Abstract
Planarian regeneration involves regionalized gene expression that specifies the body plan. After amputation, planarians are capable of regenerating new anterior and posterior poles, as well as tissues polarized along the anterior-posterior, dorsal-ventral and medial-lateral axes. Wnt and several Hox genes are expressed at the posterior pole, whereas Wnt inhibitory genes, Fgf inhibitory genes, and prep, which encodes a TALE-family homeodomain protein, are expressed at the anterior pole. We found that Smed-pbx (pbx for short), which encodes a second planarian TALE-family homeodomain transcription factor, is required for restored expression of these genes at anterior and posterior poles during regeneration. Moreover, pbx(RNAi) animals gradually lose pole gene expression during homeostasis. By contrast, pbx was not required for initial anterior-posterior polarized responses to wounds, indicating that pbx is required after wound responses for development and maintenance of poles during regeneration and homeostatic tissue turnover. Independently of the requirement for pbx in pole regeneration, pbx is required for eye precursor formation and, consequently, eye regeneration and eye replacement in homeostasis. Together, these data indicate that pbx promotes pole formation of body axes and formation of regenerative progenitors for eyes.
Keywords: Axis formation, Eye, Homeodomain, pbx, Planarians, Regeneration
INTRODUCTION
Planarians are capable of regenerating any missing body part and are an emerging system for investigation of cellular and molecular mechanisms underlying regeneration. Regeneration requires production of new cells and instructions that specify the identity of cell types to be regenerated. The planarian Schmidtea mediterranea utilizes a population of dividing regenerative cells called neoblasts (Reddien and Sánchez Alvarado, 2004), which includes pluripotent stem cells (cNeoblasts) (Wagner et al., 2011), to regenerate any missing body part. Robust regenerative mechanisms exist for restoration of the body plan, involving genes that regulate anterior-posterior (AP), medial-lateral (ML) and dorsal-ventral (DV) polarization of tissues (Reddien and Sánchez Alvarado, 2004; Reddien, 2011).
Several signaling pathways and transcription factors are essential for regulation of planarian regeneration. Wnt signaling controls AP regeneration polarity (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008; Adell et al., 2009; Petersen and Reddien, 2009b; Gurley et al., 2010; Petersen and Reddien, 2011), which is the decision to regenerate a head or tail at transverse amputation planes (Morgan, 1898; Morgan, 1905). Multiple Wnt genes and genes encoding candidate secreted inhibitors of Wnt signaling are expressed in distinct spatial domains along the AP axis (Reddien, 2011; Almuedo-Castillo et al., 2012). Several members of the Hox family, such as DjAbd-Ba and Plox4-Dj, are expressed in the planarian posterior (Orii et al., 1999; Nogi and Watanabe, 2001). Smed-prep, which encodes a TALE (three amino acid loop extension) family homeodomain protein, is expressed at the tip of planarian heads and is required for anterior pole marker expression in regeneration (Felix and Aboobaker, 2010). A LIM-homeobox gene, Djislet, is expressed in the posterior and is required for posterior pole marker expression in regeneration (Hayashi et al., 2011). Regeneration of the DV and ML axes requires Bmp signaling, with Bmp signaling components differentially expressed along the DV and/or ML axes (Orii et al., 1998; Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007; Molina et al., 2009; Gaviño and Reddien, 2011; Molina et al., 2011). In addition to their functions in regeneration, these signaling pathways and transcription factors also display constitutive expression in the adult planarian body with several being required for homeostatic maintenance of the body plan during natural tissue turnover (Reddien, 2011). For example, RNAi of the Wnt signaling component β-catenin-1 results in ectopic head appearance around the periphery of intact animals (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). These observations indicate that actively maintained expression of genes regulating body position instructs tissue turnover, but how these regional expression patterns are maintained and regenerated is poorly understood.
pbx encodes a TALE-class homeodomain protein and can regulate gene expression in a variety of developmental contexts (Moens and Selleri, 2006; Laurent et al., 2008). There are four mammalian Pbx genes (Kamps et al., 1990; Nourse et al., 1990; Monica et al., 1991; Wagner et al., 2001), five zebrafish Pbx genes (Pöpperl et al., 2000; Vlachakis et al., 2000), one Drosophila pbx homolog, extradenticle (exd) (Rauskolb et al., 1993) and three Caenorhabditis elegans pbx homologs, ceh-20 (C. elegans homeobox), ceh-40 and ceh-60 (Bürglin and Ruvkun, 1992; Bürglin, 1997; Mukherjee and Bürglin, 2007). pbx was first characterized for regulating antero-posterior patterning during embryonic development as a co-factor of the Hox genes. In Drosophila, mutations in the exd gene cause homeotic transformations without affecting the expression of corresponding Hox genes and Exd controls anterior-posterior patterning in the fly embryo by acting together with Hox proteins (Peifer and Wieschaus, 1990; Rauskolb et al., 1993; Rauskolb et al., 1995). Studies of the zebrafish lzr (pbx4) gene in AP hindbrain patterning suggest that it promotes the action of multiple Hox genes in vertebrates (Pöpperl et al., 2000). In C. elegans, Hox genes, including lin-39, mab-5 and nob-1, act with ceh-20 in a variety of AP regionalized processes, including postembryonic mesodermal differentiation, cell migration, vulval development and programmed cell death (Liu and Fire, 2000; Shemer and Podbilewicz, 2002; Van Auken et al., 2002; Arata et al., 2006; Takács-Vellai et al., 2007). Pbx genes can interact with additional transcription factors to control expression of various signaling factors. exd is required for proper expression of wingless and dpp in parasegments of the Drosophila midgut (Rauskolb and Wieschaus, 1994). Induction of Fgf during zebrafish hindbrain and fin development requires lzr (pbx4) and pbx2 (Pöpperl et al., 2000; Waskiewicz et al., 2002). Mouse Pbx proteins, together with Prep1 (also known as Pknox1) and Meis, can regulate expression of Wnt9b and Wnt3 for face morphogenesis (Ferretti et al., 2011).
We found that pbx in planarians is required for proper expression of genes implicated in control of AP or DV axis patterning. pbx is the first gene reported to be important for regeneration/formation of both poles of the AP axis. pbx is also required for eye regeneration and formation of eye progenitors. Our results suggest that pbx has an essential role in cell fate specification and pole formation during regeneration in planarians.
MATERIALS AND METHODS
Animal culture and radiation treatment
Asexual Schmidtea mediterranea strain (CIW4) animals were starved 7-14 days prior to experiments. Animals were exposed to a dose of 6000 rads of radiation using a dual Gammacell-40 137 cesium source and amputated 4 days after irradiation.
Molecular biology and RNAi
Primer sequences for constructs generated are listed in supplementary material Table S1. Clone H.110.1c was used for pbx RNAi feeding. The negative RNAi control was C. elegans unc-22. Bacteria were mixed with 67% liver paste at a 1:300 ratio to culture volume. Worms were fed on days 0, 3, 6 and 9 and amputated on day 10. RNAi feeding (Fig. 1C; Fig. 3; Fig. 4C; Figs 5, 6, 7) strongly reduced pbx expression (supplementary material Fig. S1). For long-term RNAi, animals were fed twice every week. prep RNAi by feeding utilized coding nucleotides 135-2002 and RNAi efficacy was comparable to a previous study (Felix and Aboobaker, 2010). In Fig. 1C, animals were injected with 600 μg/ml of dsRNA to create weak pbx and smedwi-2 RNAi phenotypes. RNAi by injection (Fig. 1A,B; Fig. 2B,C,E; Fig. 4A,B) of 3-5 mg/ml pbx dsRNA yielded a stronger phenotype than did RNAi by feeding. Transversely amputated fragments were injected with dsRNA within 30 minutes to 4 hours after amputation, and were injected again the next day. This procedure (amputation and injection, followed by booster injection the next day) was performed again on days 4/5 and 7/8 after the initial amputation. For assessing the neoblast wound response, amputation and injection, followed by booster injection the next day was repeated on days 3/4, injections were performed on days 6/7, and amputation was carried out on day 9.
