Abstract
Secreted and transmembrane proteins play critical roles in myocardial health and disease. Studies in non-myocytes have shown that the peri-nuclear ER is the site for synthesis, folding and quality control of most secreted and transmembrane proteins, as well as a nexus of a signal transduction system, the ER stress response, that informs the cell about the status of ER protein folding. Moreover, the dynamic physical and functional association of the ER with mitochondria has emerged as a key site of integrating ER function with mitochondrial metabolism, but is only just beginning to be understood in the myocardium. Although a great deal is known about roles played by the sarcoplasmic reticulum (SR) in contractile calcium handling in the heart, little is known about the relative locations and functions of the peri-nuclear ER and the SR in terms of secreted and membrane protein synthesis and folding. In this review we will explore the current state of knowledge of the location of secreted and membrane protein synthesis, folding, and quality control machinery in cardiac myocytes, as well as our understanding of the functional consequences of ER stress and the unfolded protein response in the heart in terms of protein synthesis, cell growth, and metabolic regulation.
Keywords: Myocardium, cardiac myocyte, sarcoplasmic reticulum, endoplasmic reticulum, protein synthesis, folding, quality control ER stress, unfolded protein response
1. Introduction: Defining the SR and ER in Cardiac Myocytes
The network of membranes called the endoplasmic reticulum (ER) is a well-studied organelle in various cell types [1]. Since its discovery and visualization by George Palade [2], the rough ER (RER), studded with ribosomes, has been shown to be the major site of secreted and membrane protein synthesis [3]. Proteins synthesized in the RER are routed to the Golgi where they are directed to their final destinations [4]. Although secreted and membrane protein synthesis in the RER has been studied extensively in many cell types [5, 6], it remains largely uncharacterized in cardiac myocytes. A network of membranes similar to the ER, called the sarcoplasmic reticulum (SR) (Fig. 1A) has been defined and studied in striated muscle cells, including in cardiac myocytes. The SR surrounds the myofilaments and operates in collaboration with deep invaginations of the sarcolemma, called transverse (t)-tubules (Fig. 1B), to regulate the release of calcium from the SR lumen into the cytoplasm, where it regulates myocyte contraction [7, 8]. However, the relative locations, protein synthetic functions, and protein expression profiles of the RER and the SR in cardiac myocytes are unclear [9, 10]. Some evidence suggests that the RER and, thus, the site for secreted and membrane protein synthesis in cardiac myocytes is in a peri-nuclear network that is contiguous with the nuclear envelop (Fig. 1C and 1D), while other evidence suggests that protein synthesis may also take place in the SR [11–14]. This latter concept is supported by findings that at least part of the SR is physically contiguous with the peri-nuclear ER (Fig. 1E), such that calcium can diffuse freely between the two membrane systems [15].
Figure 1. Sarco/endoplasmic Reticulum Network in a Cardiac Myocyte.
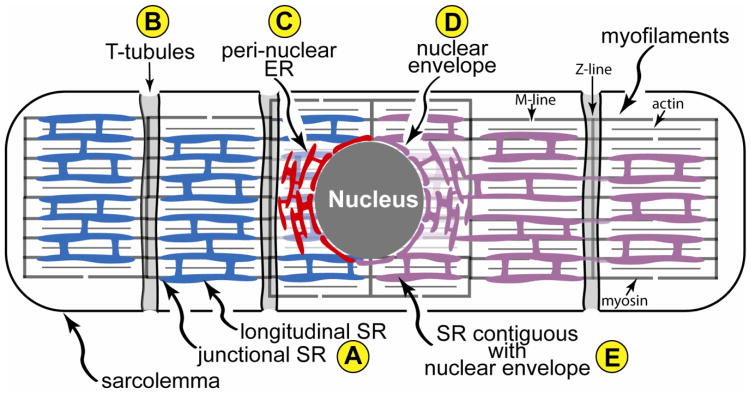
Shown is a diagram of a cardiac myocyte depicting the relationships between the region of the SR near the t-tubule, junctional SR, the longitudinal SR (A), transverse, or t-tubules (B), the peri-nuclear ER (C) and nuclear envelope (D), and a depiction of SR that is contiguous with the nuclear envelope (E) The t-tubules are invaginations of the sarcolemma that reside over the Z-line of the sarcomeres. Also shown are the actin and myosin that comprise major portions of myofilaments, as well as the M- and Z-line regions of the sarcomeres. The nuclear envelope and peri-nuclear ER are contiguous, and constitute a location for secreted and membrane protein synthesis, as well as calcium storage and release. The hypothetical localization of secreted and membrane protein synthesis to only the nuclear envelope and peri-nuclear ER, depicted in red, and not to the SR, depicted in blue, is shown on the left of the diagram. The hypothetical localization of secreted and membrane protein synthesis to the nuclear envelope, peri-nuclear ER and the SR, depicted as a contiguous membranous system, shaded purple, is shown on the right of the diagram.
