Abstract
Background
We identified a mitochondrial tRNA mutation (m.586G>A) in a patient with renal failure and symptoms consistent with a mitochondrial cytopathy. This mutation was of unclear significance because there were neither consistent reports of linkage to specific disease phenotypes nor an existing analysis of effects upon mitochondrial function.
Case-Diagnosis/Treatment
A 16-month-old girl with failure-to-thrive, developmental regression, persistent lactic acidosis, hypotonia, GI dysmotility, adrenal insufficiency and hematologic abnormalities developed hypertension and renal impairment with chronic tubulointerstitial fibrosis, progressing to renal failure with need for peritoneal dialysis. Evaluation of her muscle and blood identified a mutation of the mitochondrial tRNA for phenylalanine, m.586G>A.
Conclusions
The m.586G>A mutation is pathogenic and is a cause of end-stage renal disease in childhood. The mutation interferes with the stability of tRNAPhe and affects the translation of mitochondrial proteins and the stability of the electron transport chain.
Keywords: Mitochondrial DNA, Mitochondrial tRNA, Mitochondrial Disease, Heteroplasmy
Introduction
We report a case of a child who presented with multi-organ dysfunction, including severe neurologic and renal impairment, who ultimately succumbed due to neurological deterioration while on peritoneal dialysis. Mitochondrial cytopathies are an uncommon cause of renal failure [1], but in our patient the diagnosis was suggested by the combination of clinical features.
Mitochondrial DNA (mtDNA) sequencing identified an m.586G>A mutation affecting the mitochondrial tRNA for phenylalanine (MTTF). This mutation had been previously reported in a 57-year-old woman with movement disorder and psychiatric disease [2]. In the previous case the mutation was heteroplasmic with loads of 85% in muscle, 29% in urinary epithelia and 3% in blood. The authors showed that COX-negative fibers from this patient had a disproportionate load of mutation and concluded it was pathogenic. The disparity in severity, onset and affected tissues prompted us to consider whether m.586G>A was responsible for mitochondrial dysfunction or was merely coincidental.
It is often difficult to conclusively link mitochondrial mutations to the mechanism of disease. The evaluation of mitochondrial function in patient tissues is dependent upon biopsy and analytical techniques. The presence of variable heteroplasmic loads of mutation in ostensibly healthy family members complicates the interpretation of genetic testing. To confirm the pathogenicity of the m.586G>A mutation we examined the effect of the mutation upon oxygen consumption, mitochondrial protein levels and tRNAPhe levels in patient tissues and in transmitochondrial cybrid cell lines.
Patient and methods
Case report
The proband was aged 16 months at the time of evaluation. At 8 months she had failure to thrive, microcephaly, hypotonia and gastroparesis. Symptoms progressed to include developmental regression, adrenal insufficiency, pancreatitis, anemia and neutropenia. At 15 months she was admitted for feeding intolerance and hyponatremia. Maintenance medications upon admission included hydrocortisone, clonidine and erythropoietin. She had proteinuria (urine albumin 288mg/L), hypertension and enlarged, echogenic kidneys on ultrasound. A renal biopsy showed chronic tubulointerstitial fibrosis and electron microscopy showed dilated mitochondria without distortion of the cristae. Despite aggressive management she remained hypertensive and volume overloaded, with serum urea concentration increasing to 101 mg/dL [normal 2–19 mg/dL] and serum creatinine concentration increasing to 1.2 mg/dL [normal 0.1–0.4 mg/dL]. The development of pleural effusions and pulmonary edema necessitated continuous veno-venous hemodiafiltration (CVVHDF). She transitioned to peritoneal dialysis, which continued until her death at 24 months.
Laboratory analysis included elevated lactate (5.62–8.95mM) [normal 0.8–2mM] and pyruvate (0.18–0.23mM) [normal 0.05–0.14mM]. Serial brain MRIs demonstrated progressive volume loss with white matter changes in the periventricular and centrum semiovale white matter, bilateral hippocampi and putamina. MR spectroscopy revealed a lactate doublet within the basal ganglia, the centrum semiovale and the ventricles. Clinical laboratory evaluation of the electron transport chain in muscle was normal. The TrnF mutation (m.586G>A) was identified clinically using Sanger sequencing of muscle and blood. This mutation was not identified in her mother. A rare homoplasmic polymorphism, m.5746G>A, was identified in the proband and her mother.
Cell culture
Fibroblasts were obtained by skin biopsy and cultured in DMEM (Gibco) supplemented with 10% Fetal Calf Serum, pyruvate (110 μg/mL) and uridine (500 μg/mL). Cybrids were created from patient fibroblasts and 143B ρ0 osteosarcoma cells (a gift from Michael King) using previously described protocols [3].
