Abstract
PICKLE plays a critical role in repression of genes that regulate development identity in Arabidopsis thaliana. PICKLE codes for a putative ATP-dependent chromatin remodeler that exhibits sequence similarity to members of subfamily II of animal CHD remodelers, which includes remodelers such as CHD3/Mi-2 that also restrict expression of developmental regulators. Whereas animal CHD3 remodelers are a component of the Mi-2/NuRD complex that promotes histone deacetylation, PICKLE promotes trimethylation of histone H3 lysine 27 suggesting that it acts via a distinct epigenetic pathway. Here, we examine whether PICKLE is also a member of a multisubunit complex and characterize the biochemical properties of recombinant PICKLE protein. Phylogenetic analysis indicates that PICKLE-related proteins in plants share a common ancestor with members of subfamily II of animal CHD remodelers. Biochemical characterization of PICKLE in planta, however, reveals that PICKLE primarily exists as a monomer. Recombinant PICKLE protein is an ATPase that is stimulated by ssDNA and mononucleosomes and binds to both naked DNA and mononucleosomes. Furthermore, recombinant PICKLE exhibits ATP-dependent chromatin remodeling activity. These studies demonstrate that subfamily II CHD proteins in plants, such as PICKLE, retain ATP-dependent chromatin remodeling activity but act through a mechanism that does not involve the ubiquitous Mi-2/NuRD complex.
Keywords: chromatin remodeling, CHD proteins, ATP-dependent chromatin remodeler, SANT-SLIDE, developmental identity
1. Introduction
Chromatin structure plays a critical role in enabling developmental regulation of gene expression in eukaryotes. Plants and animals last shared a single-celled ancestor 1.6–1.9 billion years before present [1, 2], and development is likely to have evolved independently in each [3]. The machinery that determines chromatin structure and gene expression is comprised of components that are common to both kingdoms as well as factors unique to each [4]. It is of considerable interest to understand how these various molecular elements are made use of by plants and animals to determine chromatin structure and thereby facilitate their distinct developmental attributes.
CHD proteins are a family of ATP-dependent chromatin remodelers that are found in plants and animals and play a variety of roles in gene expression. The CHD proteins derive their name from the presence of three domains of sequence similarity [5]: a Chromatin organization modifier domain (chromodomain), a SWI2/SNF2 ATPase/Helicase domain, and a motif with sequence similarity to a DNA-binding domain. Three subfamilies of CHD proteins (I, II, and III) are recognized in eukaryotes [6, 7]. Members of subfamily II are distinguished by the presence of one or two copies of a PHD zinc finger domain [5, 8], as well as by the presence of two domains of unknown function.
Genetic and biochemical analyses of several animal CHD subfamily II members have revealed roles in repression of genes involved in developmental regulation [6, 9]. For example, loss of dMi-2 in D. melanogaster leads to derepression of genes that promote neural development [10, 11], whereas loss of Mi-2 in C. elegans leads to derepression of germ line-specific genes in somatic cells [12]. A related protein, CHD4, has been shown to contribute to repression of lymphoid and erythroid lineage genes in mouse hematopoietic stem cells [13] and to repression of embryonic and fetal globin genes in human adult erythroid cells [14].
Plant CHD proteins are closely related to animal members of subfamily II and also play a role in repression of developmental identity. PICKLE (PKL) codes for a CHD3/4-related protein in Arabidopsis [15, 16]. Loss of PKL leads to derepression of seed-specific genes during germination [15, 17] and to ectopic expression of meristematic genes in carpel tissue [16].
The discovery that CHD3 and CHD4 proteins are components of the Mi-2/NuRD complex provided a biochemical basis for understanding how these remodelers contribute to gene repression in vertebrates [18–21]. The Mi-2/NuRD complex contains histone deacetylase HDAC1/2 as well as the methyl CpG binding domain protein MBD2 and provides a mechanistic link between DNA methylation and transcriptional repression: DNA methylation results in targeting of the Mi-2/NuRD associated histone deacetylase activity and the resulting deacetylated histones subsequently contribute to transcriptional repression [22]. Importantly, this complex can also be recruited to loci via protein-protein interactions as well as by DNA methylation [6, 23, 24]. Mi-2/NuRD is the most abundant histone deacetylase complex in mammalian cells and has been linked to numerous developmental processes [9, 22, 25]. In addition, Mi-2/NuRD complex has been biochemically characterized in D. melanogaster [26, 27] and in C. elegans [12, 28], suggesting that CHD3-related proteins act in conjunction with histone deacetylases in invertebrates as well.
In Arabidopsis, however, PKL appears to contribute to repression not by promoting histone deacetylation but by instead promoting trimethylation of histone H3 at K27 (H3K27me3), a repressive epigenetic mark. H3K27me3-enriched loci are over-represented in the set of genes that exhibit increased transcript levels in pkl plants [29, 30]. Loss of PKL results in reduction of H3K27me3 at genomic loci, which is thought to contribute to transcriptional derepression of many of these loci [29, 30]. Although a previous study suggested that PKL acts by promoting expression of the PRC2 complex that methylates H3K27 [30], a subsequent investigation found that PKL is unlikely to act in this fashion [31]. Zhang et al. observed that PKL is not necessary for expression of the PRC2 machinery and further demonstrated that PKL protein is present at the promoters of H3K27me3-enriched genes, suggesting that PKL directly contributes to H3K27me3 at these genes. In particular, PKL is present at the promoters of LEC1 and LEC2 during germination, which is when PKL is required to repress expression of these H3K27me3-enriched loci [31].
PKL is likely to play additional roles beyond promoting H3K27me3. H3K27me3-enriched loci are also over-represented in the set of genes that exhibit decreased transcript levels in pkl plants [30, 31], suggesting that PKL also contributes to promoting expression from loci subject to this epigenetic modification. Furthermore, PKL is also present at ubiquitously expressed genes such as ACT7 and UBQ10, although a functional role has yet to be identified for PKL at this type of locus [31].
Analysis of CHD3-related proteins in animal systems similarly reveals that they can operate outside of the Mi-2/NuRD paradigm. Repression of proneural genes in D. melanogaster and of germline potential in C. elegans is likely to be mediated by a CHD3-containing complex that is distinct from the Mi-2/NuRD complex [27, 28]. CHD4 is found in a complex with the histone acetyltransferase p300 and in that context promotes expression of CD4 during T-cell development in mice [32]. In Drosophila, dMi-2 is recruited to active heat shock genes and is required for both efficient expression and transcript processing of heat shock genes [33]. Furthermore, CHD3 can also function as a co-activator for human c-Myb in a fashion that does not depend on ATPase activity [34]. Thus members of subfamily II of CHD proteins can participate in multiple remodeling pathways and can either repress or activate gene expression depending on the other factors they associate with and can also contribute to gene expression in a manner that is not dependent on remodeling activity.
Much less is known regarding the biochemical properties of plant CHD remodelers. In fact, no plant CHD remodelers have been shown to exhibit remodeling activity. Here we undertake biochemical characterization of PKL to understand how it contributes to epigenetic modifications and gene expression in plants. Phylogenetic analysis of animal and plant CHD proteins confirms that PKL-related proteins in plants share a common ancestor with animal CHD proteins that are members of subfamily II. This analysis also reveals that the existence of distinct clade of subfamily II members in plants and further reveals that subfamily III is unique to animals. We find that in contrast to CHD3-related proteins in animals, PKL primarily exists as a monomer in planta, which is consistent with its function being independent of the Mi-2/NuRD complex. Like related CHD proteins in animals, however, PKL is an ATPase and exhibits ATP-dependent chromatin remodeling activity. Our data are consistent the hypothesis that PKL restricts gene expression in plants as a result of its ability to remodel chromatin and that it does so via an epigenetic pathway that is distinct from that used by related remodeling factors in animals.