Secreted and membrane proteins made in the heart have important functions in the heart, as well as in other locations [4]. Since the folding and synthesis of secreted and membrane proteins can be impaired during some cardiac pathologies, there is heightened interest amongst cardiovascular researchers in the unfolded protein response (UPR), sometimes called the ER stress response [16]. This interest has driven studies aimed at delineating the location of the ER in cardiac myocytes [16]. Electron microscopy has been used extensively to examine the ultrastructure of cardiac myocytes. While this technique may be useful for assessing some aspects of myocyte structure, the location and extent of the RER in cardiac myocytes has been difficult to determine by electron microscopy due to technical limitations [17]. Moreover, while confocal immunocytofluorescent microscopy has thus far played a major role in localizing many SR proteins that are associated with excitation-contraction coupling, it has been used in only a few studies to localize proteins associated with ER protein synthesis, folding and quality control in myocytes. Such studies have shown that proteins involved in ER protein synthesis, folding and quality control can be found in peri-nuclear regions of cardiac myocytes, as well as in peripheral areas, where they adopt an SR-like pattern [11–14, 18, 19]. However, additional studies are required to settle fundamental questions about whether the SR serves as a site of protein synthesis, and if not, whether the separate network of membranes around the nucleus in cardiac myocytes is sufficient to fulfill the needs of cardiac myocytes to synthesize secreted and membrane proteins. But since it is clear that most secreted and membrane proteins must be synthesized within a network of cellular membranes, the definition of which, in cardiac myocytes, is a topic of ongoing investigation, for the purposes of this review, we will refer to the network in which protein synthesis and folding take place as the ER, and to the unfolded protein response emanating from the ER, as the ER stress response.
2. The ER Stress Response
Nearly all proteins must be folded into functional configurations [20]. The folding of proteins synthesized in the ER takes place co- and post-translationally, and it involves a complex cast of characters that reside in the ER, which constitute the ER protein synthesis, folding and quality control machinery [21, 22]. An important function of this machinery is sensing the status of ER protein folding by detecting slight changes in the levels of unfolded proteins, and then communicating a status report proactively to the other parts of the cell that respond by adjusting the capacity of the system, thus homeostatically balancing the protein folding demand with the capacity of the protein folding machinery [23]. Drastic changes in the status of protein folding that threaten this homeostatic mechanism initiate a more reactive version of this response, which is sometimes called the ER stress response [10, 24–27].
The ER stress response can be activated by conditions that alter the ER environment in ways that impair nascent ER protein glycosylation, disulfide bond formation, or calcium levels; such conditions are often observed in the ischemic, hypertrophic and failing heart [13, 28, 29] (Fig. 2A). Three ER-transmembrane signaling proteins are major proximal sensors of unfolded proteins in the ER: PERK (protein kinase RNA-like ER kinase) [30, 31], IRE-1 (inositol-requiring protein-1) [32–35], and ATF6 (activating transcription factor 6) [36–38] (Fig. 2B–D). When activated by misfolded proteins in the ER, these sensors facilitate the activation of the transcription factors, ATF4, XBP1, and ATF6 (Fig. 2E–G), which mediate the induction of ER stress response genes that encode ER-targeted chaperones, calcium-binding proteins, and disulfide isomerases, as well as many proteins targeted to other cellular locations. Together, these proteins enhance nascent ER protein folding. ER stress also suppresses most protein synthesis, while selectively increasing translation of selected mRNAs, most of which encode ER stress response genes (Figure 2H) [39]. This selective translational repression is thought to conserve energy and reduce demands on the ER protein folding machinery [22]. ER stress also augments the ER-associated protein degradation system (ERAD), leading to proteasome-mediated degradation of terminally misfolded ER proteins, which helps relieve ER stress (Figure 2I) [40]. Furthermore, under some conditions ER stress activates autophagy [41–43], an energy-conserving, catabolic process that can promote cell survival [44]. Together, under homeostatic conditions, these aspects of the ER stress response match ER protein folding capacity with demand, which is adaptive (Fig. 2J). However, if these aspects of the ER stress response are not sufficient to meet the demand, other ER stress signaling processes guide the cell toward apoptotic cell death, which converts the ER stress response to a maladaptive process (Figure 2K) [45]. Thus, depending on its strength and duration, ER stress can be survival-, or death-oriented [21].
Figure 2. ER Stress Response Signaling.
Shown is a diagram of the rough ER with attached ribosomes translating mRNAs that encode ER luminal proteins. Conditions that impair the folding of nascent ER proteins, which include ischemia, hypertrophy and heart failure, can result in ER stress (A). Under non-stressed conditions, the ER-luminal chaperone, glucose-regulated protein 78 kDa (GRP78) associates with the luminal domains of the 3 proximal effectors of ER stress, PERK (B), IRE-1 (C) and ATF6 (D). Upon ER stress, GRP78 relocates from the luminal domains of these proteins to misfolded proteins and either facilitates their folding, or escorts them to the degradation machinery. The disassociation of GRP78 from PERK, IRE-1 allows their oligomerization, which fosters trans-phosphorylation and activation of these effectors. In the case of ATF6, dissocation of GRP78 allows ATF6 to relocate to the Golgi, where it is cleaved by site 1 and site 2 proteases which reside in the Golgi. The resulting N-terminal fragment is liberated from the Golgi (G), translocates to the nucleus, and binds to ER stress response elements in ER stress response (ERSR) genes, and regulates their transcription. Activated PERK (C) phosphorylates the eIF2a, which fosters transient global translational repression (H) and the translation of the ATF4 mRNA from an alternate start site to generate active ATF4 using an alternate open reading frame (ORF) (E). Activated IRE-1 splices the unspliced form of XBP1 mRNA (XBP1u mRNA) to generate a splice varient form (XBP1s mRNA) which encodes the active transcription factor, XBP1 (F). Like ATF6, XBP1 and ATF6 translocate to the nucleus, and bind to various types of regulator elements in ERSRs to regulate their expression. Depending on the strength and duration of the ER stress, ERSR proteins can foster enhanced ER protein folding capacity, as well as energy conservation, which is sometimes called the adaptive ER stress response, which supports cell survival (J), or ER stress response proteins can lead to cell death, which is sometimes called the maladaptive ER stress response (K).