Fig. 1.
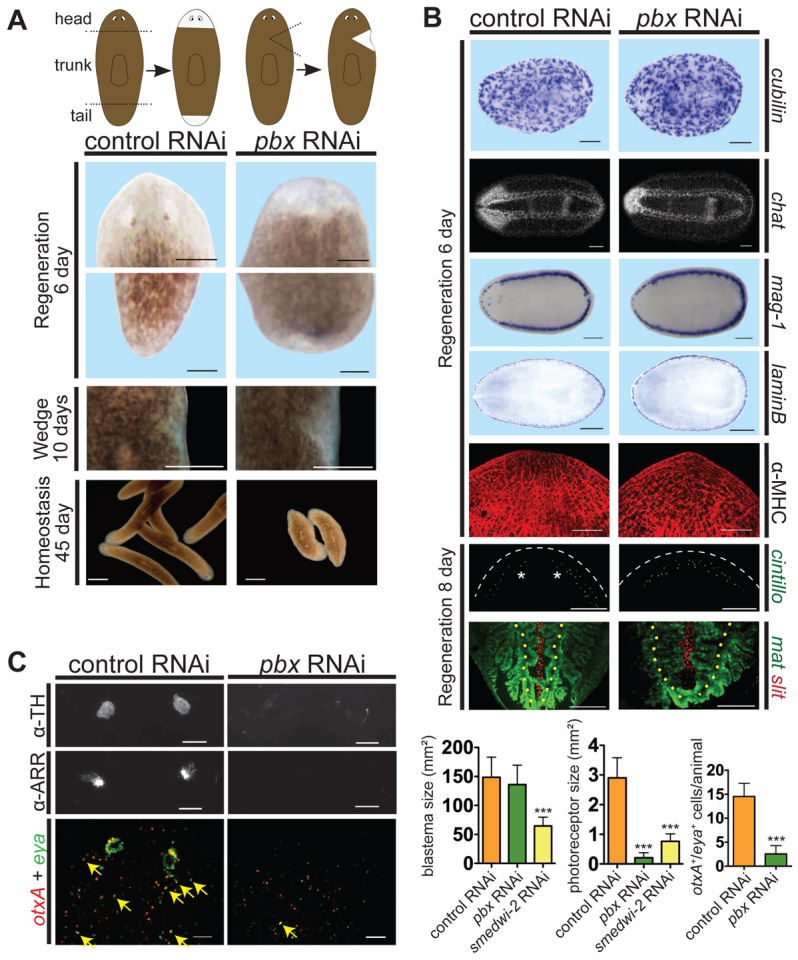
pbx(RNAi) planarians fail to regenerate eyes. (A) pbx(RNAi) animals regenerated blastemas without eyes 6 days after cutting (n=96) and displayed locomotion defects when exposed to long-term RNAi (n=20). pbx(RNAi) animals generated unpigmented tissue at the site of wedge amputations at 10 days after amputation (control, n=8/8; pbx(RNAi), n=8/8). Left cartoon depicts head and tail amputation sites; right cartoon depicts wedge regeneration sites. Scale bars: 200 μm for pole regeneration, 1 mm for wedge regeneration, 1.26 mm for homeostasis. (B) Whole-mount ISH 6 days after regeneration demonstrates that pbx(RNAi) animals regenerated protonephridia (cubilin, n=5/5), the nervous system with morphological defects (chat, n=13/13), subepidermal marginal adhesive gland cells (mag-1, n=6/6), lamin B-positive lateral tissue (n=12/12) and muscle fibers (α-MHC, n=16/16). At 8 days, pbx(RNAi) sensory cells (cintillo+) regenerated with normal cell number (P=0.325, t-test), but failed to distribute into two lateral domains (n=6/6). pbx(RNAi) intestinal branches were generated, but slight defects in medial branch length were present (mat, n=2/7), and midline cells failed to extend completely posteriorly (slit, n=6/7). Feeding RNAi was used for lamin B and α-MHC images; similar results were obtained with dsRNA injection. Anterior is to the left except for α-MHC, cintillo and mat/slit panels, which are oriented anterior to the top. Asterisks, photoreceptors; white dashed lines, head rim; yellow dotted lines, primary posterior intestinal branches. Scale bars: 200 μm except chat, 100 μm; and α-MHC, 50 μm. (C) pbx(RNAi) animals failed to regenerate pigment cup cells (α-TH immunostaining, n=0/10 were normal) and photoreceptor neurons (α-ARR immunostaining, n=0/10 were normal); eye progenitor numbers (double-labeled with otxA and eya RNA probes) were greatly reduced (n=13, right-hand graph, ***P<0.0001, t-test). Animals are oriented anterior to the top. Yellow arrows, double-positive cells. The eye regeneration defect in pbx(RNAi) animals is not explained only by reduced blastema size (n≥8 each). Left and middle graphs, one-way ANOVA tests followed by a Dunnet post-hoc test; ***P≤0.001 for experimental condition versus control. Graphs show mean±s.e.m. Scale bars: 50 μm.
Fig. 3.
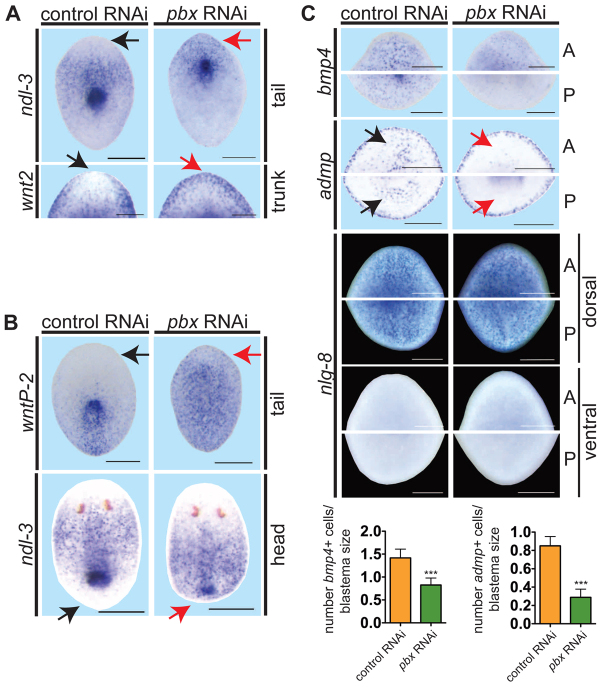
pbx(RNAi) animals display abnormal pole gene expression and fail to re-scale expression gradients during regeneration. (A-C) Whole-mount ISH of regenerating pieces of planarians at day 6. Anterior is to the top. Black arrows, normal gene expression location (control); red arrows, aberrant expression in pbx(RNAi) animals. (A) pbx(RNAi) animals exhibited aberrant anterior blastema gene expression: ndl-3 (control, n=15/15; pbx RNAi, n=1/16 display restriction from the head tip of tail fragments) and wnt2 (control, n=10/11; pbx RNAi, n=0/13 display restriction from the head tip; head blastema of trunk fragment shown). (B) pbx(RNAi) animals failed to re-scale the wntP-2 expression gradient (control, n=17/18; pbx RNAi, n=4/20 tail fragments show re-scaling) and ndl-3 expression (control, n=22/25; pbx RNAi, n=1/22 head fragments show re-scaling). (C) Dorsal bmp4 expression (control, n=18; pbx RNAi, n=20) and ventral admp expression (control, n=13; pbx RNAi, n=10) were reduced in pbx(RNAi) animals, but dorsal expression of nlg-8 was normal in both anterior (A) and posterior (P) blastemas (control, n=13/13; pbx RNAi, n=12/12). ‘Head’, ‘trunk’ and ‘tail’ refer to a head fragment regenerating a tail, a fragment with head and tail amputated transversely, and a tail fragment regenerating a new head after transverse amputation, respectively. Graphs show quantification of bmp4+ and admp+ cells normalized to blastema size (mean±s.e.m.; ***P=0.0,001, t-test). Scale bars: 200 μm.