Detection of mitochondrial heteroplasmy
The mitochondrial sequence from nt.467–828 was amplified and sequenced with forward primer CCCATACTACTAATCTCATCA and reverse primer GTTAATCACTGCTGTTTCCCG.
Oxygen consumption studies
Cybrid lines were assayed for oxygen consumption using an XF24 (Seahorse Biosciences) as previously described [4]. Cells were plated at 30,000 cells per well and sequentially treated with oligomycin (1μM), dinitrophenol (150μM) and rotenone (160nM). Samples were analyzed in quadruplicate and values are normalized to protein concentration.
Northern blotting
RNA samples were produced from nearly confluent fibroblast cultures using Trizol (Invitrogen) and were resolved on a 15% acrylamide/TBE gel and transferred to a Hybond N+ membrane (GE-Amersham). Mitochondrial tRNAs were identified by Northern blotting [4]. The probe for tRNAPhe hybridizes to positions encoded by nucleotides 594–613 to avoid interference from the m.586G>A mutation.
Western blotting
Whole cell protein extracts from cybrid cells were evaluated with the following antibodies: Complex I subunit NDUFA9 (MitoSciences MS111), Complex II subunit 70 kDa (MitoSciences MS204), Complex III subunit core 2 (MitoSciences MS304), and ATP Synthase subunit alpha (MitoSciences MS507). Actin (Santa Cruz sc-1616) was used as a loading control. Western blots were imaged with a chemiluminescent detection system (VersaDoc) as previously described [4].
Results
Patient and cybrid heteroplasmy
Resequencing of DNA derived from the proband’s fibroblasts confirmed the m.586G>A mutation. Low levels of the wild-type allele were also detected, suggesting that the mutation was heteroplasmic (data not shown). To study the mutation in isolation, mitochondrial cybrid lines were created from the fusion of patient’s fibroblasts and 143Bρ0 cells. Three distinct lines were isolated: a wild-type m.586G cybrid, a heteroplasmic cybrid with approximately 70% mutant load and a homoplasmic mutant m.586A cybrid. All of the tested lines were homoplasmic for the m.5746G>A variant, identified in both the patient and her mother. This polymorphism has been previously identified in a database of normal human mtDNA variation [5].
Oxygen consumption in cybrid cells
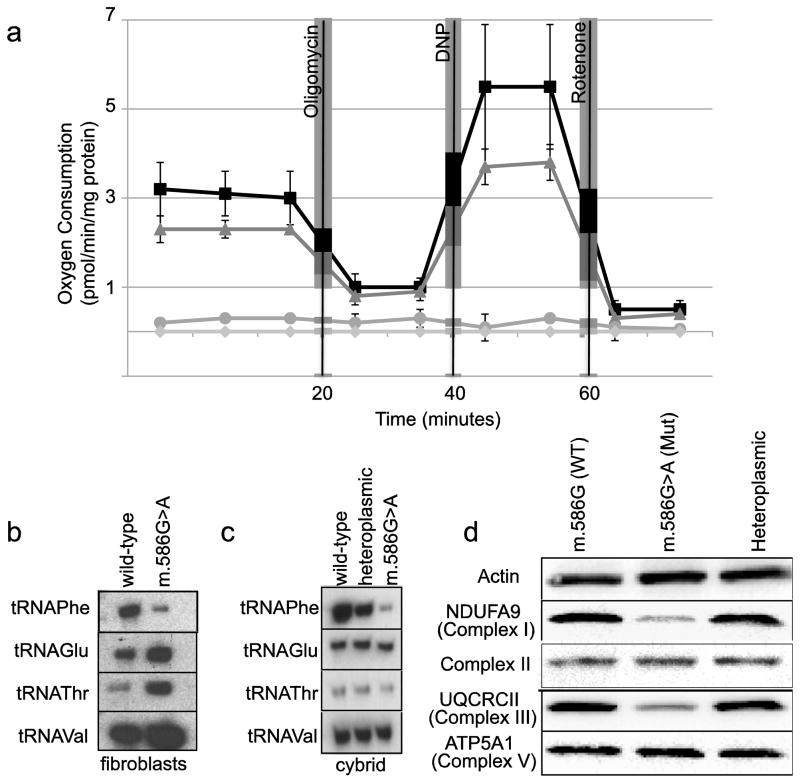
Oxygen consumption was evaluated in intact cybrid cells. Basal respiration was severely reduced in the homoplasmic mutant m.586G>A cell line, and there was a trend towards decreasing respiration in the heteroplasmic line when compared to the wild-type cybrid (Figure 1a). The addition of oligomycin, which inhibits the F0F1-ATPase, had a similar effect upon the heteroplasmic and wild-type cybrid lines. Maximal respiration induced by the addition of the uncoupler dinitrophenol was again somewhat lower in the heteroplasmic cybrid line compared with the homoplasmic wild-type cybrids. The homoplasmic mutant cybrid had very reduced oxygen uptake under all tested conditions.