3. The ER Stress Response in the Heart
Interest in the ER stress response in the heart was spawned partly by the realization that myocardial ischemia and cardiac hypertrophy might alter the ER in cardiac myocytes in ways that would be predicted to impair protein folding in this organelle [13, 46, 47]. Accordingly, studies were carried out to examine the conditions under which ER stress is activated in cardiac myocytes in culture or in vivo. For example, ER calcium depletion induces SERCA2 gene expression in cultured neonatal cardiac myocytes [48]. In addition to pharmacological inhibitors of protein glycosylation and compounds that alter ER redox status, ER stress is also activated in cultured cardiac myocytes by maneuvers that mimic pathology, such as simulated ischemia [13, 49]. Moreover, ER stress is activated in border zone cardiac myocytes in the infarcted mouse heart, in vivo, [13], in the hearts of mice subjected to pressure overload [47], and in a genetic model of heart failure [50].
Once it was apparent that ER stress was activated in cardiac myocytes under various pathological states, studies were undertaken to examine the functional effects of ER stress in the heart. For example, several studies have shown that in cultured cardiac myocytes, and in mouse heart, in vivo, the ATF6 branch of the ER stress response appears to be adaptive [11, 13, 28, 51]. Transgenic mouse models of ATF6 gain- [51] and loss-of-function [52] have demonstrated that ATF6 is cardioprotective, in vivo. Thrombospondin4 has been shown to be cardioprotective, in part because it is required for ATF6 activation in cardiac myocytes, in vivo [53]. A microarray study showed that in the mouse myocardium, ATF6 induces hundreds of genes encoding numerous SR/ER-targeted chaperones, protein disulfide isomerases, calcium binding proteins, and other proteins, some of which are targeted to the cytosol [11], as well as changes in the levels of key microRNAs [54]. Although most of these studies support adaptive, protective roles for the ATF6 and XBP1 branches of the ER stress response in the heart, other studies have shown maladaptive effects of ER stress activation in myocardial pathologies (reviewed in a series [55]). For example, -adrenergic receptor activation has been shown to activate ER stress-mediated apoptosis in cultured cardiac myocytes [56, 57], and PKC-mediated myocardial damage was shown to be mediated, partly by its effects on activating ER stress [58]. Pressure overload is thought to activate ER stress-mediated apoptosis in the mouse myocardium [47], and ER stress was shown to contribute to ischemia-induced apoptosis in cultured cardiac myocytes [59].
4. The ER as a Nexus for Metabolic Signaling and Cell Growth
In addition to ER stress-mediated activation of the canonical unfolded protein response, the ER, which accounts for more than 50% of cellular membrane [1], serves as a focal point of signaling processes, many of which are oriented toward regulating metabolic signaling and cellular growth.
Regulation of Metabolic Signaling by the ER
While there exists an extensive history of studies devoted to examining protein synthesis and quality control in the ER, as well as roles for the ER in regulated calcium release, only recently has it become apparent that the ER plays a regulatory role in cellular metabolism [60]. Indeed, under conditions of increased ER protein and lipid synthesis, which are ATP-utilizing processes, it is reasonable to assume that metabolic pathways responsible for ATP synthesis must be sufficient to meet the increased energy demands. In part, ER-mediated regulation of energy metabolism is the result of a direct interaction between mitochondria and the ER. There exists an intricate multi-organelle signaling process that involves calcium transfer between the ER and mitochondria [61–64], which is facilitated by a physical association of the two organelles in a structure known as the mitochondria-associated ER membrane, or MAM [65]. Only a small portion of the outer mitochondrial membrane is ER-associated [64], suggesting that a relatively small proportion of ER-derived calcium is transferred to mitochondria. Nevertheless, this direct calcium transfer serves as a mechanism by which mitochondria can sense and respond to conditions in the ER that require adjustments in metabolism [60, 66].
Calcium is released from the ER through the ryanodine and IP3 receptors (RyR; IP3R) [67]. A portion of ER-derived calcium enters a microdomain of the cytosol that is in direct juxtaposition with mitochondria. This positioning, which is the result of physical tethering of the two organelles, involves the binding of the cytosolic chaperone, GRP75, to the ER-associated IP3 receptor and mitochondrial-associated voltage-dependent ion selective channel (VDAC) [66]. Studies using non-cardiac myocyte cells established that the ER/mitochondrial tethering also involves the mitochondrial and ER transmembrane GTPase, mitofusin 2 [68]. Thus, calcium released from the RyR or IP3R enters the ER/mitochondrial space, passes through voltage-dependent ion selective channel (VDAC) in the outer mitochondrial membrane, and then through the mitochondrial calcium uniporter (MCU) located in the inner mitochondrial membrane [69]. In the mitochondrial matrix, calcium influences the activities of the Krebs cycle enzymes, -ketogluarate, pyruvate, and isocitrate dehydrogenases, in ways that increase mitochondrial ATP production [70]. During certain ER stress conditions, calcium release from the ER and, thus, calcium entry into mitochondria is increased, which enhances ATP production and, in so doing, provides a metabolic response that can support adaptive, ATP-requiring aspects of the ER stress response [65].