Fig. 4.
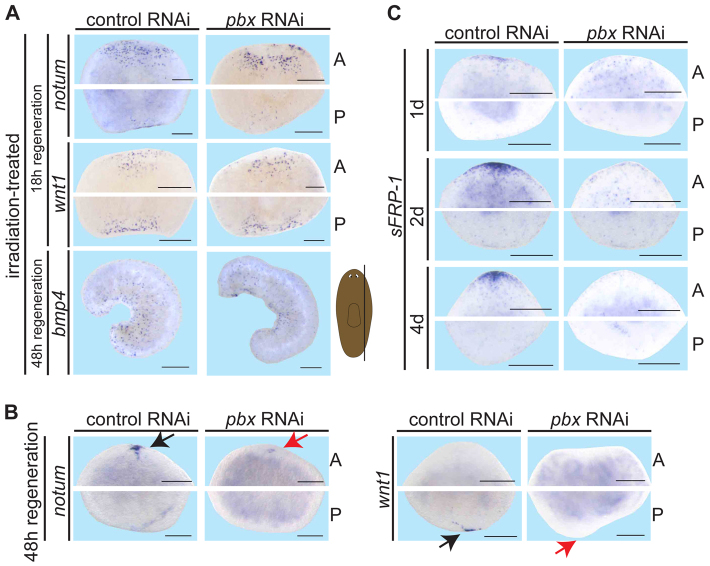
Wound-induced patterning gene expression is not affected in pbx(RNAi) planarians. (A) Anterior (A) and posterior (P) blastemas of irradiated pbx(RNAi) trunk pieces exhibited normal irradiation-insensitive, wound-induced expression of notum (control, n=5/7; pbx RNAi, n=6/6; transversely amputated), wnt1 (control, n=7/7; pbx RNAi, n=8/8; transversely amputated) and bmp4 (control, n=8/8; pbx RNAi, n=13/13; parasagitally amputated). Cartoon depicts amputation site; thin fragments were used. (B) notum expression (control, n=6/6; pbx RNAi, n=2/6 no expression, n=3/6 reduced expression) in the anterior blastema and wnt1 expression (control, n=6/6; pbx(RNAi), n=0/7 trunk pieces) in the posterior blastema of transversely cut animals 48 hours post-cutting. Black and red arrows indicate normal and reduced/absent expression, respectively. (C) sFRP-1 expression in transversely cut trunk pieces during regeneration; 1, 2 and 4 days (d) shown (pbx RNAi, n=14 trunk pieces for each condition). Scale bars: 200 μm.
Fig. 5.
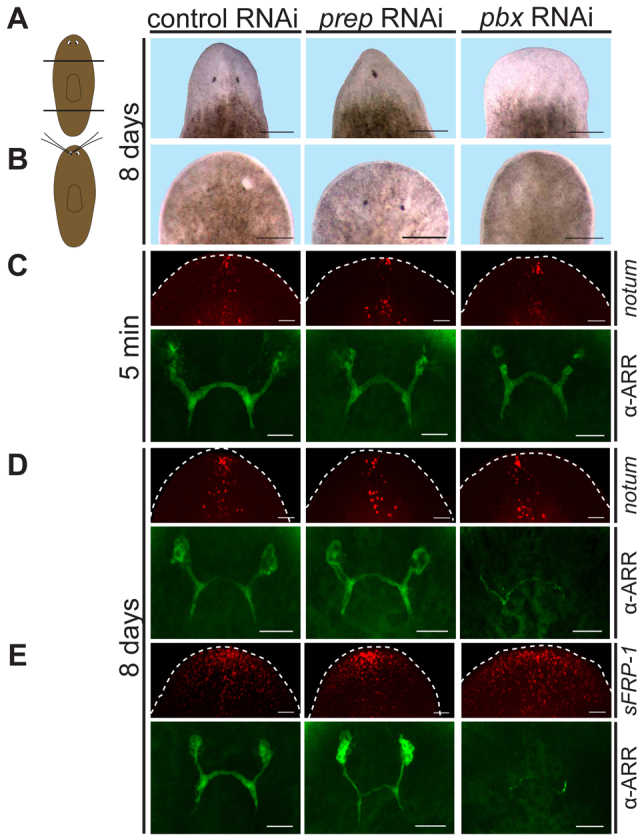
The requirement of pbx for eye regeneration is separable from its requirement for pole regeneration. (A,B) Cartoons depict head or eye removal. (A) pbx (n=0/20 no eyes) and prep (n=18/19 one eye; n=1/19 no eyes) RNAi planarians exhibited different eye regeneration phenotypes after transverse amputation. (B) Both control (n=25/25) and prep RNAi (n=24/24) animals regenerated eyes but pbx(RNAi) animals (n=0/25) did not 8 days after eye removal with glass needles. (C-E) Double-fluorescent labeling of notum or sFRP-1 FISH with α-ARRESTIN immunostaining were used to assess anterior pole gene expression and eye regeneration in each animal. (C) Photoreceptor cell bodies were mostly absent 5 minutes after eye removal, with axons (green fluorescence) remaining (control, n=9/9; prep RNAi, n=10/10; pbx RNAi, n=10/10). notum expression was normal. (D,E) Eye regeneration defects were observed in pbx(RNAi) animals but not in prep(RNAi) or control animals. The size of the cluster of photoreceptor cells increased in prep(RNAi) (n=24/24) and control (n=25/25) animals with time, but did not in pbx(RNAi) (n=25/25) animals with only sparse pre-existing axon fragments remaining. notum and sFRP-1 expression were normal. Dashed line depicts animal boundary. Anterior is to the top. Scale bars: in A,B, 200 μm; in C-E, 50 μm.
Fig. 6.
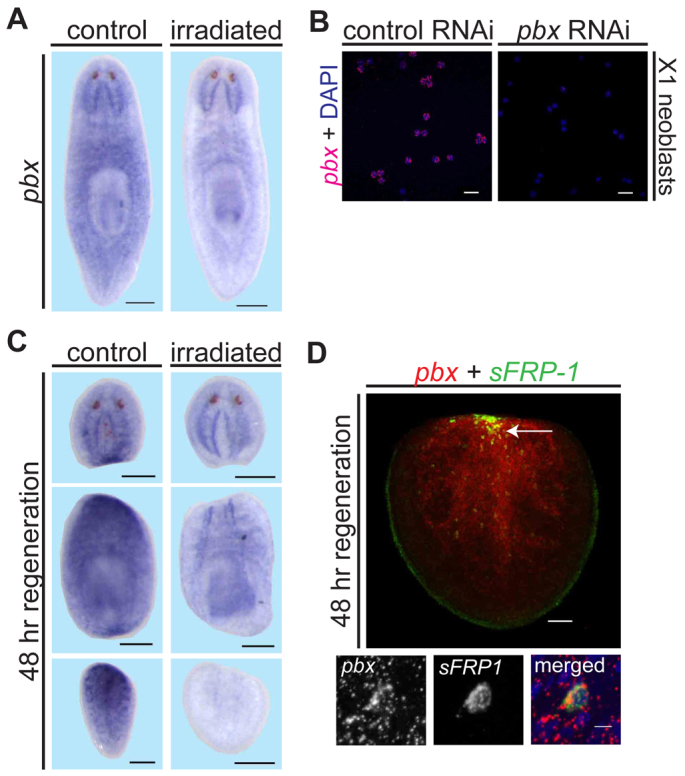
pbx displays broad expression, including in neoblasts. (A) Whole-mount ISH showing pbx expression in non-irradiated (control) and irradiated planarians (n=6 for each). Scale bars: 200 μm. (B) pbx expression in neoblasts (referred to as X1 cells) sorted using FACS (Hayashi et al., 2006) was detected using FISH. Expression was absent following pbx RNAi. Scale bars: 20 μm. (C) pbx mRNA levels were strong and irradiation sensitive near wounds, 48 hours after amputation (n=6 for each). Scale bars: 200 μm. (D) Double FISH of pbx (red) and sFRP-1 (green) with DAPI (blue) in a regenerating tail (n=10) fragment at 48 hours. Some cells displayed both pbx and sFRP-1 signal, indicated by an arrow. Scale bars: upper panel, 50 μm; lower panels, 5 μm.