2. Materials and methods
2.1. Expression of recombinant PKL
The PKL coding sequence was fused at the C-terminus to the FLAG epitope using PCR and appropriate primers and cloned into the pVL1392 expression vector generating pJO1252. Sf9 cells were transfected with pJO1252 and linearized baculovirus DNA (BD Biosciences, Cat# 554739) using transfectin II reagent (Life Technologies, Cat# 10362–100) to generate recombinant baculovirus according to the manufacturer’s instructions. For protein production, SF9 insect cells were infected with the recombinant baculoviruses and maintained at 27°C in TNMF-H medium [35] supplemented with 10% fetal bovine serum and 10 μg/ml gentamycin. Three days after infection, the cells were harvested, washed 1× with phosphate buffered saline (PBS) and spun down at 1000 rpm for 10 min. The cell pellets were stored at −80°C before subsequent use. All subsequent steps were conducted either on ice or at 4°C. The frozen cell mass was thawed and resuspended in a 10-fold volume of lysis buffer of 25 mM Hepes, pH 7.6, 0.5 mM EDTA, 0.5 mM EGTA, 25 mM MgCl2, 500 mM NaCl, 1 mM DTT, 10% glycerol containing Roche complete, mini protease inhibitor cocktail (1 tablet/10 ml). The cell suspension was subjected to two cycles of freezing and thawing, and then several strokes of Dounce homogenizer. After douncing, the cell lysate was centrifuged for 30 min at 15,000 rpm and the resulting supernatant was diluted with an equal volume of the lysis buffer devoid of salts and metal chelators, and mixed with anti-Flag M2 conjugated agarose beads (Sigma, 10 μl/106 cells). After rotating overnight, the beads was spun down for 10 min at 5000 rpm and washed six times (each time 15 min of rotation) in 1.5 ml of wash buffer of 25 mM Hepes, pH 7.6, 250 mM NaCl and 10% glycerol. PKL-FLAG was eluted with 100 μl of the wash buffer containing 500 μg/ml Flag peptide (Sigma). The elution step was repeated thrice. The eluted proteins were either used within a couple of days or kept at −20°C in 50% glycerol and 1 mM DTT. The recombinant PKL-FLAG used in Figure 3D was prepared using wash buffer containing 0.1% Tween 20 and exhibited decreased ATPase activity relative to PKL-FLAG prepared in the absence of Tween 20.
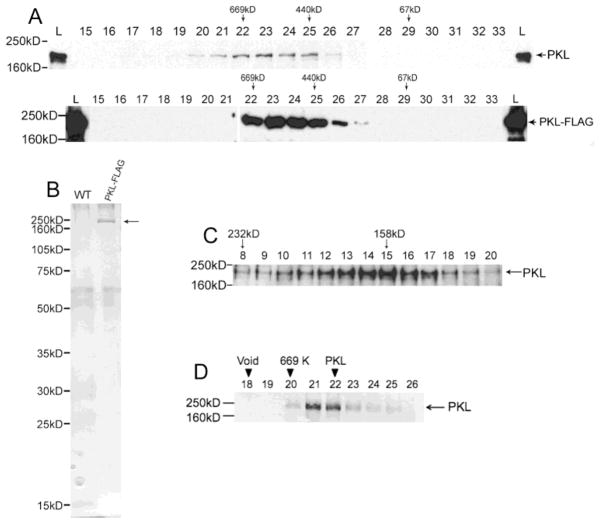
Figure 3.
PKL exists as a monomer in plants. (A) Western analysis was used to examine fractions from a Superose 6 gel-filtration column. The westerns on top are from an extract of wild-type plants whereas the bottom westerns are from an extract of PKL-FLAG plants. A polyclonal antibody to PKL was used on the top blots, and an anti-FLAG antibody was used on the bottom blots. The fraction number is indicated by the number above each lane, L = total lysates, and MW standards for column (top) and western (left side) are indicated. PKL has been previously noted to migrate on an SDS-PAGE gel at a MW that is larger than predicted [17]. (B) Total protein from WT and PKL-FLAG plants was first passed over an anti-FLAG affinity resin and then subsequently fractionated on a sizing column as described in panel A. Fractions corresponding to lanes 22–26 of panel A were then separated on an SDS-PAGE gel and analyzed by silver staining. The arrow to right of the gel indicates PKL-FLAG protein. (C) Western analysis of fractions from a sucrose gradient using polyclonal anti-PKL antibodies. Crude cell extracts were prepared in buffer containing 350 mM NaCl. MW standards for the gradient (top) and western (left side) are indicated. (D) Recombinant PKL-FLAG protein elutes with a peak of about 650 KDa. Western analysis was used to examine fractions from a Superose 6 gel-filtration column. The fraction number is indicated by the number above each lane, L = total lysates, and MW standards for column (top) and western (left side) are indicated. The migration of the 440 kDa marker is in lanes 27–28 (not included on this blot).
2.2. Characterization of PKL in planta
All the steps were performed at 4°C or on ice unless otherwise noted. 3-day-old PKL-FLAG or wild-type (as negative control) seedlings grown on MS plates (supplemented with 1% sucrose) were harvested, washed and ground into homogeneous lysate in 2 ml/g cold protein extraction buffer (PEB; 40 mM HEPES·KOH, pH 7.5, 350 mM NaCl, 0.1% Tween 20, 10% glycerol) supplemented with 1% protease inhibitor cocktail (Sigma P9599). Cell debris was then pelleted by centrifugation at 4°C at 16,000 × g for 15 min and the supernatant was used for subsequent experiments. For anti-FLAG purification, 20 μl anti-FLAG M2 resin is incubated with every 1 ml supernatant with rotation for 1–3 hours, washed with cold PEB for 3 times and eluted in 10 μl (per ml lysate) SDS-PAGE sample buffer. For gel filtration analysis, cleared lysate were filtered through a 0.22-μm syringe filter (Pall Corporation). Protein concentration was then determined using a Bio-Rad RC-DC kit (Bio-Rad) and ~10 mg of total protein (or unknown amount of purified PKL-FLAG protein) were resolved on a Superose 6 10/300 GL column (GE Healthcare). For sucrose density gradient centrifugation, 10–30% gradients (5 ml total) were poured with a gradient maker. 10% and 30% sucrose solutions were made in 50 mM NaPO4 pH 7.5, 400 mM NaCl, 0.1% Triton X-100. Supernatant from wild-type plants prepared as described above were diluted ~2 fold in this buffer w/o sucrose to lower the glycerol concentration to 5%. 50 μl of wild-type supernatant was loaded onto the top of two gradients. Two identical but separate gradients were loaded with 100 μg each of ferritin, catalase, aldolase, and BSA to serve as standards. Gradients were spun at 48,000 rpm for 18 hours at 4 °C using an SW55 Ti rotor. 14-drop fractions (~150 μl) were collected from bottom of tubes using a peristaltic pump. These fractions were then characterized by SDS-PAGE and western analysis.
2.3. Reconstitution of mononucleosomes
Mononucleosomes were assembled on 277 or 343-bp of fluorescent DNA fragments using recombinant histones of Xenopus laevis. DNA fragments (Table S2) were generated as PCR products with fluorescent primers (5′-Alex fluor 488 or 647-N-hydroxysuccinimide) and pGEM3z-601 [36] templates according to Thompson et al. [37] with the exception that the labeled primers were not diluted 10-fold. Histones were prepared according to Luger et al. [38]. For reconstitution, different molar ratios of DNA-to-octamer were mixed in 2M NaCl and subjected to gradient dialysis as described by Dyer et al. [39]. After dialysis, reconstitutions were verified by loading reactions onto a 4–5% non-denaturing acrylamide/bisacrylamide (37.5:1, 2.6% C) 0.5X Tris borate-EDTA gel. Labeled nucleosomes were detected using a Typhoon 8600 Imager (GE Healthcare). Mononucleosomes with the least amount of free DNA were used either directly or after further purification by gel electrophoresis [40] as indicated.
2.4. ATPase assays
Standard assays contained 20 μl of 2 mM ATP, 50 mM NaCl, 0.5 mM dithiothreitol, 5 mM MgCl2, 25 mM Tris (pH 7.9) and were conducted in the presence of cofactors at a concentration that was several fold in excess of over the concentration of recombinant PKL as described in [41]. Reaction mixtures containing 80 nM cofactor were prepared on ice and started by addition of 14 nM PKL as the final component. A negative control was prepared for each reaction mixture using a preparation of mock PKL from uninfected insect cells. After incubating in a thermocycler at 30°C for 45 min, ATPase activity was stopped with a phosphate detection solution of malachite-green dye (Biomol Green, Enzo Life Sciences) and measured colorimetrically at 620 nm [42].