A physical connection between the SR and mitochondria has recently been established in cardiac myocytes [71]. The functional consequences of physical interactions of the ER, and perhaps the SR, with mitochondria in the heart have been examined in several papers that employed targeted disruption of the mitofusin 2 gene in mice [72, 73]. Taken together, these studies provide evidence that in cardiac myocytes, calcium transfer from the ER into mitochondria can regulate mitochondrial metabolic function in the mouse heart, in vivo. An additional example of how changes in the ER environment could affect metabolic function is the recent finding that mitofusin 2 is an ER stress-inducible protein [74], suggesting that ER stress can enhance the extent of ER/mitochondrial tethering. This is somewhat expected, since increased demands for ER protein synthesis and folding can lead to expansion of the ER and increases in membrane lipid synthesis, both of which are ATP-requiring.
In the heart, one pathological condition that increases ER stress, and may therefore affect calcium transport from the ER and SR to mitochondria, is ischemia [13, 28]. Under these conditions, decreased mitochondrial ATP production, which impairs ER protein folding, partly by depleting ER calcium. Decreased ER calcium activates proximal sensors of ER stress, because ER chaperones require calcium to fold nascent proteins [1]. A study that investigated the effects of simulated ischemia on endothelial cell injury found that the main pathway of the simulated ischemia-induced apoptosis consisted of the calcium leak from the ER, followed by activation of caspase-12 and caspase-3 [75]. Thus, changes in cellular and ER calcium dynamics have global effects on cellular viability, which could involve signals that arise from a calcium-depleted ER and/or SR, depending on the cell type.
Abnormal calcium cycling is known to cause arrhythmia, especially in the setting of transient cardiac ischemia followed by reperfusion. In isolated beating mouse heart, three minutes of ischemia increases diastolic and systolic calcium with a decrease in calcium transient amplitude [76] and increased decay time constant of calcium transient, consistent with inhibition of SR calcium release and re-uptake due to ischemia and acidosis. At the onset of reperfusion, calcium overload is thought to be further exacerbated by uncontrolled influx from the extracellular space [77, 78]. A slight but consistent transient increase in diastolic calcium occurs when the amplitude of the calcium transient is decreased. This increase in calcium may have important functional consequences. Interestingly, Valverde et al have shown in intact beating hearts that SR calcium release contributes to the transient cytosolic increase of calcium at the onset of reperfusion [76]. This is particularly important in light of interactions between mitochondria and the SR, and could be a potential cause of ER stress during ischemia/reperfusion.
ER Protein Synthesis and Cell Growth
Among the basic principles of protein synthesis is the concept that translation of proteins bound for secretion and transmembrane proteins occurs on ER-associated ribosomes, while all other proteins are made on free ribosomes. However, puzzling observations from studies carried out using microarray approaches to identify mammalian cell, yeast, and fly mRNAs associated with free and ER-associated ribosomes challenged this principle by finding that some mRNAs that encode cytosolic proteins were found on the RER [79]. A more recent study used cell fractionation and ribosome profiling to sequence all mRNAs associated with cytosolic and/or ER-associated ribosomes [80]. In that study, it was found that, in addition to secreted and membrane proteins, ER-associated ribosomes were also engaged in translating mRNAs that encode proteins targeted to other cellular locations. Moreover, it was found that, in vivo, ER-associated ribosomes are more efficient at translating mRNAs, and that they contribute to the synthesis of nearly all proteins. This finding not only expands the scope of protein synthesis on the RER to include the translation of nearly all cytosolic, secreted, and transmembrane proteins, it redefines the importance of signal transduction systems that monitor the quantity and quality of protein synthesis in the ER as regulators of the proteome under growth-promoting as well as growth-limiting conditions.
Under growth-limiting conditions many aspects of the ER stress response program the cell to conserve. As such, the adaptive ER stress response detects potential proteotoxicity due to misfolded proteins, and rebalances the proteome by decreasing overall cellular protein synthesis, and focusing cellular resources on synthesizing essential proteins [23]. Thus, stress signaling from the ER in response to misfolded proteins is predicted to shift metabolism toward maintaining essential cellular functions and modulating cellular growth. Consistent with this prediction are the results of a microarray study that characterized changes in the transcriptome in mouse hearts upon activation of the ATF6 branch of the ER stress response, which is considered to be adaptive [11]. In terms of growth and metabolism, many growth-promoting genes were downregulated, while genes related to metabolic conservation were upregulated. Thus, one way in which ATF6 is adaptive is that it reprograms cardiac myocytes in ways designed to conserve energy and limit growth.