Fig. 7.
pbx is required for maintenance of pole gene expression and eyes during homeostatic tissue turnover. (A) Six weeks of prep and pbx RNAi caused reduced sFRP-1 expression (control, n=0/13; prep RNAi, n=13/13; pbx RNAi, n=10/10) and aberrant wnt-2 (control, n=0/4; prep RNAi, n=3/3; pbx RNAi, n=3/3) and notum (control, n=0/6; prep RNAi, n=6/6; pbx RNAi, n=3/3; white circle) expression. pbx RNAi, but not prep RNAi, caused wnt1+ cell reduction at the posterior pole (control, n=7; prep RNAi, n=7; pbx RNAi, n=6; shown are mean±s.e.m.; one-way ANOVA test followed by a Dunnet post-hoc test; *P≤0.05 between experimental condition and control). pbx(RNAi) animals, labeled with α-ARRESTIN antibody gradually lost photoreceptors and prep(RNAi) animals exhibited extra photoreceptors (white arrow) (control, n=6; prep RNAi, n=6; pbx RNAi, n=3). Asterisks in images indicate eyes. Scale bars: 50 μm. (B) Unamputated pbx(RNAi) animals failed to homeostatically maintain photoreceptor neurons after 8 weeks of RNAi (labeled with α-ARRESTIN antibody; pbx RNAi, n=8/10; control, n=0/10). Scale bars: 50 μm. (C) Control, prep or pbx RNAi animals at 6 days of regeneration. Blastema size quantification is shown at right (one-way ANOVA test followed by a Dunnet post-hoc test; ***P≤0.001 between experimental condition and control; n=10 for each condition). Dorsal expression of nlg-8 and ventral expression of netrin in trunk anterior blastemas (n=14/14 for all conditions). Both prep and pbx RNAi animals exhibited aberrant wnt-2 expression at the anterior tip of all regenerating pieces (control, n=0/16; prep RNAi, n=16/16; pbx RNAi, n=11/12). However, only pbx(RNAi) animals showed aberrant wnt-2 expression at posterior blastemas (control, n=1/16; prep RNAi, n=1/16; pbx RNAi, n=11/12). Black arrows, normal expression; red arrows, aberrant expression. Neoblasts (smedwi-1+) were similar in distribution among all RNAi treatments (control RNAi n=15/15; prep RNAi n=16/16; pbx RNAi, n=14/14 regenerating pieces; shown are mean±s.e.m.). Scale bars: 100 μm. (D) Data summary. pbx is required for expression of anterior and posterior pole genes in head and tail blastemas, respectively. pbx is also required for eye progenitor formation. Dashed lines indicate animal boundaries. All animals are oriented with anterior to the top.
Fig. 2.
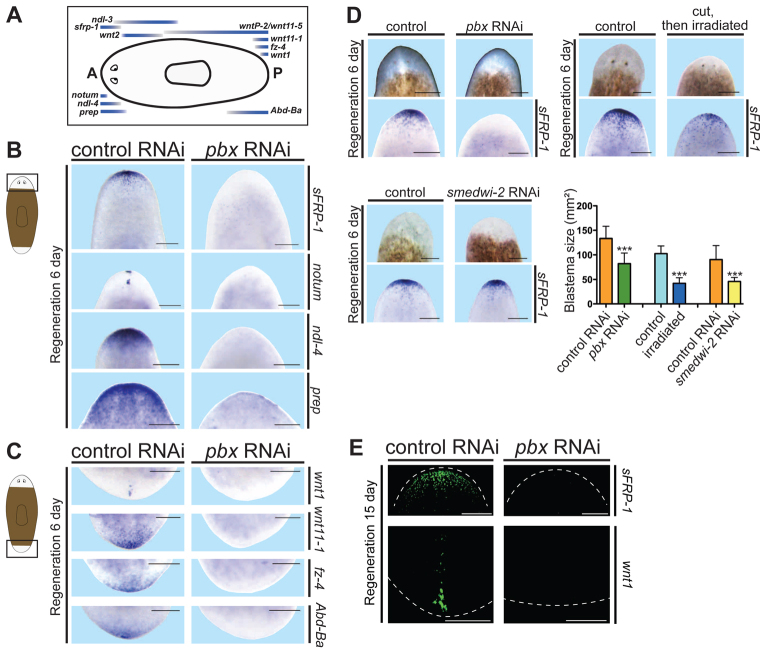
pbx is required for expression of genes at AP poles in regeneration. (A) Cartoon shows AP expression locations of genes used in this study to characterize body axis regionalization; eyes (in the head) and the pharynx (centrally located) are depicted. A, anterior; P, posterior. (B) Whole-mount ISH showing missing or reduced gene expression in the anterior blastema of pbx(RNAi) planarians: sFRP-1 (control n=14/14; pbx n=0/15), notum (control n=6/6; pbx n=4/6 greatly reduced and mislocalized, n=2/6 not present), ndl-4 (control n=5/6; pbx n=0/6) and prep (control n=12/12; pbx n=0/12). Cartoon depicts area shown in images. (C) Expression of posterior markers, wnt1 (control n=6/6; pbx n=1/6), wnt11-1 (control n=4/6; pbx n=0/6), fz-4 (control n=6/6; pbx n=1/6) and Abd-Ba (control n=6/6; pbx n=0/5) was absent in pbx(RNAi) animals. Cartoon depicts area shown in images. (D) Animals with reduced anterior blastema size displayed more sFRP-1 expression than did pbx(RNAi) animals. Upper left panel: pbx RNAi involved dsRNA injection with 3-5 mg/ml dsRNA (n=10 for both conditions); upper right panel: small blastemas were generated by exposing cut worms to 6000 rads of γ-radiation 24 hours after cutting (control n=10; irradiated n=8); lower left panel: small blastemas were generated by weak RNAi of smedwi-2 (one feeding with amputation 3 days later) (n=10). Quantification of blastema size is shown in the bottom graph (mean±s.e.m.; ***P=0.0001, left pair; ***P<0.0001, middle pair; ***P=0.0002, right pair; t-test for each pair of conditions). (E) Pole markers sFRP-1 (control, n=6/6, pbx RNAi, n=0/6) and wnt1 (control, n=6/6, pbx RNAi, n=1/6) were absent in pbx(RNAi) animals after 15 days of regeneration. All animals are at regeneration day 6 except for panel E and are oriented anterior at the top. Scale bars: in E, 100 μm; in B,C,D, 200 μm.