2.5. Restriction Enzyme Accessibility Assays
Accessibility assays were based on Smith et al. (2005) [43] and conducted in 15 μl reactions with mononucleosomes reconstituted on fluorescently labeled 343-bp DNA. Reaction mixtures were prepared on ice and initiated with the addition of HhaI (0.6 U/μl). Each reaction contained 1X NEBuffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1mM DTT, pH 7.9, New England Biolabs), 0.1 mg/ml bovine serum albumin, 2.67 mM ATP, 8 nM PKL and 47 nM mononucleosomes. After incubating in a thermocycler at 30°C for 30 min, reactions were deproteinated with 7.5 μl of stop buffer (1.5% SDS, 100 mM EDTA, 15% glycerol and 300 ng proteinase K) and kept at 55°C for 25 min. For time course studies, aliquots were withdrawn at specific time points from a master mix. Cut and uncut DNA were separated on 8% non-denaturing acrylamide/bisacrylamide (37.5:1, 2.6% C) Tris Tricine-gel. Gels were visualized on a Typhoon 8600 Imager (GE Healthcare) and quantified using ImageQuant TL software. Values represented averages of three trials. Between trials, the values at each time point varied within ±6–7%.
2.6. Electrophoretic mobility shift assays
Fluorescently labeled 277-bp DNA fragments and mononucleosomes reconstituted on the same DNA fragments were used for the band- shift assays. DNA fragments were generated using the forward and reverse primers as described by Thompson et al. (2008) [37] and the mononucleosomes were gel purified before use. Positioned mononucleosomes were purified by 4% polyacrylamide gel electrophoresis with 0.5X TBE. Bands were visualized on a Typhoon 8600 imager (GE Health) and then excised. Gel pieces were washed for 5 min in about 5 volumes of elution buffer containing 10 mM Tris/HCl (pH 7.5), 50 mM KCL, 10% glycerol and 0.4 μg/μl BSA and transferred to same volumes of fresh buffer. After overnight at 4°C, the gel pieces were spun down at 14000 × g for 5 minutes and the supernatants were further concentrated using centrifugal concentrator (100K MW cut-off, Amicon). The eluted mononucleosomes were checked on a 4% polyacrylamide gel with 0.5% TBE and stored at 4°C.
DNA fragments (4 nM) or mononucleosomes (4.7 nM) and PKL (18.5 −115.7 nM) were prepared in 15 μl Tris-HCl buffer (20 mM, pH 7.6), containing 50 mM NaCl, 1.5 mM MgCl2, 1mM DTT, 0.1 mg/ml bovine serum albumin and 10% glycerol. After incubating on ice for 10 min, the reaction products were analyzed on a 4% non-denaturing acrylamide/bisacrylamide (37.5:1, 2.6% C) 0.5% Tris borate-EDTA-gel. Bands were visualized on a Typhoon 8600 Imager (GE Healthcare).
2.7. Nucleosome mobility assays
Centrally or end positioned mononucleosomes reconstituted on fluorescently labeled 277-bp DNA fragments were used for the mobility assays. DNA fragments were generated using the forward/reverse and slid forward/slid reverse primers as described by Thompson et al. (2008) [37] and the mononucleosomes were gel-purified before use. Mononucleosomes (66.6 nM) and PKL (1× = 115.7 nM) were prepared in the same Tris buffer as described for the electrophoretic mobility shift assays plus 2.67 mM ATP. Reactions were incubated at 30°C for 40 min and stopped by adding 1 ug of competitor DNA (pGEM7z). After incubating for another 10 min at 30°C the reaction products were analyzed on a 4% non-denaturing acrylamide/bisacrylamide (37.5:1, 2.6% C) 0.5% Tris borate-EDTA-gel. Bands were visualized on a Typhoon 8600 Imager (GE Healthcare).
2.8. Phylogenetic analysis and homology modeling
The list of CHD proteins along with the starting and ending number of the amino acid residue used for sequence analysis is provided in Table S1. The sequences were aligned using MAFFT version 6 (http://mafft.cbrc.jp/alignment/server/) using the E-INS-i setting (% mafft --reorder --ep 0.0 --maxiterate 1000 --retree 1 --genafpair input) [44]. Neighbor joining was then used to create a phylogenetic tree at the above MAFFT server for which all ungapped sites were included, the WAG substitution model was used, heterogeneity among sites was estimated, and the bootstrap resampling was 1000 [45]. This tree was visualized using Archaeopteryx [46, 47].
Homology modeling was performed using the SWISS-MODEL server http://swissmodel.expasy.org/. The coordinates of aa 952–1107 of PKL were generated by threading using the yeast CHD1 DNA-binding domain as a template structure (PDB code: 3TED). The threaded model was manually superposed on the coordinates of the yeast CHD1 DNA-binding domain (PDB code: 3TED) using PyMOL v1.5.0.3 to generate a working model for the interaction of PKL with a DNA ligand.
3. Results
3.1. PKL is most closely related to subfamily II of animal CHD remodelers
Previous phylogenetic characterization of CHD remodelers has focused on animal CHD proteins and on the presence or absence of specific domains such as the PHD domain [6, 7]. To determine which CHD remodelers in plants and animals share common ancestors and thus perhaps common remodeling activities, we undertook a phylogenetic analysis based on sequence conservation that included selected organisms from both kingdoms. In particular, we included CHD remodelers from both vertebrates (Homo sapiens and Danio rerio) and invertebrates (Drosophila melanogaster and Caenorhabditis elegans) from animals and two dicots (Arabidopsis thaliana and Populus trichocarpa) and a monocot (Brachypodium distachyon) as well a lycophyte (Selaginella moellendorffii) and a moss (Physcomitrella patens) from plants. For this analysis, we specifically selected the region of the CHD protein that spans the first chromodomain to the recently identified SLIDE domain [48, 49] in the D domain (see Table S1 for list of proteins and aa residues used), which is present in almost all CHD proteins. The chromodomains, ATPase domain, and SANT-SLIDE domains contained within this region all have been implicated in interacting with the chromatin substrate of CHD remodelers [48–52].
Our analysis revealed that there are three distinct clades of plant CHD remodelers. These three clades are related to the previously identified CHD remodeler subfamilies I and II, but not subfamily III (Figure 1). For the sake of convenience, we have denoted the three clades of plant CHD remodelers by the Arabidopsis CHD proteins included in each (PKL, PKR1, and PKR3) and denoted the three subfamilies of animal CHD remodelers using the previous nomenclature (I, II, and III).
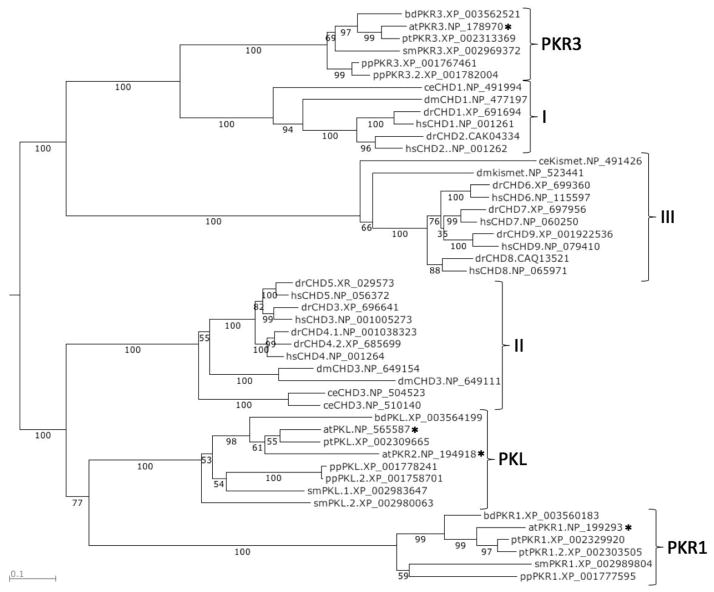
Figure 1.
PKL shares a common ancestor with CHD3-related proteins in animals. The region of CHD proteins spanning from the N-terminal chromodomain to the C-terminus of the D domain were aligned using MAFFT and a phylogenetic tree was generated using neighbor joining. The numbers beneath the branches indicate bootstrap values (percentage of 1000 replications). The Genbank accession number for the protein is listed to the right of the name. CHD proteins were included from A. thaliana (at), B. distachyon (bd), C. elegans (ce), D. melanogaster (dm), D. rerio (dr), H. sapiens (hs), P. patens (pp), P. trichocarpa (pt), and S. moellendorffii (sm). CHD proteins from Arabidopsis are marked with asterisks.
PKR3-related proteins in plants clearly share a common ancestor with animal CHD proteins belonging to subfamily I and thus can be regarded as plant members of subfamily I. The organization of domains of PKR3-related proteins is consistent with this assignment (Figure S1).