Under growth-promoting conditions the mammalian target of rapamycin (mTOR) signaling pathway is a nodal sensor of cellular protein synthesis and, thus, a central regulator of cell growth and metabolism [81]. The effects of mTOR signaling in the myocardium have been well studied and shown to be protective and growth promoting [82–84]. Accordingly, it seems reasonable to assume that cross-talk between ER stress and mTOR signaling might contribute to balancing the proteome with nutritional availability and cellular growth requirements. Generally, mTOR is activated by conditions that are favorable for growth, which makes it an anabolic pathway [81]. On the other hand, ER stress is usually activated by conditions that do not favor growth, which, for the most part, makes it an energy conserving, catabolic pathway [21, 22, 39]. In keeping with their opposing effects on metabolism, although not well established in the heart, many studies of ER stress and mTOR cross-talk in other cell types have shown that the two pathways are often activated in opposition [85]. For example, ER stress is activated by hypoxia or nutrient starvation, while mTOR is inhibited under these conditions [85]. However, the reciprocal activity status of ER stress and mTOR signaling is not always observed, and in some cases, ER stress and mTOR are co-activated [85]. Thus, while there exists crosstalk between ER stress and mTOR signaling, the effect of the cross-talk is apparently conditional, and the molecular mechanisms, as well as the functional consequences of the cross-talk are not well understood.
In addition to being regulated by growth-promoting conditions, the magnitude and effects of mTOR signaling are also dependent on the location of mTOR signaling components. For example, activation of mTOR complex 1, or mTORC1, by the small GTP binding protein, Rheb, occurs on the cytosolic surface of lysosomes, where mTOR signaling is integrated with the availability of amino acids from lysosome-mediated proteolysis [86]. Also, recent studies have shown that mTOR binds to ribosomes [87] and localizes to the ER [88, 89], and that disruption of this localization reduces the anabolic effects of mTOR on protein synthesis [90]. Thus, it is possible that the effects of ER stress and mTOR signaling on each other are not only dependent on the nature of the cellular growth activator or inhibitor, but also on the subcellular localization of signaling complexes. Moreover, it is feasible that the co-localization of mTOR components to the ER, where we now know that most protein synthesis takes place, may provide an important spatial dimension to the cross-talk between the ER and mTOR signaling pathways.
5. Therapeutic Potential of Targeting ER Stress Signaling in the Heart
The knowledge about ER stress signaling in the heart and other tissues that has been acquired over the last decade has provided an important foundation that is required to evaluate the therapeutic potential of this complex signaling pathway in treating heart disease. With the understanding that some aspects of ER stress are adaptive, while others are maladaptive has come the realization that broadly activating or inactivating ER stress signaling is not likely to be a fruitful therapeutic approach. Instead, it has become clear that selectively targeting specific aspects of ER stress signaling, such as enhancing adaptive ER stress signaling is a likely requirement in order to achieve the desired effects. An examination of the changes in gene expression exerted by ATF6 in the heart indicates that ATF6 upregulates many protective genes and downregulates numerous potentially damaging genes [11]. Thus, selectively activating the ATF6 branch of the ER stress response is predicted to be adaptive. In support of this hypothesis are studies that have shown that activating ATF6 in cardiac myocytes protects the heart from ischemic damage, as well as dysfunction resulting from overload hypertrophy, while inhibiting ATF6 has the opposite effects [51–53]. Moreover, other studies in which genes that lie downstream of ATF6, such as GRP94, GRP78, Derlin-3, PDI, PDIA6, RCAN1, and MANF are expressed in cardiac myocytes, they protect against models of ischemic damage [11, 91–96]. Additionally, the use of chemical chaperones which mimic the effects of upregulating ER-targeted chaperones during adaptive ER stress, are also effect in improving functional outcomes in animal models of cardiac pathology [97]. Presumably, inhibiting the maladaptive aspects of ER stress signaling would also be expected to be a fruitful therapeutic approach. In support of this approach is a study in which targeted deletion of the gene encoding the maladaptive ER stress-inducible protein, PUMA, improves cardiac function in mouse model of myocardial ischemia [98]. While studies such as these showcase the therapeutic potential of altering ER stress signaling pathways as a means to treat ischemic and hypertrophic heart diseases, they also demonstrate that strategies that target specific aspects of ER stress will probably be required in order to develop viable treatments.
6. Conclusions
Protein synthesis is the major regulator of cell growth, which is a critical parameter of cardiac myocyte function in the healthy and diseased heart. The ER, which is the largest organelle, serves as the site for most secreted and membrane protein synthesis, as well as the synthesis of many other proteins, even those that are targeted to other locations. Thus, the ER is a major integration site of cell growth signaling. In cardiac myocytes, the ER membrane network is potentially more expansive than many other cell types, due to the role played by the SR in contractile calcium handling. The potential overlap in function between the SR and the ER in terms of protein synthesis and folding, as well as ER stress and mTOR signal transduction, suggests that the SR and ER membrane system is a macro-organelle that plays critical roles in cardiac myocyte contraction, growth and metabolism, all of which are dominant contributors to myocardial function.
Highlights.
Secreted and transmembrane proteins have important functions in the heart.
Secreted and transmembrane proteins are made on the ER of cardiac myocytes.
The ER is a major protein synthesis site and integrator of cell growth signaling.
Folding of proteins made on the ER is impaired during cardiac pathology.
The adaptive ER stress response augments ER protein folding and reduces pathological damage.
Acknowledgments
Research in the Glembotski lab is supported by National Institutes of Health, grants HL-075573, HL-085577, and HL104535 to CCG, and by grants and fellowships from the Rees-Stealy Research Foundation, the San Diego Chapter of the Achievement Rewards for College Scientists (ARCS) Foundation, the American Heart Association (Predoctoral Fellowship 10PRE3410005), and the Inamori Foundation, to SD.