In situ hybridization, immunostaining and imaging
In situ hybridizations (ISH) and fluoresence ISH (FISH) were performed as described (Pearson et al., 2009) with two additional 30-minute prehybridization buffer washes after hybridization. For immunostaining, animals were fixed as for in situ hybridization, then treated as described (Newmark and Sánchez Alvarado, 2000). α-Myosin heavy chain, TMUS-13 antibody (Romero et al., 1991), rabbit α-tryptophan hydroxylase (α-TH) antibody against peptide HPSDPFYTPEPDCC, and α-ARRESTIN (α-ARR) were used. Fluorescent images were taken with a Zeiss LSM710 Confocal Microscope. For blastema measurements, animals were imaged with a Zeiss Discovery microscope and the head blastema area (unpigmented area) was determined using the measurement tool in Axiovision software. Numbers of cells labeled with RNA probes were determined by manual inspection and Axiovision software.
RESULTS
pbx is required for normal regeneration and locomotion
We identified a single Schmidtea mediterranea gene predicted to encode a protein containing PBC (PBX-containing) homeodomains similar to PBX proteins in other species (supplementary material Fig. S2); we named this gene Smed-pbx (‘pbx’ hereafter). RNAi of pbx resulted in multiple regeneration defects. pbx(RNAi) animals exhibited variable anterior blastema size, depending upon RNAi conditions (see Materials and methods), ranging from normal to 50% smaller than the control. These animals were capable of regenerating interior tissues following removal of a lateral tissue wedge, and lateral tissue following parasagittal amputation (Fig. 1A; supplementary material Fig. S3A). pbx(RNAi) animals failed to regenerate eyes and displayed uncoordinated movement, involving flat body posture and little cilia-mediated propulsion, while twisting in place (Fig. 1A; supplementary material Movie 2).
Despite slightly reduced blastema size and failed eye regeneration, pbx(RNAi) blastemas generated other differentiated cell types (Fig. 1B). For example, the nervous system (choline acetyltransferase, chat+) (Wagner et al., 2011), the protonephridia system (cubilin+) (Scimone et al., 2011), muscle fibers (myosin heavy chain+) (Cebrià and Vispo, 1997), subepidermal marginal adhesive gland cells (mag-1+) (Sánchez Alvarado et al., 2002; Zayas et al., 2010), laminB+ lateral tissue (Kato et al., 1999), cintillo+ sensory neurons (Oviedo et al., 2003) and mat+ (Wenemoser and Reddien, 2010) intestine were all regenerated. The central nervous system of pbx(RNAi) animals displayed morphological defects: whereas animals normally have separate cephalic ganglia, there were no separate ganglia in pbx(RNAi) head blastemas. This phenotype is similar to that caused by RNAi of Smed-prep, which is required for anterior pole formation during head regeneration (Felix and Aboobaker, 2010). However, prep RNAi only results in partial loss of photoreceptors during regeneration and does not cause overt locomotion abnormalities, indicating that pbx has distinct roles from prep. The abnormalities in differentiated tissue pattern in pbx(RNAi) blastemas (e.g. the cephalic ganglia) were also present at later time points following amputation, suggesting that the defects observed do not simply reflect delayed regeneration (supplementary material Fig. S3B). Neoblasts respond to wounds with a body-wide increase in proliferation, followed by a second increase in proliferation near wounds (Wenemoser and Reddien, 2010). This neoblast wound response pattern still occurred in pbx(RNAi) animals, with a slightly lower second increase in proliferation (supplementary material Fig. S3C,D). Therefore, pbx(RNAi) regeneration abnormalities are not associated with robust neoblast loss or a gross defect in neoblast capacity to respond to wounds.
Planarian eyes, which contain photoreceptive neurons and pigmented optic cup cells, failed to regenerate in pbx(RNAi) animals (Fig. 1C). To determine whether eye absence is simply a consequence of reduced pbx(RNAi) blastema size, we compared eye formation in pbx(RNAi) and smedwi-2(RNAi) animals following partial inhibition by RNAi. Weak smedwi-2 inhibition resulted in head blastemas smaller than pbx(RNAi) head blastemas, but with more prominent eye regeneration, indicating simply reduced blastema size does not explain the pbx RNAi eye phenotype (Fig. 1C; supplementary material Fig. S4). Planarian eye regeneration involves migratory precursor cells that express transcription factors promoting eye formation (Lapan and Reddien, 2011). otxA+ eya+ eye precursor cells were significantly reduced in pbx(RNAi) animals at day 6 of regeneration, indicating that pbx is required for eye progenitor formation (Fig. 1C).
pbx is required for expression of genes at AP poles in regeneration
Given the known roles for pbx genes in tissue patterning, we assessed whether regulatory genes involved in regeneration and adult body plan maintenance were expressed normally in pbx(RNAi) blastemas. Genes with candidate roles in regulating planarian positional information will be referred to here as ‘patterning genes’ and are utilized as markers of body regionalization in the experiments described below (Fig. 2A). Patterning genes were selected from previously reported data as (1) displaying regionalized expression and (2) either displaying a patterning-abnormal RNAi phenotype or encoding a protein predicted to regulate a pathway (Wnt, Bmp or Fgf signaling) important for patterning planarian body axes. RNAi phenotypes have not been reported for all patterning genes that will be described. The expression of four patterning genes that normally display strong and restricted expression in animal head tips, sFRP-1 (Gurley et al., 2008; Petersen and Reddien, 2008), notum (Petersen and Reddien, 2009b), ndl-4 (Rink et al., 2009) and prep (Felix and Aboobaker, 2010), was strongly reduced or absent in pbx(RNAi) anterior blastemas (Fig. 2B). sFRP-1 encodes a predicted secreted frizzled-related protein, which can antagonize Wnt signaling in other organisms (Leyns et al., 1997). notum encodes a secreted hydrolase that antagonizes Wnt signaling in planarian regeneration (Petersen and Reddien, 2011) and in Drosophila (Gerlitz and Basler, 2002; Giráldez et al., 2002). ndl-4 encodes a Nou darake-family FGF-receptor-like protein; the related planarian nou darake gene is expressed anteriorly and is required for anterior cephalic ganglia restriction (Cebrià et al., 2002). prep is required for normal head regeneration (Felix and Aboobaker, 2010). These observations suggest that pbx is required for anterior pole regeneration.
Similarly, expression of patterning genes normally restricted to the posterior pole was abnormal in pbx(RNAi) regenerating tails (Fig. 2C). The posterior pole expression of wnt1 (Petersen and Reddien, 2008), wnt11-1 (Petersen and Reddien, 2008), frizzled-4 (Gurley et al., 2008) and Abd-Ba (Nogi and Watanabe, 2001) was absent in pbx(RNAi) animals. These data indicate that pbx is also required for posterior pole regeneration. The reduced AP pole expression of patterning genes in pbx(RNAi) animals is not simply explained by slightly small blastema size. Irradiation after amputation, or partial inhibition with RNAi of 12 genes required for blastema formation (e.g. the smedwi-2 gene) (Reddien et al., 2005a; Reddien et al., 2005b), caused regeneration of smaller head blastemas than did pbx RNAi, but these small blastemas displayed more sFRP-1 expression than did pbx(RNAi) blastemas (Fig. 2D; supplementary material Fig. S5). Furthermore, patterning gene expression at poles was not present even at a late time point after amputation of pbx(RNAi) animals, indicating that pole formation is not simply delayed (Fig. 2E). These data suggest that pbx is required for expression of polarized markers in anterior and posterior blastemas.