In contrast, there appear to be 2 clades of plant CHD proteins that are derived from a common ancestor to animal CHD proteins belonging to subfamily II. Our phylogenetic analysis suggests that PKL- and PKR1-related proteins diverged early during plant evolution from a common precursor that belonged to subfamily II. In support of this conjecture, representatives of both PKL- and PKR1-related CHD remodelers are found in the genome of the moss Physcomitrella patens. In agreement with pairwise analyses, analysis of the branch lengths of the tree reveals that PKL-related proteins exhibit sequence conservation with CHD3-related proteins that is substantially greater than that exhibited by PKR1-related proteins. Similarly, the domain organization of PKL-related proteins is more closely related to CHD3-related proteins (Figure S1). Thus our analysis suggests that although both PKL- and PKR1-related remodelers are members of subfamily II of CHD remodelers, PKL-related proteins have retained more characteristics in common with animal members of subfamily II whereas PKR1-related remodelers have evolved into a specialized plant-specific clade of this subfamily.
Our phylogenetic analysis also provides clear support for classifying PKR2 from Arabidopsis as a member of subfamily II of CHD remodelers despite the absence of the PHD domain that is characteristic of this group of proteins (Figure S1). In addition, no plant CHD remodelers were identified by our database searches that were grouped with subfamily III of animal CHD proteins, which includes the four mammalian remodelers CHD6–9. Thus subfamily III is specific to animals. Intriguingly, our phylogenetic analysis suggests that the divergence of this subfamily of remodelers occurred independently of the split of subfamilies I and II into animal- and plant-specific clades, raising the possibility that this subfamily was retained in animals but not plants.
3.2. DUF1086 contains a putative SANT domain
Our sequence analysis also provides new insights into the domain architecture of CHD proteins. Analysis of the recently solved crystal structure of the DNA-binding domain of CHD1 revealed the presence of both a SANT and a SLIDE domain [48, 49]. SANT domains have structural similarity to MYB-like domains and are commonly found in proteins that interact with chromatin [53, 54], whereas SLIDE domains had previously only been identified in ISWI remodelers [55]. Subsequent database searches revealed the presence of the SLIDE domain in CHD remodelers that were members of subfamily III as well, but did not uncover strong candidates for SANT or SLIDE domains in other CHD proteins.
Intriguingly, our alignment of CHD remodelers revealed substantial sequence conservation in all CHD proteins in regions that correspond to the yeast CHD1 protein SLIDE domain (Figure 2A). In addition, we found that both animal CHD3-related proteins and plant PKL-related proteins contain a region with considerable sequence similarity to the yeast CHD1 SANT domain. Importantly, the region of the CHD3- and PKL-related proteins that corresponds to the SANT domain overlaps a previously noted domain of unknown function, DUF1086. Specifically, of the 144 amino acids that lie within DUF1086 in PKL, the 106 amino acid C-terminal region exhibits sequence similarity to the DNA binding domain of the yeast CHD1 protein. This overlap includes the 57 amino acid SANT domain, suggesting that this portion of DUF1086 folds into a SANT domain.
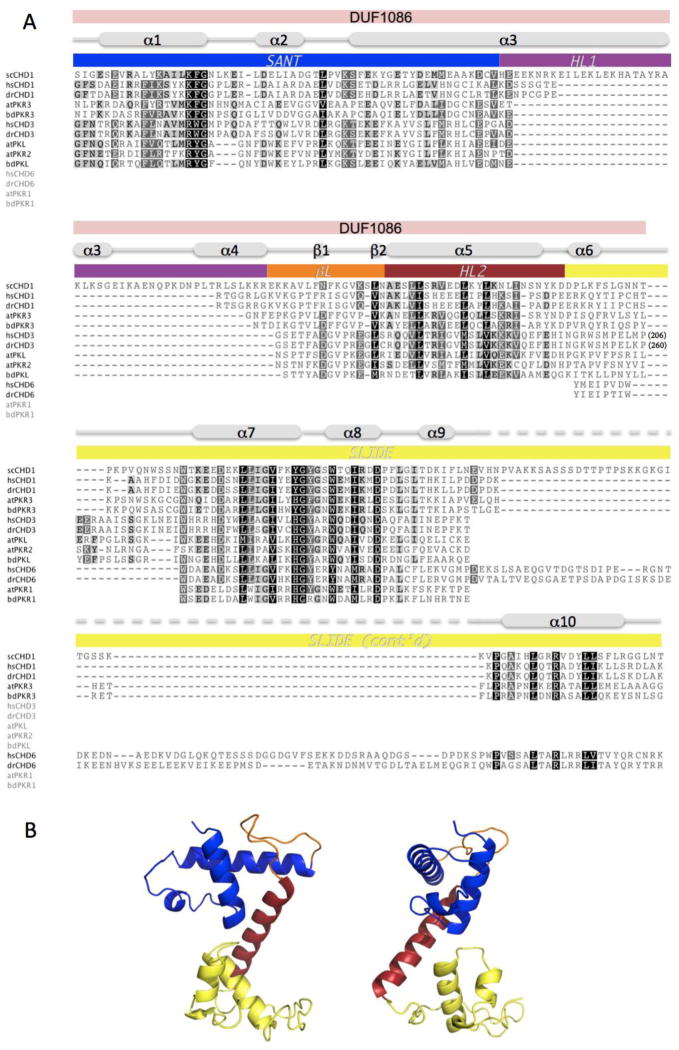
Figure 2.
PKL possesses a predicted DNA-binding domain that is similar to the DNA-binding domain of S. cerevisiae CHD1. (A) Alignment of scCHD1 with corresponding regions of CHD proteins from H. sapiens (hs), D. rerio (dr), A. thaliana (at), and the model grass species B. distachyon (bd). Specific regions of scCHD1 are marked as described in Ryan et al. (2011): SANT (blue), SLIDE (yellow), helical linker-1 (purple; HL1) and helical linker-2 (red; HL2) and β-linker (orange; βL). The region of CHD3 proteins that corresponds to DUF1086 is also marked. The extent of conservation of conserved residues is indicated by shading, and numbers in parentheses for CHD3 proteins indicate numbers of residues found in these remodelers that are not included in the region of conservation. (B) A cartoon representation of the predicted crystal structure of the putative DNA-binding domain of PKL is shown in two orientations. The domains are colored to reflect analogous domains indicated in scCHD1 in (A). Accession numbers for proteins used are provided in Table S1.
To examine the possibility that CHD3- and PKL-related proteins adopt a three dimensional structure similar to the structure of the CHD1 DNA-binding domain, we used the SWISS-MODEL server to perform homology modeling [56, 57]. A 3D model of the relevant region of PKL was generated using the yeast CHD1 DNA-binding domain as a template structure (PDB code: 3TED). We chose PKL for this analysis because our multiple sequence alignment revealed that animal CHD3-related proteins possess a sizable insertion between helices α6 and α7 of the yeast CHD1 DNA-binding domain that is lacking in PKL-related proteins and thus the architecture of PKL and CHD1 is likely to be the most similar. The predicted structure of PKL closely resembles the crystal structure of the DNA binding domain from yeast CHD1 (Figure 2B and Figure S2). This observation suggests that the putative DNA-binding domains of other CHD3- and PKL-related proteins are also likely to adopt a SANT-SLIDE architecture. Conserved residues are located at similar positions in the two structures and at key positions, such as in the hydrophobic core and at tight turns. This conservation suggests that the predicted structure of the putative PKL DNA-binding domain is likely to reflect the actual structure (Figure S2).
3.3. PKL primarily exists as a monomer in plants
CHD3 and related proteins from animals are incorporated into the multiprotein Mi-2/NuRD complex [18–21], the most abundant histone deacetylase complex in mammalian systems [22]. Given that PKL shares a common ancestor with CHD3-related proteins from animals (Figure 1), we examined whether PKL is similarly a component of the Mi-2/NuRD complex or an alternate complex. A crude protein extract was prepared from 3-day-old wild-type seedlings and subsequently applied to a Superose 6 gel filtration column. Fractions from the column were analyzed by Western blotting using an antibody that specifically recognizes native PKL protein. We observed that native PKL protein elutes in a peak corresponding to ~650 KDa (Figure 3A). To verify these results, one copy of the FLAG epitope was fused to the full-length PKL ORF to generate a PKL-FLAG translational fusion. This construct was placed under the control of PKL regulatory sequences [17] and introduced into pkl-1 plants where it rescued all mutant phenotypes associated with loss of PKL including root development, trichome architecture, leaf development, flowering time, and height (data not shown). When a crude protein extract prepared from 3-day-old PKL-FLAG seedlings was applied to a gel filtration column, we again observed that the PKL-FLAG protein elutes with a peak corresponding to ~650 KDa (Figure 3A).