Footnotes
Disclosures
None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERNCES
- 1.Simmen T, Lynes EM, Gesson K, Thomas G. Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2010;1798:1465–73. doi: 10.1016/j.bbamem.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palade GE, Siekevitz P. Liver microsomes; an integrated morphological and biochemical study. J Biophys Biochem Cytol. 1956;2:171–200. doi: 10.1083/jcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter KR, Kallman FL. Significance of cell particulates as seen by electron microscopy. Ann N Y Acad Sci. 1952;54:882–91. doi: 10.1111/j.1749-6632.1952.tb39963.x. [DOI] [PubMed] [Google Scholar]
- 4.Doroudgar S, Glembotski CC. The cardiokine story unfolds: ischemic stress-induced protein secretion in the heart. Trends Mol Med. 2011;17:207–14. doi: 10.1016/j.molmed.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caro LG, Palade GE. Protein Synthesis, Storage, and Discharge in the Pancreatic Exocrine Cell. An Autoradiographic Study. J Cell Biol. 1964;20:473–95. doi: 10.1083/jcb.20.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingappa VR, Blobel G. Early events in the biosynthesis of secretory and membrane proteins: the signal hypothesis. Recent Prog Horm Res. 1980;36:451–75. doi: 10.1016/b978-0-12-571136-4.50018-8. [DOI] [PubMed] [Google Scholar]
- 7.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 8.Sobie EA, Lederer WJ. Dynamic local changes in sarcoplasmic reticulum calcium: physiological and pathophysiological roles. J Mol Cell Cardiol. 2012;52:304–11. doi: 10.1016/j.yjmcc.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesaeli N, Nakamura K, Opas M, Michalak M. Endoplasmic reticulum in the heart, a forgotten organelle? Mol Cell Biochem. 2001;225:1–6. doi: 10.1023/a:1012209923231. [DOI] [PubMed] [Google Scholar]
- 10.Michalak M, Opas M. Endoplasmic and sarcoplasmic reticulum in the heart. Trends Cell Biol. 2009;19:253–9. doi: 10.1016/j.tcb.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Belmont PJ, Tadimalla A, Chen WJ, Martindale JJ, Thuerauf DJ, Marcinko M, et al. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J Biol Chem. 2008;283:14012–21. doi: 10.1074/jbc.M709776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaisto T, Metsikko K. Distribution of the endoplasmic reticulum and its relationship with the sarcoplasmic reticulum in skeletal myofibers. Exp Cell Res. 2003;289:47–57. doi: 10.1016/s0014-4827(03)00231-3. [DOI] [PubMed] [Google Scholar]
- 13.Thuerauf DJ, Marcinko M, Gude N, Rubio M, Sussman MA, Glembotski CC. Activation of the unfolded protein response in infarcted mouse heart and hypoxic cultured cardiac myocytes. Circ Res. 2006;99:275–82. doi: 10.1161/01.RES.0000233317.70421.03. [DOI] [PubMed] [Google Scholar]
- 14.Volpe P, Villa A, Podini P, Martini A, Nori A, Panzeri MC, et al. The endoplasmic reticulum-sarcoplasmic reticulum connection: distribution of endoplasmic reticulum markers in the sarcoplasmic reticulum of skeletal muscle fibers. Proc Natl Acad Sci U S A. 1992;89:6142–6. doi: 10.1073/pnas.89.13.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, et al. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J Clin Invest. 2006;116:675–82. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knollmann BC. A “rough” journey to the sarcoplasmic reticulum--implications of altered calsequestrin trafficking for cardiac arrhythmia. J Mol Cell Cardiol. 2010;49:554–5. doi: 10.1016/j.yjmcc.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes A. Cardiomyocyte Ultrastructure. In: Hill JaO EN, editor. Muscle: Fundamental Biology and Mechanisms of Disease. San Diego: Academic Press; 2012. pp. 47–55. [Google Scholar]
- 18.McFarland TP, Milstein ML, Cala SE. Rough endoplasmic reticulum to junctional sarcoplasmic reticulum trafficking of calsequestrin in adult cardiomyocytes. J Mol Cell Cardiol. 2010;49:556–64. doi: 10.1016/j.yjmcc.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vangheluwe P, Louch WE, Ver Heyen M, Sipido K, Raeymaekers L, Wuytack F. Ca2+ transport ATPase isoforms SERCA2a and SERCA2b are targeted to the same sites in the murine heart. Cell Calcium. 2003;34:457–64. doi: 10.1016/s0143-4160(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 20.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–30. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 21.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 22.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 23.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–94. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101:975–84. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 25.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44:453–9. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107:1071–82. doi: 10.1161/CIRCRESAHA.110.227819. [DOI] [PubMed] [Google Scholar]
- 27.Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- 28.Doroudgar S, Thuerauf DJ, Marcinko MC, Belmont PJ, Glembotski CC. Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J Biol Chem. 2009;284:29735–45. doi: 10.1074/jbc.M109.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groenendyk J, Sreenivasaiah PK, Kim do H, Agellon LB, Michalak M. Biology of endoplasmic reticulum stress in the heart. Circ Res. 2010;107:1185–97. doi: 10.1161/CIRCRESAHA.110.227033. [DOI] [PubMed] [Google Scholar]
- 30.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 33.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–24. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–17. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa M, Sidrauski C, Kaufman RJ, Walter P. A role for presenilin-1 in nuclear accumulation of Ire1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–66. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–9. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 38.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–43. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plemper RK, Wolf DH. Endoplasmic reticulum degradation. Reverse protein transport and its end in the proteasome. Mol Biol Rep. 1999;26:125–30. doi: 10.1023/a:1006913215484. [DOI] [PubMed] [Google Scholar]