Some patterning gene expression fails to be restricted from pbx(RNAi) poles
Because of the anterior and posterior pole defects of pbx(RNAi) animals, we assessed patterning gene expression that is normally restricted from poles. wnt2 and ndl-3 are expressed in the anterior, but not strongly at the head tip of intact animals (Petersen and Reddien, 2008; Rink et al., 2009). In contrast to other AP polarized markers, wnt2 and ndl-3 were expressed in pbx(RNAi) anterior blastemas, but expression extended abnormally to the regenerating head tip (Fig. 3A). Therefore, pbx is not required for expression of all patterning genes in regeneration. Furthermore, these observations indicate that the wild-type anterior-most gene expression domain is absent in pbx(RNAi) heads, with head tips instead displaying expression typical for the posterior region of heads.
pbx is required for re-scaling of AP gene expression gradients during regeneration
Blastema formation can be accompanied by re-scaling and re-patterning of pre-existing tissues (a process known as morphallaxis) (Morgan, 1898; Reddien and Sánchez Alvarado, 2004). Accordingly, patterning genes can change expression during regeneration through new expression (in fragments initially lacking expression) or by re-scaling an existing expression domain to accommodate changing dimensions of the regenerating fragment. For example, wntP-2 (also known as wnt11-5 as wntP-2 has been proposed to be a wnt11-family member) (Gurley et al., 2010) is normally expressed in a posterior-to-anterior gradient (Petersen and Reddien, 2008) and can be involved in regeneration polarity (Petersen and Reddien, 2009b). wntP-2 expression is initially uniform in tail fragments and recedes towards the posterior pole during regeneration (Petersen and Reddien, 2009a; Gurley et al., 2010). pbx(RNAi) animals failed to rescale wntP-2 expression 6 days after amputation (Fig. 3B). Irradiation kills neoblasts and consequently blocks regeneration (Bardeen and Baetjer, 1904; Dubois, 1949; Reddien and Sánchez Alvarado, 2004). pbx(RNAi) tail fragments are similar to irradiated tail fragments (supplementary material Fig. S6), in that wntP-2 expression is initially restricted from the wound but returns to the anterior within 6 days (Gurley et al., 2010). Similarly, ndl-3 expression (normally prepharyngeal) was restricted from the posterior-facing wound of control head fragments, but failed to restrict away from the regenerating posterior blastemas of pbx(RNAi) head fragments (Fig. 3B). These results are consistent with the possibility that pbx-dependent patterning gene expression at regenerating AP poles is required for stable re-scaling of patterning gene expression gradients.
Reduced bmp4 and admp expression in pbx(RNAi) animals
Given the patterning gene expression defects at anterior and posterior poles, we next assessed whether regenerating pbx(RNAi) animals had defects in polarized gene expression on the DV axis. bmp4 expression in unamputated animals is normally strongest medially on the dorsal side and graded laterally; expression is also strong in the dorsal head region (Orii et al., 1998; Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007). The dorsally restricted bmp4 expression pattern is regenerated in control blastemas. In pbx(RNAi) animals, bmp4 expression in blastemas was reduced but not eliminated (Fig. 3C). Because bmp4 is normally expressed more strongly in the anterior than the posterior blastema, AP defects might contribute to the bmp4 expression reduction observed in pbx(RNAi) animals. Furthermore, because the effect of pbx RNAi on bmp4 expression was weaker than the effect on AP pole gene expression, it is possible that pbx has a more prominent role in AP rather than DV axis regeneration. A second Bmp family-encoding gene, admp, is expressed laterally and in a ventral, medial domain (Molina et al., 2009; Gaviño and Reddien, 2011). In transversely amputated pbx(RNAi) animals regenerating heads and tails, the ventral domain of admp expression was reduced, whereas the lateral domain appeared normal (Fig. 3C). By contrast, nlg-8, a noggin-like gene involved in Bmp pathway regulation and normally expressed evenly over the dorsal side of animals and blastemas (Molina et al., 2009), was expressed normally in regenerating pbx(RNAi) animals. Therefore, not all patterning genes with DV polarized expression require pbx for their expression (Fig. 3C). These data indicate that some DV patterning gene expression levels at the dorsal and ventral midlines also require pbx.
pbx is not required for wound-induced patterning gene expression
The patterning gene wnt1 is induced rapidly at virtually all wounds (Petersen and Reddien, 2009b), and is required for regeneration polarity (Adell et al., 2009; Petersen and Reddien, 2009b). wnt1 is expressed at wounds of irradiated animals, indicating that wound-induced wnt1 expression occurs even in the absence of new cell production (Petersen and Reddien, 2009b). notum is also induced at many wound types, at anterior-facing wound edges (Petersen and Reddien, 2011). Both wnt1 and notum wound-induced expression were detected in irradiated pbx(RNAi) animals (Fig. 4A). Similarly, bmp4 is expressed rapidly (within 12 hours) following parasagittal amputation in lateral fragments, even in irradiated animals (Reddien et al., 2007). This irradiation-insensitive bmp4 expression observed 48 hours after amputation was normal in pbx(RNAi) animals (Fig. 4A). Therefore, early wound-induced patterning gene expression involved in axis polarization in regeneration did not require pbx.
By 48 hours after transverse amputation, wnt1 expression is normally clustered at regenerating tail tips and notum expression is clustered at regenerating head tips. By contrast, this polarized wnt1 and notum expression was greatly reduced in pbx(RNAi) animals (Fig. 4B). To determine more precisely when pole formation abnormalities appear in pbx(RNAi) animals, we analyzed sFRP-1 expression, which is specific to anterior-facing blastemas and largely (but not completely) irradiation-sensitive (Petersen and Reddien, 2009b; Gurley et al., 2010). A low level of sFRP-1 expression appears in the first 3-9 hours of regeneration, but significant numbers of sFRP-1+ cells are not visible until 24 hours after amputation (Gurley et al., 2010), later than the robust wound-induced expression phases of wnt1 and notum. sFRP-1 expression was reduced, but initially present at 24 hour and 48 hour anterior-facing wounds in pbx(RNAi) animals, and was absent by 4 days of regeneration (Fig. 4C). Together, these data indicate that pbx is not required for the first step in regeneration of tissue pattern (wound-induced expression of AP and DV polarizing genes) but is required after this wound-induced gene expression phase for further development of poles.
The requirement of pbx for eye regeneration is separable from its requirement in pole regeneration
Several lines of evidence indicate that failed eye regeneration in pbx(RNAi) animals is not simply explained by anterior pole absence in these animals. First, whereas pole gene expression defects are observed in both pbx and prep RNAi animals, pbx RNAi caused a more severe eye regeneration defect than did prep RNAi. For instance, prep(RNAi) head blastemas are cyclopic (Felix and Aboobaker, 2010); 18/19 prep trunk fragments had one eye whereas 0/20 pbx(RNAi) animals regenerated eyes (Fig. 5A). Second, we investigated whether pbx(RNAi) animals could regenerate surgically removed eyes, while leaving the anterior pole intact. notum and sFRP-1 had similar expression patterns in control, prep(RNAi) and pbx(RNAi) animals at this time point after RNAi, indicating that expression of patterning genes was intact (Fig. 5C-E). Both control and prep(RNAi) animals regenerated removed eyes within 8 days, whereas pbx(RNAi) animals had no detectable eye regeneration (Fig. 5B). Finally, we also investigated the requirement for pbx in eye progenitor formation 48 hours after amputation. Eye progenitors initially appear and are dispersed at the wound site/early blastema at this time (Lapan and Reddien, 2011). The number of these initial eye progenitors was severely reduced in pbx(RNAi) animals (supplementary material Fig. S7). These results suggest that pbx has a requirement in eye regeneration that is not caused by the absence of poles during regeneration.