The elution profile exhibited by PKL using gel filtration chromatography suggests that PKL is a member of a multisubunit complex. We therefore biochemically characterized affinity-purified PKL-FLAG protein from plants to test the hypothesis that PKL is a member of multisubunit complex and to enable subsequent identification of any proteins that interact with PKL. We first affinity purified PKL-FLAG protein under native conditions from crude protein extract prepared from 3-day-old PKL-FLAG seedlings. This purified PKL-FLAG protein was then analyzed by gel filtration chromatography. We observed that the PKL-FLAG protein from purified extracts eluted with a profile that is indistinguishable from PKL-FLAG protein from crude extracts (and thus also elutes with a peak that corresponds to ~650 KDa). We then collected and pooled fractions from the gel filtration column that corresponded to lanes 22–26 of Figure 3A, ran the concentrated sample out on an SDS-PAGE gel under denaturing conditions, and then used silver staining to examine the abundance of proteins present in the sample (Figure 3B). Our control for this analysis is a sample that was obtained from a wild-type plant that was treated in an identical fashion to the sample obtained from the PKL-FLAG plant. This analysis does not reveal a protein in the gel filtration fractions that is present in amounts that are stoichiometric to PKL and is thus inconsistent with the hypothesis that the observed molecular weight of PKL using gel filtration is a result of the incorporation of PKL into a multisubunit complex.
Our data thus suggest that PKL is the major (if not sole) protein that contributes to the peak that elutes at a molecular weight corresponding to 650 KDa on the gel filtration column. The predicted molecular weight of PKL is 158 KDa. These data are consistent with possibility that PKL exists as a homo-multimer or that PKL exhibits an anomalously large Stokes’ radius characteristic of proteins that adopt a highly elongated structure [58]. In order to address these possibilities, we carried out a sucrose gradient sedimentation experiment with crude extracts from WT plants. Determination of molecular weight by sucrose gradient sedimentation exhibits a different dependence on the shape of a protein than determination of molecular weight using gel filtration [58]. Western blot analysis of sucrose gradient fractions with a polyclonal antibody to PKL revealed that PKL sediments with an apparent molecular weight of 158 KDa (Figure 3C), which is the predicted MW of PKL. PKL behaves in a similar fashion in this type of analysis whether the extract is prepared with a buffer that contains 350 mM NaCl or with a buffer that contains only 150 mM NaCl (data not shown). In addition, we undertook co-immunoprecipitation experiments in transgenic lines that express both PKL-FLAG and PKL-STREP-HA to examine self-association of PKL protein. This analysis indicates that PKL does not exist as a homo-multimer in planta (Figure S3). Taken together, our data strongly suggest that PKL predominantly exists as a monomer in plants and has an atypically large Stokes radius. Another chromatin-related protein, the methyl-CpG binding protein MeCP2, has also been shown to exhibit an unusually large Stokes radius and exhibits an apparent molecular mass of 400–500 KDa even though its monomeric molecular weight is 53 KDa [59].
3.4. PKL is a nucleosome-stimulated ATPase
The ATP-dependent chromatin remodeling activity of animal CHD3-related proteins is well established [6, 22]. If, as suggested by our phylogenetic analysis, PKL and PKL-related proteins have a similar function to CHD3-related proteins, they should also exhibit ATP-dependent chromatin remodeling activity. To test this hypothesis, we generated recombinant C-terminal FLAG epitope-tagged PKL using the baculovirus expression system. Given that transgenic pkl plants expressing an identical PKL-FLAG protein are rescued for all mutant phenotypes, the addition of the FLAG epitope does not appear to affect the function of PKL protein in vivo. Immunoaffinity purification of the baculovirus-generated recombinant PKL-FLAG protein resulted in a single visible band on a Coomassie-stained gel (Figure 4A), and this band was recognized by anti-FLAG antiserum in a western blot analysis (Figure 4B). Importantly, the recombinant PKL-FLAG protein also elutes with a peak that corresponds to ~650 KDa when analyzed by gel filtration chromatography (Figure 3D). This result provides strong additional evidence that the anomalous elution profile of the native PKL protein is not due to association with other plant proteins and further indicates that the recombinant protein exhibits a large Stokes radius in a similar fashion to the native protein.
Figure 4.
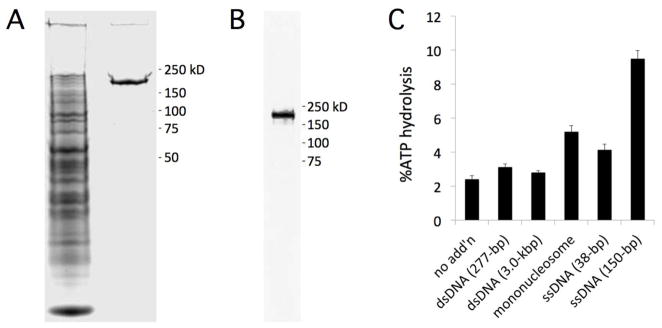
Recombinant PKL-FLAG is a nucleosome-stimulated ATPase. (A) Extract from infected S2 cells was treated with anti-FLAG M2 affinity resin and applied to SDS-PAGE and stained with Coomassie Blue. The left lane is the unbound fraction and the right lane is purified PKL-FLAG. (B) Purified PKL-FLAG was analyzed by western blotting using an a-FLAG antibody. (C) Recombinant PKL-FLAG was incubated with ATP in the presence or absence of dsDNA, mononucleosomes, or ssDNA (indicated on x-axis). Hydrolysis of ATP was measured using a colorimetric assay and buffer alone was used as a reference for each sample. MW standards for panels A and B are indicated to right. Error bars in panel C represent SE calculated from 3 replicates.
We examined the ATPase activity of the purified recombinant PKL-FLAG protein in the presence of DNA and mononucleosomes using a colorimetric assay [42]. Buffer was used as a reference. We found that PKL exhibited modest ATPase activity that was increased more than twofold by the presence of mononucleosomes and more than five-fold by addition of 150-bp of ssDNA (Figure 4C). In contrast, dsDNA had little if any effect on ATPase activity. Thus as observed for animal CHD3 proteins [18, 26], nucleosomes simulate PKL ATPase activity whereas free dsDNA has a negligible effect. Although the ability of ssDNA to increase the ATPase activity of animal CHD3 proteins was not reported in these studies, it has previously been demonstrated that ssDNA stimulates the ATPase activity of the SWI/SNF-class yeast remodeler STH1 [60]. It is worth noting, however, that this study also found that dsDNA and mononucleosomes were about as effective as ssDNA at increasing the ATPase activity of STH1. Thus PKL ATPase activity is stimulated by a different spectrum of substrates than that of STH1.
3.5. PKL interacts with free DNA and mononucleosomes
The ability of mononucleosomes to stimulate the ATPase activity of PKL suggests that PKL can bind to mononucleosomes. Further, although the presence of dsDNA had a negligible effect on the ATPase activity (Figure 4), our sequence analysis suggested the presence of a putative DNA-binding domain in PKL (Figure 2). We therefore used gel shift assays to examine the ability of PKL to interact with DNA and/or mononucleosomes.
Our analyses indicated that PKL binds to either free DNA or mononucleosomes. Gel shift assays were carried out in 50 mM NaCl using a 277-bp fluorescently end-labeled DNA fragment containing either the 601 nucleosome positioning sequence alone [36] or the same fragment incorporated into a mononucleosome generated using recombinant histones from Xenopus laevis. A gel shift assay with free DNA and increasing amounts of PKL revealed decreasing amounts of free DNA as the concentration of PKL increased (Figure 5A). At higher concentrations of PKL, a slower migrating broadly smeared band appeared, indicating that PKL associated non-specifically with multiple sites in the DNA. Similarly, a gel shift assay with mononucleosomes and increasing amounts of PKL revealed decreasing amounts of free mononucleosome as the concentration of PKL increased (Figure 5B). Again, no distinct band appeared at higher concentrations of PKL but rather a smear near the top of the gel, indicating the formation of more than one species of complex.
Figure 5.