- 41.Clarke R, Cook KL, Hu R, Facey CO, Tavassoly I, Schwartz JL, et al. Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Cancer Res. 2012;72:1321–31. doi: 10.1158/0008-5472.CAN-11-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–7. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 43.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gustafsson AB, Gottlieb RA. Recycle or die: the role of autophagy in cardioprotection. J Mol Cell Cardiol. 2008;44:654–61. doi: 10.1016/j.yjmcc.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–64. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277:20734–9. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- 47.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, et al. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–12. doi: 10.1161/01.CIR.0000137836.95625.D4. [DOI] [PubMed] [Google Scholar]
- 48.Thuerauf DJ, Hoover H, Meller J, Hernandez J, Su L, Andrews C, et al. Sarco/endoplasmic reticulum calcium ATPase-2 expression is regulated by ATF6 during the endoplasmic reticulum stress response: intracellular signaling of calcium stress in a cardiac myocyte model system. J Biol Chem. 2001;276:48309–17. doi: 10.1074/jbc.M107146200. [DOI] [PubMed] [Google Scholar]
- 49.Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, et al. AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol. 2005;25:9554–75. doi: 10.1128/MCB.25.21.9554-9575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bousette N, Chugh S, Fong V, Isserlin R, Kim KH, Volchuk A, et al. Constitutively active calcineurin induces cardiac endoplasmic reticulum stress and protects against apoptosis that is mediated by alpha-crystallin-B. Proc Natl Acad Sci U S A. 2010;107:18481–6. doi: 10.1073/pnas.1013555107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, et al. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–93. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 52.Toko H, Takahashi H, Kayama Y, Okada S, Minamino T, Terasaki F, et al. ATF6 is important under both pathological and physiological states in the heart. J Mol Cell Cardiol. 2010;49:113–20. doi: 10.1016/j.yjmcc.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 53.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, et al. A Thrombospondin-Dependent Pathway for a Protective ER Stress Response. Cell. 2012;149:1257–68. doi: 10.1016/j.cell.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belmont PJ, Chen WJ, Thuerauf DJ, Glembotski CC. Regulation of microRNA expression in the heart by the ATF6 branch of the ER stress response. J Mol Cell Cardiol. 2012;52:1176–82. doi: 10.1016/j.yjmcc.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitakaze M, Tsukamoto O. What is the role of ER stress in the heart? Introduction and series overview. Circ Res. 2010;107:15–8. doi: 10.1161/CIRCRESAHA.110.222919. [DOI] [PubMed] [Google Scholar]
- 56.Ni L, Zhou C, Duan Q, Lv J, Fu X, Xia Y, et al. beta-AR blockers suppresses ER stress in cardiac hypertrophy and heart failure. PLoS One. 2011;6:e27294. doi: 10.1371/journal.pone.0027294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dalal S, Foster CR, Das BC, Singh M, Singh K. Beta-adrenergic receptor stimulation induces endoplasmic reticulum stress in adult cardiac myocytes: role in apoptosis. Mol Cell Biochem. 2012;364:59–70. doi: 10.1007/s11010-011-1205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qi X, Vallentin A, Churchill E, Mochly-Rosen D. deltaPKC participates in the endoplasmic reticulum stress-induced response in cultured cardiac myocytes and ischemic heart. J Mol Cell Cardiol. 2007;43:420–8. doi: 10.1016/j.yjmcc.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szegezdi E, Duffy A, O’Mahoney ME, Logue SE, Mylotte LA, O’Brien T, et al. ER stress contributes to ischemia-induced cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2006;349:1406–11. doi: 10.1016/j.bbrc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 60.Bravo R, Gutierrez T, Paredes F, Gatica D, Rodriguez AE, Pedrozo Z, et al. Endoplasmic reticulum: ER stress regulates mitochondrial bioenergetics. Int J Biochem Cell Biol. 2012;44:16–20. doi: 10.1016/j.biocel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–7. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- 62.Giacomello M, Drago I, Bortolozzi M, Scorzeto M, Gianelle A, Pizzo P, et al. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol Cell. 2010;38:280–90. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 63.Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–32. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–21. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–8. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 67.Marks AR. Intracellular calcium-release channels: regulators of cell life and death. Am J Physiol. 1997;272:H597–605. doi: 10.1152/ajpheart.1997.272.2.H597. [DOI] [PubMed] [Google Scholar]
