1 with alditol oxidase. FAD is shown in cyan sticks bound to His46. The substrate xylitol is shown in purple sticks and xylitol-coordinating residues are shown in cyan sticks. Residues approximately equivalent to the flexible loop regions are colored in blue.
d. Close-up view comparing the active site region of DprE1 with cytokinin dehydrogenase. FAD is shown in green sticks bound to His105. The cytokinin dehydrogenase product N6-isopentenyladenine is shown in purple sticks. Residues approximately equivalent to the flexible loop regions are colored in dark green.
e. N-terminal region absent in the DprE1 structure as observed in the DprE1 structure from M. tuberculosis (colored in blue, PDB code 4FDN). FAD and the DprE1 inhibitor CT325 in DprE1_tb are shown in cyan and blue sticks, respectively. The structure of DprE1_sm is colored as before.
f. Comparison of residues in the DprE1 active site (shown in white sticks) with DprE1 from M. tuberculosis (cyan sticks). The conformational change of the active site loop between residues 121 and 128 is indicated with an arrow.
A very recent report describing the crystal structure of the DprE1 homologue from M. tuberculosis15 further confirms our findings. The M. tuberculosis DprE1 homologue (DprE1_mt) shares 81.9 % sequence identity with M. smegmatis. The two structures superimpose with an rmsd of 1.2 Å for 336 Cα atoms. In contrast to the structures presented here, the FAD cofactor remains bound in DprE1_tb. The structural superposition confirms that the N-terminal structural elements absent in our M. smegmatis structure are similar to alditol oxidase (Fig. 2e). Furthermore, the comparison of the FAD-bound active site reveals that cofactor binding induces a rearrangement of the active site loop between residues 121 and 128 and adjustments of FAD-coordinating residues such as Tyr422 (Fig. 2f). The inhibitor-bound DprE1_tb structure further shows that inhibitor binding displaces both lysine residues (Lys374 and Lys141) that likely engage the substrate phosphate group. These observations provide strong support for our model of cofactor- and substrate binding to the M. smegmatis DprE1 homologue.
The observation of two disordered regions in the DprE1 core structure was unexpected. Both loop regions are disordered in the two crystal forms, and both are also disordered in the DprE1_tb homologue structure, indicating that this flexibility is not an artifact of the N-terminal truncation introduced in this study. Both loops are flanking the access to the active site and could conceivably assume an ordered conformation upon contact with the bacterial membrane surface to access the membrane bound substrate. The inhibitor-bound DprE1_tb structure shows that one of the loops can assume an ordered conformation that interacts with the inhibitor molecule in the active site. Thus, a functional contribution of these flexible loop regions to mediate the interaction with the membrane and to further stabilize substrate binding in the active site seems plausible.
In summary, the DprE1 core structure described here reveals a unique active site conformation in a conserved vanillyl-alcohol oxidase fold and identifies two flexible segments that are likely to facilitate access to the membrane-bound substrates. Analysis of the active site cavity together with computational studies will identify the key binding interactions of currently known inhibitor molecules and potentially enable the selection of new compounds that do not require prior activation by enzymatic reduction of a nitroso group. Together with the DprE1_mt study, these data provide a structural basis for further development of benzothiazinone and dinitrobenzamide compounds as tuberculosis drug candidates.
Supplementary Material
Acknowledgments
This work was supported by grant P20RR15578 from the U.S. National Institutes of Health. We thank John Schwanof and Randy Abramowitz for access to the X4A and X4C beamlines.
References
- 1.Makarov V, Manina G, Mikusova K, Mollmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324(5928):801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikusova K, Huang H, Yagi T, Holsters M, Vereecke D, D'Haeze W, Scherman MS, Brennan PJ, McNeil MR, Crick DC. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J Bacteriol. 2005;187(23):8020–8025. doi: 10.1128/JB.187.23.8020-8025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K. Benzothiazinones: prodrugs that covalently modify the decaprenylphosphoryl-beta-D-ribose 2'-epimerase DprE1 of Mycobacterium tuberculosis. J Am Chem Soc. 2010;132(39):13663–13665. doi: 10.1021/ja106357w. [DOI] [PubMed] [Google Scholar]
- 4.Trefzer C, Skovierova H, Buroni S, Bobovska A, Nenci S, Molteni E, Pojer F, Pasca MR, Makarov V, Cole ST, Riccardi G, Mikusova K, Johnsson K. Benzothiazinones are suicide inhibitors of mycobacterial decaprenylphosphoryl-beta-D-ribofuranose 2'-oxidase DprE1. J Am Chem Soc. 2012;134(2):912–915. doi: 10.1021/ja211042r. [DOI] [PubMed] [Google Scholar]
- 5.Musa TL, Ioerger TR, Sacchettini JC. The tuberculosis structural genomics consortium: a structural genomics approach to drug discovery. Advances in protein chemistry and structural biology. 2009;77:41–76. doi: 10.1016/S1876-1623(09)77003-8. [DOI] [PubMed] [Google Scholar]
- 6.Leferink NG, Heuts DP, Fraaije MW, van Berkel WJ. The growing VAO flavoprotein family. Archives of biochemistry and biophysics. 2008;474(2):292–301. doi: 10.1016/j.abb.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Shi W, Robinson H, Sullivan M, Abel D, Toomey J, Berman LE, Lynch D, Rosenbaum G, Rakowsky G, Rock L, Nolan B, Shea-McCarthy G, Schneider D, Johnson E, Sweet RM, Chance MR. Beamline X29: a novel undulator source for X-ray crystallography. Journal of synchrotron radiation. 2006;13(Pt 5):365–372. doi: 10.1107/S0909049506027853. [DOI] [PubMed] [Google Scholar]
- 8.Kabsch W. XDS. Acta Crystallog D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 10.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallog D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallog D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 14.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batt SM, Jabeen T, Bhowruth V, Quill L, Lund PA, Eggeling L, Alderwick LJ, Futterer K, Besra GS. Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(28):11354–11359. doi: 10.1073/pnas.1205735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.