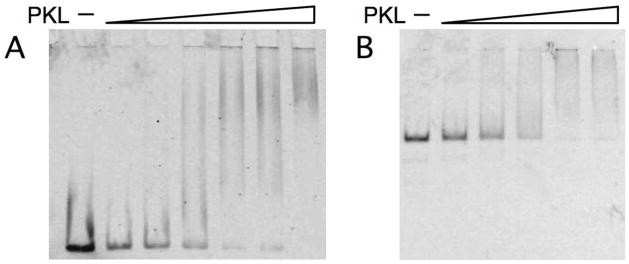
Recombinant PKL-FLAG binds to free DNA and mononucleosomes. (A) Recombinant PKL (44–275 ng) was incubated with 10 ng of 277-bp DNA end-labeled with Alex Fluor 647 and the mixture was analyzed by electrophoresis on a 2% agarose gel. (B) Recombinant PKL (44–275 ng) was incubated with 20 ng of mononucleosomes containing the same end-labeled 277-bp DNA fragment as in panel A and then analyzed by electrophoresis on a 4% PAGE gel.
3.6. PKL is an ATP-dependent chromatin remodeler
The ability of PKL to interact with free DNA and mononucleosomes (Figure 5) and the stimulation of PKL ATPase activity by mononucleosomes (Figure 4) suggested that PKL could remodel chromatin in an ATP-dependent fashion. We first tested the hypothesis that PKL exhibits ATP-dependent remodeling activity by examining the ability of PKL to increase the restriction enzyme accessibility of DNA incorporated within a mononucleosome [43, 61]. Mononucleosomes were reconstituted on a fluorescently end-labeled 343-bp fragment of DNA containing the 601 nucleosome positioning sequence and a unique HhaI site. The presence of PKL in a reaction containing these mononucleosomes and HhaI greatly increased accessibility of the DNA to HhaI cleavage, and this increase was dependent on the presence of ATP (Figure 6A). These reactions were all allowed to proceed for 30 minutes. We performed a time course to examine the kinetics with which PKL altered the access of HhaI to the nucleosomal DNA and observed a linear increase in digested DNA over time (Figure 6B), suggesting that PKL continuously remodels the nucleosome during this time.
Figure 6.
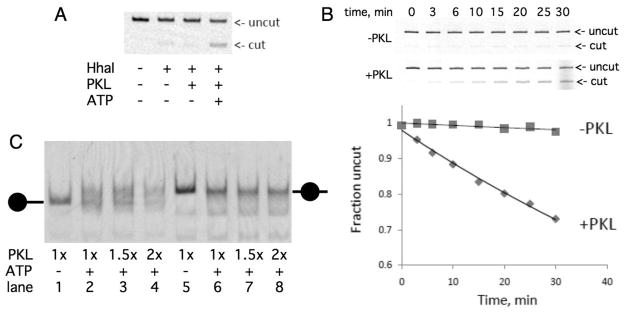
PKL is a chromatin remodeling factor. (A) Recombinant PKL increases the accessibility of DNA in mononucleosomes to restriction enzyme HhaI in an ATP-dependent fashion. The top of the panel depicts a PAGE/agarose gel analysis of 277-bp DNA fragment incorporated within a mononucleosome that was incubated with the components indicated underneath. Position of uncut and cut DNA is indicated to right of panel. (B) Time course of cleavage of DNA in mononucleosome by HhaI in absence and presence of PKL. The graph depicts quantification of cut DNA. (C) Recombinant PKL increases the mobility of positioned mononucleosomes in an ATP-dependent fashion. Recombinant PKL was incubated with end-positioned (lanes 1–4) or center-positioned (lanes 5–8) mononucleosomes in the absence (lanes 1 and 5) or presence of ATP, which were then separated by native gel electrophoresis. The positions of center-positioned and end-positioned nucleosomes are indicated on the sides of the graph.
We also examined the ability of PKL to remodel chromatin by using the ‘nucleosome sliding’ assay [62, 63]. This assay examines the ability of a remodeler to reposition an otherwise stably positioned nucleosome. The position of the nucleosome on the DNA is determined by its migration on a native polyacrylamide gel; center-positioned nucleosomes migrate slower than end-positioned nucleosomes. We incubated PKL with a fluorescently end-labeled 277-bp fragment of DNA with nucleosomes positioned on the end or on the center. We observed that PKL clearly remobilized end-positioned nucleosomes to a center position in an ATP-dependent fashion (Figure 6C). In contrast, center-positioned nucleosomes largely failed to be repositioned to the ends of DNA in the presence of PKL. Thus PKL preferentially positions nucleosomes toward the center of DNA fragments, much like animal CHD3 and CHD1 proteins [26, 64, 65].
4. Discussion
In Arabidopsis, PKL, like animal CHD3 remodelers, plays an important role in repression of developmentally regulated genes. Initial characterization of PKL in Arabidopsis revealed strong similarities to CHD3 proteins in animals. PKL exhibits considerable sequence similarity to CHD3 remodelers and a similar organization of conserved domains of sequence homology [15, 16]. Subsequent characterization of PKL, however, has revealed significant differences from CHD3 remodelers. In particular, PKL promotes the repressive epigenetic mark H3K27me3 [29, 31] whereas animal CHD3 proteins promote deacetylation of histones as components of the Mi-2/NuRD complex [18–21].
Although the biochemical properties of animal CHD3 proteins have been extensively characterized, very little is known regarding the biochemical properties of PKL. These studies are of particular interest because PKL appears to operate through a different biochemical pathway than its animal CHD remodeler counterparts. Here we present phylogenetic and biochemical analyses that provide greater insight into the relationship between PKL remodelers in plants and CHD3 remodelers in animals.
4.1. Plants possess a repertoire of CHD remodelers that is distinct from that of animals
Previous phylogenetic analyses of CHD remodelers have focused on animal CHD proteins and frequently focus on the presence or absence of various domains of sequence conservation [5, 6, 66]. These analyses revealed the existence of three subfamilies of CHD remodelers and placed CHD3-related proteins into subfamily II. We undertook an extensive phylogenetic analysis that included both animal and plant CHD remodelers to gain insight into the relationship between them (Figure 1). This analysis focused on the region of CHD proteins that is common to the vast majority of this family of remodelers: from the N-terminus of the first chromodomain to the C-terminus of the DNA-binding domain. There are a number of observations that suggest our analysis was robust. Conserved sequence motifs are consistently identified and aligned in our alignment in all domains contained within this region (both chromodomains, ATPase domain, D domain, and also two domains of unknown function DUF1086 and DUF1087). Further, the various clades identified by our analysis predict the domain architecture of the full-length CHD remodelers grouped within the clade (for example PKL-related proteins all have a PHD domain immediate adjacent to the chromodomain whereas PKR1-related proteins all have a PHD domain that is located closer to the N-terminus of the protein). In addition, the subfamilies of animal CHD remodelers that were identified by previous analyses (I, II, and III) are also identified by our analysis. Finally, our analyses provide new insight into the putative DNA-binding domain of CHD3- and PKL-related proteins (discussed in more detail below).
Our phylogenetic analysis reveals that plants have three clades of CHD remodelers that can be delineated by the CHD remodelers found in Arabidopsis: PKR3-related, PKL-related, and PKR1-related remodelers (Figure 1). In agreement with previous comparative sequence analyses, our phylogenetic analysis indicates that PKL-related proteins share a common ancestor with CHD3-related proteins in animals and are members of subfamily II of CHD remodelers. Our analysis further indicates that PKR3-related proteins in plants and CHD1-related proteins in animals have a common ancestor and accordingly fall into subfamily I of CHD remodelers. The third clade of plant CHD remodelers, PKR1-related proteins, also share a common ancestor with CHD3-related proteins and thus also fall within subfamily II of CHD remodelers with PKL-related proteins. Importantly, however, our phylogenetic analysis indicates that PKR1-related proteins have diverged substantially from PKL-related proteins, suggesting that they have evolved to undertake distinct chromatin remodeling roles in plants. The distinct domain architecture of PKR1-related proteins and the absence of the SANT domain in these remodelers are consistent with this possibility.