- 68.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–10. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 69.De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szabadkai G, Duchen MR. Mitochondria: the hub of cellular Ca2+ signaling. Physiology (Bethesda) 2008;23:84–94. doi: 10.1152/physiol.00046.2007. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Perez C, Schneider TG, Hajnoczky G, Csordas G. Alignment of sarcoplasmic reticulum-mitochondrial junctions with mitochondrial contact points. Am J Physiol Heart Circ Physiol. 2011;301:H1907–15. doi: 10.1152/ajpheart.00397.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papanicolaou KN, Khairallah RJ, Ngoh GA, Chikando A, Luptak I, O’Shea KM, et al. Mitofusin-2 maintains mitochondrial structure and contributes to stress-induced permeability transition in cardiac myocytes. Mol Cell Biol. 2011;31:1309–28. doi: 10.1128/MCB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Csordas G, Jowdy C, Schneider TG, Csordas N, Wang W, et al. Mitofusin 2-Containing Mitochondrial-Reticular Microdomains Direct Rapid Cardiomyocyte Bioenergetic Responses via Inter-Organelle Ca2+ Crosstalk. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ngoh GA, Papanicolaou KN, Walsh K. Loss of mitofusin 2 promotes endoplasmic reticulum stress. J Biol Chem. 2012;287:20321–32. doi: 10.1074/jbc.M112.359174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Kasseckert S, Kostin S, Abdallah Y, Schafer C, Kaminski A, et al. Ischemic acidosis causes apoptosis in coronary endothelial cells through activation of caspase-12. Cardiovasc Res. 2007;73:172–80. doi: 10.1016/j.cardiores.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 76.Valverde CA, Kornyeyev D, Ferreiro M, Petrosky AD, Mattiazzi A, Escobar AL. Transient Ca2+ depletion of the sarcoplasmic reticulum at the onset of reperfusion. Cardiovasc Res. 2010;85:671–80. doi: 10.1093/cvr/cvp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Piper HM, Abdallah Y, Schafer C. The first minutes of reperfusion: a window of opportunity for cardioprotection. Cardiovasc Res. 2004;61:365–71. doi: 10.1016/j.cardiores.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 78.Kusuoka H, Porterfield JK, Weisman HF, Weisfeldt ML, Marban E. Pathophysiology and pathogenesis of stunned myocardium. Depressed Ca2+ activation of contraction as a consequence of reperfusion-induced cellular calcium overload in ferret hearts. J Clin Invest. 1987;79:950–61. doi: 10.1172/JCI112906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicchitta CV, Lerner RS, Stephens SB, Dodd RD, Pyhtila B. Pathways for compartmentalizing protein synthesis in eukaryotic cells: the template-partitioning model. Biochem Cell Biol. 2005;83:687–95. doi: 10.1139/o05-147. [DOI] [PubMed] [Google Scholar]
- 80.Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–27. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem. 2010;285:14071–7. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Proud CG. Ras, PI3-kinase and mTOR signaling in cardiac hypertrophy. Cardiovasc Res. 2004;63:403–13. doi: 10.1016/j.cardiores.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Hedhli N, Pelat M, Depre C. Protein turnover in cardiac cell growth and survival. Cardiovasc Res. 2005;68:186–96. doi: 10.1016/j.cardiores.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 84.Aoyagi T, Matsui T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Curr Pharm Des. 2011;17:1818–24. doi: 10.2174/138161211796390976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 2012;22:274–82. doi: 10.1016/j.tcb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 86.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–68. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 88.Drenan RM, Liu X, Bertram PG, Zheng XF. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–8. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 89.Boulbes DR, Shaiken T, Sarbassov dos D. Endoplasmic reticulum is a main localization site of mTORC2. Biochem Biophys Res Commun. 2011;413:46–52. doi: 10.1016/j.bbrc.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu X, Zheng XF. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Mol Biol Cell. 2007;18:1073–82. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu HY, Minamino T, Tsukamoto O, Sawada T, Asai M, Kato H, et al. Overexpression of endoplasmic reticulum-resident chaperone attenuates cardiomyocyte death induced by proteasome inhibition. Cardiovasc Res. 2008;79:600–10. doi: 10.1093/cvr/cvn128. [DOI] [PubMed] [Google Scholar]
- 92.Vekich JA, Belmont PJ, Thuerauf DJ, Glembotski CC. Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J Mol Cell Cardiol. 2012;53:259–67. doi: 10.1016/j.yjmcc.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tadimalla A, Belmont PJ, Thuerauf DJ, Glassy MS, Martindale JJ, Gude N, et al. Mesencephalic astrocyte-derived neurotrophic factor is an ischemia-inducible secreted endoplasmic reticulum stress response protein in the heart. Circ Res. 2008;103:1249–58. doi: 10.1161/CIRCRESAHA.108.180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Glembotski CC, Thuerauf DJ, Huang C, Vekich JA, Gottlieb RA, Doroudgar S. Mesencephalic astrocyte-derived neurotrophic factor protects the heart from ischemic damage and is selectively secreted upon sarco/endoplasmic reticulum calcium depletion. J Biol Chem. 2012;287:25893–904. doi: 10.1074/jbc.M112.356345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vitadello M, Penzo D, Petronilli V, Michieli G, Gomirato S, Menabo R, et al. Overexpression of the stress protein Grp94 reduces cardiomyocyte necrosis due to calcium overload and simulated ischemia. FASEB J. 2003;17:923–5. doi: 10.1096/fj.02-0644fje. [DOI] [PubMed] [Google Scholar]
- 96.Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U, et al. Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J Am Coll Cardiol. 2007;50:1029–37. doi: 10.1016/j.jacc.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 97.Park CS, Cha H, Kwon EJ, Sreenivasaiah PK, Kim do H. The chemical chaperone 4-phenylbutyric acid attenuates pressure-overload cardiac hypertrophy by alleviating endoplasmic reticulum stress. Biochem Biophys Res Commun. 2012;421:578–84. doi: 10.1016/j.bbrc.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 98.Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, et al. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006;291:H52–60. doi: 10.1152/ajpheart.01046.2005. [DOI] [PubMed] [Google Scholar]