Figure 1.
Oxygen consumption studies were performed in cybrid cells. Lines homoplasmic for m.568G>A (circles), heteroplasmic (triangles) or homoplasmic wild-type (squares) were analyzed, along with parental strain 143Bρ0 (diamonds). Oligomycin, dinitrophenol and rotenone were added at the times indicated. The m.568G>A strain had significantly diminished oxygen consumption under both basal and DNP-stimulated conditions (10% and 5% respectively; p<.01 by t-test; bars indicate standard error) (a). Levels of mitochondrial tRNAs were analyzed by Northern blotting. Total RNA was extracted from proband fibroblasts (b) and cybrid lines (c), resolved on acrylamide gels and blotted with probes for the mitochondrial tRNAs for phenylalanine, glutamine, threonine and valine. Western blotting for subunits of the electron transport chain in cybrid whole cell extracts (d).
tRNA and protein levels in cybrids
Since the m.586G>A mutation may destabilize the tRNA, we investigated the levels of MTTF by Northern blotting. There was a significant reduction of MTTF in patient fibroblasts compared to an unaffected control, with increased expression of the other tRNAs tested (Figure 1b). Similarly, Northern blotting of the three cybrid lines showed a dose dependent decrease of MTTF in the heteroplasmic and mutant cybrids (Figure 1c). Other mitochondrial tRNAs were unaffected or even increased in the m.586G>A cybrid.
Western blotting of mitochondrial extracts from all three cybrid lines was performed to look at the effect of the mutation on the electron transport chain subunits (Figure 1d). The homoplasmic mutant cybrid line had reduced levels of complex I and III subunits, as compared to the wild-type and heteroplasmic cybrids.
Discussion
We have studied the m.586G>A mutation which has now been observed in 2 unrelated probands. Our patient is a 16-month-old girl with chronic kidney disease, lactic acidosis, developmental delay and multi-organ involvement. The previously reported patient with this mutation was a 57-year-old woman with psychiatric problems, dementia and akinesia-rigidity [2]. Despite the differences in these presentations, we have confirmed that m.586G>A is pathogenic and not a neutral polymorphism. Basal and maximally stimulated oxygen consumption in cybrids harboring the mutation is impaired in a dose dependent manner. Levels of the tRNAPhe were significantly reduced in both the patient and in cybrid lines bearing the m.586G>A mutation. Cybrids carrying the m.586G>A mutation had reduced levels of electron transport chain subunits, likely due to the mutation’s deleterious effects on translation and the subsequent turnover of incompletely formed complexes.
Renal involvement is observed in several mitochondrial disorders such as the Pearson, Leigh and Kearns-Sayre syndromes (for more complete reviews see [6,7]). The common MELAS mutation m.3243A>G has been implicated in steroid-resistant focal segmental glomerulosclerosis [8], an Alport syndrome-like phenotype [9], maternally-inherited diabetes and deafness [10] and in a patient with renal cancer and nephrotic syndrome [11]. Patients with both isolated and syndromic renal disease have been found to have other, novel mtDNA mutations, such as a 7.3kB mtDNA deletion in a patient with renal Fanconi syndrome [12] and a frame-shift deletion of m.12425delA in a patient with Complex I deficiency, renal failure and myopathy [13]. Here, we add a new mtDNA mutation, m.586G>A, to the list of mitochondrial DNA mutations associated with renal disease and as a cause of chronic tubulointerstitial nephritis.
Based on clinical history of the proband, the evolutionary conservation of the base-pair created by m.586G>A and cybrid studies, the mutation scores as definitely pathogenic using a scale for determining the impact of mitochondrial tRNA variation [14]. In light of our patient’s phenotype, the absence of renal pathology in the m.586A>G-affected patient described by Young and colleagues is intriguing [2]. One possibility is that the low levels of m.586A>G in urine-derived DNA, presumably from renal epithelium, reflect a lower level of heteroplasmy in the kidneys, with subsequent organ-specific escape from mitochondrial injury.
In summary we describe a child with a novel mitochondrial mutation causing tubulointerstitial fibrosis and neurological deterioration that was progressive. The phenotype of this tRNAPhe mitochondrial mutation should be added to the multiple ways in which kidney disease can manifest in children with mitochondrial disorders.
Acknowledgments
This work was supported by NIH grant HD58022 to NS. The authors thank Michael King for the gift of the 143Bρ0 cell line and guidance on making cybrids, Marni Falk for her participation in the care of our patient, and the family that participated in this research study.