pbx is broadly expressed, including in neoblasts
pbx displays broad, diffuse expression throughout the entire planarian body and pharynx, including prominent expression in the central nervous system (CNS) (Fig. 6A). Four days after irradiation, pbx+ signal was reduced, with the exception of expression in the CNS and pharynx (Fig. 6A). Because neoblasts are specifically eliminated by irradiation, this result suggests that some pbx expression might occur in neoblasts. Neoblasts can be isolated using fluorescence-activated cell sorting (FACS) based upon their >2N DNA content (Hayashi et al., 2006). We isolated neoblasts and found these cells to display pbx expression (Fig. 6B). pbx expression was strong at anterior and posterior wounds 24-48 hours after amputation, correlating with the time pbx is required for patterning gene expression (Fig. 6C; supplementary material Fig. S8). pbx expression near wounds during regeneration was also irradiation sensitive (Fig. 6C). Cells expressing both pbx and the patterning gene sFRP-1 were detected at 48 hours after amputation. However, the diffuse nature of the pbx signal made it difficult to resolve expression at the cellular level, and pbx expression appeared to be present in most cells of the blastema (Fig. 6D). These results suggest that pbx is expressed in neoblasts that give rise to many newly forming blastema cells where it can regulate expression of pole markers and formation of eye progenitors.
pbx inhibition causes gradual loss of patterning gene expression and eyes during tissue turnover
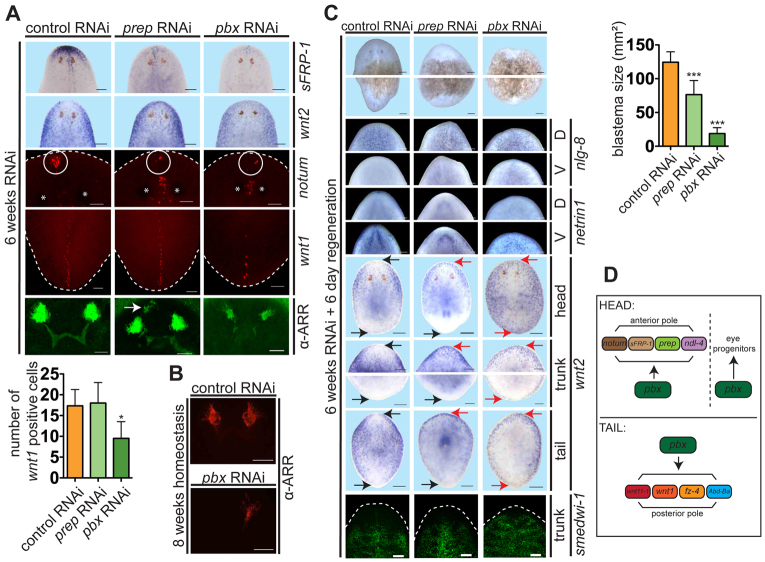
Does pbx have a regeneration-specific role in pole formation? We investigated this question by inhibiting pbx during homeostatic tissue turnover. Unamputated pbx(RNAi) animals displayed uncoordinated movement (by 20 days of RNAi), whereas neither the control nor prep(RNAi) animals displayed a mobility defect (supplementary material Movies 1-3). By more than 6 weeks of RNAi, both prep and pbx RNAi animals displayed decreased anterior sFRP-1 and notum expression, and increased anterior wnt2 expression, suggesting that pbx and prep are both required to maintain normal patterning gene expression at the anterior pole (Fig. 7A). By contrast, only pbx(RNAi) animals exhibited reduced wnt-1+ cell numbers, suggesting that pbx functions independently of prep in maintenance of posterior patterning gene expression (Fig. 7A). Additionally, photoreceptor neuron cluster size was reduced by pbx but not prep RNAi (Fig. 7A; supplementary material Fig. S9). By 8 weeks of pbx inhibition, most animals had completely lost one or both eyes (Fig. 7B). We conclude that the roles of pbx in regeneration and tissue turnover are similar for the regulation of gene expression at poles and for eye formation.
Because pole marker expression decreased in pbx(RNAi) animals during tissue turnover, we tested whether these animals could regenerate. This experiment differs from prior RNAi experiments because amputation occurred after pole marker expression had decreased. Animals amputated after 6 weeks of pbx RNAi produced very small blastemas with 100% penetrance (Fig. 7C). These blastemas displayed normal DV polarized nlg-8 (dorsally) and netrin-1 (ventrally) (Cebrià and Newmark, 2005) expression, and generated differentiated cells (Fig. 7C; supplementary material Fig. S10). wnt2 was expressed at the anterior pole of pbx(RNAi) tail fragments, similar to results described above, but was also expressed around the entire animal periphery. Similar wnt2 levels were present from the anterior to the posterior pole around the periphery of these pbx(RNAi) tail fragments (Fig. 7C). In pbx(RNAi) head fragments, wnt2 expression failed to scale away from the posterior pole (Fig. 7C). Control and prep(RNAi) trunks and tails had some, but weaker, wnt2 expression in the posterior periphery. These data indicate that significant blastema formation failed to occur in long-term pbx RNAi animals and that these animals display defects in the AP-polarized character of the primary body axis. Very small blastemas could in principle be caused by defects with neoblasts manifesting after long-term pbx RNAi. However, approximately normal distribution of and numbers of smedwi-1+ neoblasts and agat-1+ neoblast progeny were present in these RNAi animals. Whereas the reason for the severe blastema growth defect in these long-term pbx(RNAi) animals is unknown, these data suggest that regeneration failure is not caused by loss of the neoblast population or of the general capacity of neoblasts to differentiate (Fig. 7C; supplementary material Fig. S10).
DISCUSSION
The recent application of cellular and molecular methods to the study of planarian regeneration has allowed identification of genes controlling multiple aspects of regeneration and tissue turnover. Many conserved genes and signaling pathways with central roles in metazoan development have crucial roles during planarian regeneration, such as involvement in stem cell regulation, organ regeneration and body patterning (Sánchez Alvarado, 2007; Forsthoefel and Newmark, 2009; Adell et al., 2010; Shibata et al., 2010; Aboobaker, 2011; Reddien, 2011). The Smed-pbx RNAi phenotype described here identifies pbx as a new player in multiple steps of planarian regeneration and homeostatic tissue turnover.
pbx and pole regeneration
Planarian regeneration involves molecular mechanisms that direct restoration of tissue identity (Reddien, 2011). Regeneration polarity (the choice to regenerate a head or tail at transverse amputation planes) is initiated by wound-induced wnt1 expression (Petersen and Reddien, 2009b). Feedback inhibition of Wnt signaling by notum at anterior-facing wounds controls the regeneration polarity switch (Petersen and Reddien, 2011). Once the polarity decision is made at wounds, neoblast-dependent blastema formation is necessary for expression of additional patterning genes (e.g. other Wnt genes and Wnt inhibitory genes) at anterior and posterior poles (Gurley et al., 2010). Despite emerging data regarding signaling molecule expression for polarity initiation, little is understood about the molecules that control expression of pole-specific gene programs.
Our data indicate that pbx acts in regeneration at a step after initial expression of wnt1 and notum to control pole-specific patterning gene expression (Fig. 7D). The anterior pole defect in pbx(RNAi) animals is similar to that in prep(RNAi) animals (Felix and Aboobaker, 2010). pbx is required for prep expression in regeneration, suggesting that pbx functions a step prior to prep expression in promoting anterior pole gene expression. The posterior pole defect is similar to that of Djislet(RNAi) animals (Hayashi et al., 2011). However, pbx is unique in being required for regeneration of both poles. pbx is also required for expression during regeneration of bmp4 medially on the dorsal side and admp medially on the ventral side, indicating that the patterning role for pbx is not restricted to anterior and posterior extremities.
Several observations suggest that the requirement for pbx in regulating gene expression at poles reflects a specific role for pbx in pole development rather than being explained as a non-specific consequence of a defect in blastema growth. First, smaller blastemas than those present in pbx(RNAi) animals can express pole markers. Second, the effects of pbx RNAi on pole expression are robust; little to no expression is observed for some genes, such as sFRP-1, in pbx(RNAi) blastemas that are only slightly small. Third, pbx(RNAi) animals show defects in pole gene expression by 48 hours after amputation and substantial blastema growth and differentiation occurs subsequently, indicating that growth is not arrested before the pole should have formed. Finally, pbx is required for homeostatic maintenance of pole gene expression rather than this defect being observed only in blastemas.