4.2. CHD remodelers in subfamilies I and II are likely to have similar DNA-binding domains
The structure of the DNA-binding domain of yeast CHD1 has been determined and contains both a SANT and a SLIDE domain that both contribute to binding of DNA [48, 49]. Sequence conservation in animal CHD1-related remodelers from subfamily I strongly suggests that this SANT-SLIDE module is also present in animal CHD1 proteins. Our sequence alignment reveals that this SANT-SLIDE module is also likely to be present in PKR3-related proteins that are plant members of subfamily I (Figure 2A) in concordance with sequence analysis presented in supplementary data by Ryan and colleagues [49]. Inclusion of PKL-related proteins in our phylogenetic analysis further reveals strong sequence conservation in regions of both CHD3- and PKL-related proteins that correspond to the SANT and SLIDE domains of yeast CHD1 (Figure 2A). This result differs from previous sequence analyses, which did not suggest the presence of SANT or SLIDE domains in animal CHD3-related proteins [48, 49]. The region of CHD3- and PKL-related proteins that exhibits sequence similarity to the yeast CHD1 SANT domain was previously annotated as a domain of unknown function, DUF1086. Thus our analysis strongly suggests that DUF1086 is integral to the DNA binding domain of CHD remodelers in subfamily II (Figure 2).
Homology modeling of the portion of PKL that corresponds to the yeast CHD1 DNA-binding domain strongly suggests that PKL and CHD3 proteins adopt the same SANT-SLIDE architecture found in yeast CHD1 (Figure 2B and 2C). Adjacent SANT and SLIDE domains were first identified in ISWI chromatin remodelers [55], and both domains were subsequently found to contribute to DNA binding by ISWI [67] and by yeast CHD1 [48, 49]. Both CHD1 and ISWI have the ability to position mononucleosomes in the center of short DNA fragments and also to generate evenly spaced nucleosomal arrays [68, 69]. The structural similarity of the DNA-binding domain of the two remodelers has been proposed to contribute these shared activities. The predicted presence of the SANT-SLIDE module in CHD3- and PKL-related proteins in combination with the ability of both to center mononucleosomes on short DNA fragments [26, 65] (Figure 6C) is consistent with this hypothesis. It is also worth noting that the SANT-SLIDE module has also been proposed to play an important role in targeting of remodelers in vivo [70]. Our analyses thus raise the possibility that a SANT-SLIDE module also contributes to targeting of CHD3 and PKL remodelers.
We specifically selected PKL instead of an animal member of subfamily II for homology modeling because it does not contain the long intervening stretch of amino acids between regions that correspond to helices α6 and α7 of yeast CHD1 that is found in animal CHD3-related proteins (Figure 2A). One notable difference in the generated model of PKL from the determined structure of yeast CHD1 is the absence of an alpha helix that corresponds to α10 of yeast CHD1. As α10 contributes in a significant fashion to the ability of the DNA-binding domain of yeast CHD1 to interact with DNA [48, 49], we propose that an analogous helix exists in CHD3- and PKL-related proteins that is not identified based on primary sequence conservation to yeast CHD1. In fact, a difference in primary sequence is to be expected if the DNA-binding specificity is different between the two proteins. Analysis of the predicted secondary structure of the C-termini of CHD3 and PKL indicates the existence of amphipathic alpha helixes that could play an equivalent role to α10 in yeast CHD1, but these predictions remain to be tested biochemically. In this regard, however, it is interesting to note that both PKL (KKP, unpublished observations) and the yeast CHD1 DNA-binding domain [48] can bind to a 12-bp fragment of dsDNA.
4.3. PKL exists as a monomer in planta
Our phylogenetic analysis reveals that PKL shares a common ancestor with animal CHD3 proteins. Animal CHD3-related proteins have been demonstrated to be components of Mi-2/NuRD and several other multisubunit complexes [18–21, 27, 28, 32, 71]. The ability of CHD3-related proteins to participate in multiple remodeling complexes that contribute in markedly distinct fashions to transcription suggests that they function as remodeling engines that can be harnessed in multiple manners to help determine transcriptional output. Given that PKL promotes H3K27me3 in Arabidopsis [29, 31], we initially hypothesized that PKL would associate with one or more PRC2 complexes or form a distinct complex with a histone methyltransferase to promote methylation of H3K27. Instead, our combined gel filtration and sucrose sedimentation analyses strongly suggest that PKL predominantly exists in monomeric form in planta (Figure 3). Thus despite the similarity of PKL to CHD3 proteins, our analyses do not provide support for the idea that PKL participates in one or more multisubunit complexes. Thus in this regard, PKL behaves more like the Drosophila CHD3 protein dCHD3, which also primarily exists as a monomer [72]. It is important to note, however, that our results do not preclude the possibility that PKL may participate in one or more multisubunit complexes that are not stable under the given isolation conditions and/or are present in low abundance (e.g. because of tissue-specific accumulation).
4.4. Recombinant PKL exhibits ATP-dependent chromatin remodeling activity
We used the baculovirus expression system to generate PKL and characterized the biochemical properties of this recombinant protein. Our biochemical analyses revealed that PKL exhibits the hallmarks of an ATP-dependent chromatin remodeler, including the ability to increase accessibility of a restriction enzyme to DNA incorporated in a mononucleosome in an ATP-dependent fashion and the ability to reposition mononucleosomes on a short fragment of DNA in an ATP-dependent fashion (Figure 6). Our analyses further revealed that the biochemical properties of PKL are highly reminiscent of CHD3-related proteins from animals. PKL binds to both dsDNA and mononucleosomes (Figure 5), but the ATPase activity is stimulated only by mononucleosomes, not dsDNA (Figure 4). In addition, PKL preferentially mobilizes end-positioned mononucleosomes to the center position of DNA and largely fails to reposition center-positioned mononucleosomes (Figure 6C). Animal CHD3 proteins behave similarly in these types of assays [26, 65]. Thus our biochemical characterization of PKL supports the prediction from our phylogenetic analysis that PKL- and CHD3-related remodelers share a common ancestor and are thus likely to exhibit similar biochemical properties.
It is worth noting that in our nucleosome remobilization assays we observe PKL-dependent production of additional minor products that are likely to correspond to alternatively positioned nucleosomes (Figure 6C lanes 2–4,6–8). Thus although PKL preferentially positions nucleosomes in the center of the DNA fragment in our assays, this does not appear to be the exclusive product. In particular, we sometimes observe a band that migrates slightly faster than the end-positioned nucleosome. This type of product has also been observed in reactions containing the Drosophila CHD3 variant dCHD3 [72] and has been hypothesized to represent a partially evicted histone octamer that has been positioned “over the edge” of the DNA fragment.
Although these data highlight the similarity between CHD remodelers in animals and plants, it is important to note that our analyses also underscore the reality that plants and animals each have a distinct repertoire of chromatin remodeling factors. Although both plants and animals have members of subfamily I and subfamily II of CHD remodelers, only animals have members of subfamily III whereas only plants have members of subfamily IV. Among the different chromatin constituents found in plants and animals are unique populations of histone variants [4, 73–75]. Thus the chromatin substrate that PKL acts on in planta is distinct from that used in our in vitro assay, which is derived from Xenopus histones. In this regard, it may not be surprising that related CHD3 and PKL remodelers act in a similar fashion on an analogous substrate. Previous characterization of another ATP-dependent remodeler from plants, DDM1, reveals that it exhibits different activities than PKL [76], demonstrating that different plant remodelers can exhibit distinct biochemical characteristics when using animal chromatin. Nevertheless, the possibility remains that PKL may exhibit different biochemical properties when assayed using chromatin substrates prepared using plant histones.
4.5. Role of PKL-related proteins in plants
Functional and transcriptome analysis of pkl plants indicates that PKL contributes to repression of developmental identity genes via promoting the repressive epigenetic modification H3K27me3 [29, 31]. ChIP analysis further reveals that PKL is associated with H3K27me3-enriched genes, suggesting that PKL acts directly to promote H3K27me3 at these loci [31]. Our biochemical data revealing that PKL primarily exists as a monomer in planta is consistent with these and other data that indicate that PKL does not participate in the plant equivalent of a Mi-2/NuRD complex, but do not suggest a mechanism by which PKL contributes to H3K27me3. In this regard, ChIP also reveals that PKL is present at ubiquitously expressed loci such as those coding for actin and ubiquitin that do not exhibit PKL-dependent expression, suggesting that PKL may play a more general role in chromatin structure in Arabidopsis [31]. Such a general role has recently been identified for animal CHD3 remodeling proteins. The Drosophila CHD3 protein dMi-2 associates throughout the genome as a component of the NuRD complex and is implicated in contributing to genome-wide nucleosome positioning [77]. Further characterization of the biochemical activity of PKL protein, particularly using chromatin templates derived from plant components, in conjunction with genome-wide analysis of PKL targeting is likely to greatly clarify the epigenetic pathway or pathways by which PKL contributes to chromatin architecture and gene expression in plants.