Abbreviations
- MTTF
mitochondrial tRNAPhe
- mtDNA
mitochondrial DNA
- ESRD
end-stage renal disease
- CVVHDF
continuous veno-venous hemodiafiltration
References
- 1.Hall AM, Unwin RJ, Hanna MG, Duchen MR. Renal function and mitochondrial cytopathy (MC): more questions than answers? QJM. 2008;101:755–766. doi: 10.1093/qjmed/hcn060. [DOI] [PubMed] [Google Scholar]
- 2.Young TM, Blakely EL, Swalwell H, Carter JE, Kartsounis LD, O’Donovan DG, Turnbull DM, Taylor RW, de Silva RN. Mitochondrial transfer RNA(Phe) mutation associated with a progressive neurodegenerative disorder characterized by psychiatric disturbance, dementia, and akinesia-rigidity. Arch Neurol. 2010;67:1399–1402. doi: 10.1001/archneurol.2010.283. [DOI] [PubMed] [Google Scholar]
- 3.King MP, Koga Y, Davidson M, Schon EA. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol Cell Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glatz C, D’Aco K, Smith S, Sondheimer N. Mutation in the mitochondrial tRNA(Val) causes mitochondrial encephalopathy, lactic acidosis and stroke-like episodes. Mitochondrion. 2011;11:615–619. doi: 10.1016/j.mito.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–751. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emma F, Bertini E, Salviati L, Montini G. Renal involvement in mitochondrial cytopathies. Pediatr Nephrol. 2012;27:539–550. doi: 10.1007/s00467-011-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finsterer J. Overview on visceral manifestations of mitochondrial disorders. Neth J Med. 2006;64:61–71. [PubMed] [Google Scholar]
- 8.Lowik MM, Hol FA, Steenbergen EJ, Wetzels JFM, van den Heuvel LPWJ. Mitochondrial tRNALeu(UUR) mutation in a patient with steroid-resistant nephrotic syndrome and focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2005;20:336–341. doi: 10.1093/ndt/gfh546. [DOI] [PubMed] [Google Scholar]
- 9.Fujii H, Mori Y, Kayamori K, Igari T, Ito E, Akashi T, Noguchi Y, Kitamura K, Okado T, Terada Y, Kanda E, Rai T, Uchida S, Sasaki S. A familial case of mitochondrial disease resembling Alport syndrome. Clin Exp Nephrol. 2008;12:159–163. doi: 10.1007/s10157-007-0022-5. [DOI] [PubMed] [Google Scholar]
- 10.Massin P, Dubois-Laforgue D, Meas T, Laloi-Michelin M, Gin H, Bauduceau B, Bellanne-Chantelot C, Bertin E, Blickle J-F, Bouhanick B, Cahen-Varsaux J, Casanova S, Charpentier G, Chedin P, Dupuy O, Grimaldi A, Guerci B, Kaloustian E, Lecleire-Collet A, Lorenzini F, Murat A, Narbonne H, Olivier F, Paquis-Flucklinger V, Virally M, Vincenot M, Vialettes B, Timsit J, Guillausseau PJ GEDIAM (Mitochondrial Diabetes French Study Group) . Retinal and renal complications in patients with a mutation of mitochondrial DNA at position 3,243 (maternally inherited diabetes and deafness). A case-control study. Diabetologia. 2008;51:1664–1670. doi: 10.1007/s00125-008-1073-1. [DOI] [PubMed] [Google Scholar]
- 11.Piccoli GB, Bonino LD, Campisi P, Vigotti FN, Ferraresi M, Fassio F, Brocheriou I, Porpiglia F, Restagno G. Chronic kidney disease, severe arterial and arteriolar sclerosis and kidney neoplasia: on the spectrum of kidney involvement in MELAS syndrome. BMC Nephrol. 2012;13:9. doi: 10.1186/1471-2369-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Au KM, Lau SC, Mak YF, Lai WM, Chow TC, Chen ML, Chiu MC, Chan AYW. Mitochondrial DNA deletion in a girl with Fanconi’s syndrome. Pediatr Nephrol. 2006;22:136–140. doi: 10.1007/s00467-006-0288-y. [DOI] [PubMed] [Google Scholar]
- 13.Alston CL, Morak M, Reid C, Hargreaves IP, Pope SAS, Land JM, Heales SJ, Horváth R, Mundy H, Taylor RW. A novel mitochondrial MTND5 frameshift mutation causing isolated complex I deficiency, renal failure and myopathy. Neuromuscul Disord. 2010;20:131–135. doi: 10.1016/j.nmd.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Yarham JW, Al-Dosary M, Blakely EL, Alston CL, Taylor RW, Elson JL, McFarland R. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat. 2011;32:1319–1325. doi: 10.1002/humu.21575. [DOI] [PubMed] [Google Scholar]