Supplementary Material
Highlights.
PICKLE---related proteins share a common ancestor with CHD3 chromatin remodelers.
Homology modeling predicts that PICKLE has a SANT-SLIDE DNA-binding domain.
Unlike animal CHD3 proteins, PICKLE primarily exists as a monomer.
Recombinant PICKLE is an ATPase that is stimulated by ssDNA and mononucleosomes.
Recombinant PICKLE exhibits ATP-dependent chromatin remodeling activity.
Acknowledgments
We thank Craig Peterson and Manisha Sinha for extensive technical assistance with regards to remodeling assays. We thank Mark Hall for assistance in analyzing PKL protein using sucrose gradient sedimentation. We thank Scott Briggs, Ann Kirchmaier and Beth Tran for sharing equipment and reagents. We thank Michael Zanis, Elisabeth Svedin, and Yi Li for advice on phylogenetic analyses. We thank members of the laboratory for assistance in generating figures and tables and in critical reading of the manuscript. This work was supported by the National Institutes of Health [GM59770 to J.O.]; and the National Science Foundation [MCB-0918954 to J.O.].
Abbreviations
- PHD
plant homeodomain
- PKL
PICKLE
- HDAC
histone deacetylase
- MBD2
methyl-CpG-binding domain protein 2
- H3K27me3
trimethylation of lysine 27 histone H3
- LEC
LEAFY COTYLEDON
- ACT7
ACTIN7
- UBQ10
UBIQUITIN10
- PKR
PICKLE-RELATED
- DUF
domain of unknown function
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- WT
wild type
- PRC2
Polycomb Repressive Complex 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang DY, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proceedings Biological sciences/The Royal Society. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyerowitz EM. Plants compared to animals: the broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- 4.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 7.Sims JK, Wade PA. SnapShot: Chromatin remodeling: CHD. Cell. 2011;144:626–626. e621. doi: 10.1016/j.cell.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Bienz M. The PHD finger a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murawsky CM, Brehm A, Badenhorst P, Lowe N, Becker PB, Travers AA. Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodelling complex. EMBO Rep. 2001;2:1089–1094. doi: 10.1093/embo-reports/kve252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamasaki Y, Nishida Y. Mi-2 chromatin remodeling factor functions in sensory organ development through proneural gene repression in Drosophila. Dev Growth Differ. 2006;48:411–418. doi: 10.1111/j.1440-169X.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 12.Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a Homolog of the NURD Complex Component Mi-2 Act Together to Maintain Germline-Soma Distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnanapragasam MN, Scarsdale JN, Amaya ML, Webb HD, Desai MA, Walavalkar NM, Wang SZ, Zu Zhu S, Ginder GD, Williams DC., Jr p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7487–7492. doi: 10.1073/pnas.1015341108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/s0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- 17.Li HC, Chuang K, Henderson JT, Rider SD, Jr, Bai Y, Zhang H, Fountain M, Gerber J, Ogas J. PICKLE acts during germination to repress expression of embryonic traits. Plant J. 2005;44:1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 19.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 20.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 21.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 22.Wolffe AP, Urnov FD, Guschin D. Co-repressor complexes and remodelling chromatin for repression. Biochem Soc Trans. 2000;28:379–386. [PubMed] [Google Scholar]
- 23.Gao Z, Huang Z, Olivey HE, Gurbuxani S, Crispino JD, Svensson EC. FOG-1-mediated recruitment of NuRD is required for cell lineage re-enforcement during haematopoiesis. The EMBO journal. 2010;29:457–468. doi: 10.1038/emboj.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilera C, Nakagawa K, Sancho R, Chakraborty A, Hendrich B, Behrens A. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- 25.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 26.Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. Embo J. 2000;19:4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunert N, Wagner E, Murawska M, Klinker H, Kremmer E, Brehm A. dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. Embo J. 2009;28:533–544. doi: 10.1038/emboj.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passannante M, Marti CO, Pfefferli C, Moroni PS, Kaeser-Pebernard S, Puoti A, Hunziker P, Wicky C, Muller F. Different Mi-2 Complexes for Various Developmental Functions in Caenorhabditis elegans. PLoS ONE. 2010;5:e13681. doi: 10.1371/journal.pone.0013681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Rider SD, Jr, Henderson JT, Fountain M, Chuang K, Kandachar V, Simons A, Edenberg HJ, Romero-Severson J, Muir WM, Ogas J. The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem. 2008;283:22637–22648. doi: 10.1074/jbc.M802129200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Kohler C. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J. The CHD3 Remodeler PICKLE Associates with Genes Enriched for Trimethylation of Histone H3 Lysine 27. Plant Physiology. 2012;159:418–432. doi: 10.1104/pp.112.194878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A. Stress-Induced PARP Activation Mediates Recruitment of Drosophila Mi-2 to Promote Heat Shock Gene Expression. PLoS Genet. 2011;7:e1002206. doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. The chromatin remodeling factor Mi-2alpha acts as a novel co-activator for human c-Myb. J Biol Chem. 2007;282:13994–14005. doi: 10.1074/jbc.M700755200. [DOI] [PubMed] [Google Scholar]
- 35.Hink WF. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 36.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 37.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 39.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 40.Linxweiler W, Horz W. Reconstitution of mononucleosomes: characterization of distinct particles that differ in the position of the histone core. Nucleic acids research. 1984;12:9395–9413. doi: 10.1093/nar/12.24.9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durr H, Hopfner KP. Structure-function analysis of SWI2/SNF2 enzymes. Methods in enzymology. 2006;409:375–388. doi: 10.1016/S0076-6879(05)09022-1. [DOI] [PubMed] [Google Scholar]
- 42.Lewis R, Durr H, Hopfner KP, Michaelis J. Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic acids research. 2008;36:1881–1890. doi: 10.1093/nar/gkn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith CL, Peterson CL. A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol Cell Biol. 2005;25:5880–5892. doi: 10.1128/MCB.25.14.5880-5892.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 48.Sharma A, Jenkins KR, Heroux A, Bowman GD. Crystal structure of the chromodomain helicase DNA-binding protein 1 (Chd1) DNA-binding domain in complex with DNA. The Journal of biological chemistry. 2011;286:42099–42104. doi: 10.1074/jbc.C111.294462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. The EMBO journal. 2011;30:2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouazoune K, Mitterweger A, Langst G, Imhof A, Akhtar A, Becker PB, Brehm A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. Embo J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 52.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends in biochemical sciences. 1996;21:87–88. [PubMed] [Google Scholar]
- 54.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 55.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 56.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nature protocols. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 57.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 58.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klose RJ, Bird AP. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. The Journal of biological chemistry. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- 60.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 62.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 63.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 64.Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc Natl Acad Sci U S A. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guschin D, Wade PA, Kikyo N, Wolffe AP. ATP-Dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry. 2000;39:5238–5245. doi: 10.1021/bi000421t. [DOI] [PubMed] [Google Scholar]
- 66.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 68.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of Nucleosome Repositioning by Yeast ISWI and Chd1 Chromatin Remodeling Complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lusser A, Urwin DL, Kadonaga JT. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol. 2005;12:160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 70.Hopfner KP, Gerhold CB, Lakomek K, Wollmann P. Swi2/Snf2 remodelers: hybrid views on hybrid molecular machines. Current opinion in structural biology. 2012;22:225–233. doi: 10.1016/j.sbi.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shimono K, Shimono Y, Shimokata K, Ishiguro N, Takahashi M. Microspherule Protein 1, Mi-2{beta} and RET Finger Protein Associate in the Nucleolus and Up-regulate Ribosomal Gene Transcription. J Biol Chem. 2005;280:39436–39447. doi: 10.1074/jbc.M507356200. [DOI] [PubMed] [Google Scholar]
- 72.Murawska M, Kunert N, van Vugt J, Langst G, Kremmer E, Logie C, Brehm A. dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol Cell Biol. 2008;28:2745–2757. doi: 10.1128/MCB.01839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jerzmanowski A. SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta. 2007;1769:330–345. doi: 10.1016/j.bbaexp.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nature reviews. Molecular cell biology. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 75.Ingouff M, Berger F. Histone3 variants in plants. Chromosoma. 2010;119:27–33. doi: 10.1007/s00412-009-0237-1. [DOI] [PubMed] [Google Scholar]
- 76.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 77.Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WF, Dekkers DH, Demmers JA, Travers AA, Verrijzer CP. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Molecular and cellular biology. 2012;32:675–688. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.