Tissue polarization still occurred in pbx(RNAi) animals, despite decreased pole marker expression. For example, although blastemas were abnormal following pbx RNAi, cephalic ganglia still formed at anterior-facing wounds and DV polarized gene expression (nlg-8 and netrin-1) remained restricted in the normal manner. However, markers that are normally strongest in the pre-pharyngeal region or the posterior base of the head (wnt2 and ndl-3) extended to the anterior head tip of pbx(RNAi) animals. These observations indicate that reduced pole marker expression in pbx RNAi did not ablate regeneration polarity, but can be associated with axial patterning abnormalities.
Normally posterior features of heads were also present at the anterior end of pbx(RNAi) head blastemas. These features include expression domains for wnt2 and ndl-3, a decreased gap between neoblasts (smedwi-1+) and the head tip, and medial/anterior compression of brain (chat+) and cintillo+ domains. One possible interpretation for these defects is that pbx specifically promotes anterior pole formation, with pole absence resulting in failed restriction of the posterior head blastema domain. A similar, but alternative explanation is that pbx RNAi causes a Hox-like phenotype involving replacement of anterior regions with more posterior ones. However, pbx RNAi also resulted in failed posterior pole regeneration. If pbx RNAi caused expansion of posterior tail blastema regions at the expense of normally anterior tail blastema regions, tails with expanded, rather than absent, poles would be predicted. Therefore, posterior pole absence in pbx(RNAi) animals suggests that pbx has a specific role in pole formation and not simply a role in preventing posterior regions from expanding anteriorly. The pole-absence phenotype described in this article and the molecular mechanisms involved in pole formation will be an important area of continued investigation for understanding regeneration.
Multiple roles for pbx in regeneration
pbx is required for normal animal locomotion and eye regeneration. Although the cellular basis for the locomotion defect remains to be identified, we determined that pbx is required for formation of eye progenitor cells. Planarian eyes are an attractive system for studying eye biology and regeneration, because regenerative eye precursor cells have recently been identified (Lapan and Reddien, 2011). These precursors originate in the neoblasts, migrate into the head blastema, and coalesce to form the regenerating eye. pbx was required for otxA+ eya+ eye progenitor formation during regeneration, and ultimately for formation of photoreceptor neurons and pigmented optic cups, indicating that pbx is a new regulator of planarian eye formation (Fig. 7D). Involvement of pbx genes in eye development has been reported in mouse, zebrafish and Drosophila, although the role of pbx in these cases can be different and its mechanism of action remains to be further elucidated. Conditional Pbx1 knockout in mouse corneal epithelial cells causes corneal clouding, probably because of failure in proper corneal tissue turnover (Murphy et al., 2010). In zebrafish, pbx2/4 null embryos have a disorganized photoreceptor layer and retinal ganglion cells that fail to innervate the optic tectum (French et al., 2007). By contrast, exd suppresses eye formation in Drosophila (Pai et al., 1998). The requirement of pbx in eye development in planarians, a member of the Lophotrochozoan superphylum, raises the possibility of a broad use of pbx genes in animal eye biology. Further comparison of pbx roles in animal eye formation will be important for testing this possibility.
The multiple aspects of the pbx(RNAi) phenotype appear to be explained by separable roles for pbx in regeneration. For example, pbx(RNAi) animals display eye replacement defects following specific eye removal in animals with pole gene expression still present. Therefore, pole absence does not appear to explain the requirement for pbx in eye regeneration. Similarly, the eye and locomotion defects observed in pbx(RNAi) animals were not observed in prep(RNAi) animals, suggesting that these defects are not explained by the pole and brain abnormalities seen under both RNAi conditions. Furthermore, pbx is required for the earliest stage of eye progenitor formation, when these progenitors are broad at the wound site (48 hours after amputation). Finally, during tissue turnover in unamputated animals, pbx and prep RNAi animals lose anterior pole gene expression, but only pbx(RNAi) animals lose eyes. Mutants in Pbx genes in other organisms, including in mouse, zebrafish, Drosophila and C. elegans, also exhibit a pleiotropic phenotype, consistent with the multiple patterning and cell type specification roles for pbx in planarian regeneration (Kurant et al., 1998; Pöpperl et al., 2000). Expression data suggest that pbx is expressed in neoblasts and their progeny in blastemas to regulate patterning and eye regeneration (Fig. 7D). A detailed molecular investigation of the role of pbx in eye regeneration will be an important future direction.
PBX proteins can interact with other transcription factors, such as Hox, non-Hox homeodomain-containing proteins, or basic helix-loop-helix (bHLH) proteins, for controlling distinct developmental processes (Moens and Selleri, 2006). Therefore, it is possible that SMED-PBX functions with different partner proteins in the control of gene expression important for pole regeneration, eye regeneration and locomotion. Some of the genes for which expression requires pbx in planarians, including Wnt and Hox genes, are targets of pbx gene action in vertebrate limb development (Capellini et al., 2011). Further analysis of the targets and partner proteins of SMED-PBX during planarian regeneration will be important for understanding its conserved and divergent roles in patterning and cell type specification.
In conclusion, we report that Smed-pbx is an important component of multiple steps of planarian regeneration and homeostatic tissue turnover. Formation of new poles of body axes is an essential step in regeneration, and no gene was previously known to be required for this process at both AP poles. pbx is therefore an attractive target for molecular dissection of the mechanisms by which re-establishment of tissue pattern occurs in regeneration.
Supplementary Material
Acknowledgments
We thank members of the Reddien lab for discussions; Josien van Wolfswinkel, Lucila Scimone, Mansi Srivastava, Jessica Witchley and Mirjam Mayer for manuscript comments; Jessica Witchley, Sylvain Lapan, Mike Gavino, Dan Wagner, Lucila Scimone and Jared Owen for providing RNA probe templates; and Mansi Srivastava for phylogenetics assistance.
Footnotes
Funding
P.W.R. is an HHMI Early Career Scientist. We acknowledge support from the National Institutes of Health (NIH) [R01GM080639] and the Keck Foundation. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.083741/-/DC1
References
- Aboobaker A. A. (2011). Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304-311 [DOI] [PubMed] [Google Scholar]
- Adell T., Salò E., Boutros M., Bartscherer K. (2009). Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905-910 [DOI] [PubMed] [Google Scholar]
- Adell T., Cebrià F., Saló E. (2010). Gradients in planarian regeneration and homeostasis. Cold Spring Harb. Perspect. Biol. 2, a000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almuedo-Castillo M., Sureda-Gomez M., Adell T. (2012). Wnt signaling in planarians: new answers to old questions. Dev. Biol. 56, 53-65 [DOI] [PubMed] [Google Scholar]
- Arata Y., Kouike H., Zhang Y., Herman M. A., Okano H., Sawa H. (2006). Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev. Cell 11, 105-115 [DOI] [PubMed] [Google Scholar]
- Bardeen C. R., Baetjer F. H. (1904). The inhibitive action of the Roentgen rays on regeneration in planarians. J. Exp. Zool. 1, 191-195 [Google Scholar]
- Bürglin T. R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173-4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin T. R., Ruvkun G. (1992). New motif in PBX genes. Nat. Genet. 1, 319-320 [DOI] [PubMed] [Google Scholar]
- Capellini T. D., Zappavigna V., Selleri L. (2011). Pbx homeodomain proteins: TALEnted regulators of limb patterning and outgrowth. Dev. Dyn. 240, 1063-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F., Newmark P. A. (2005). Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132, 3691-3703 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Vispo M. (1997). Myocyte differentiation and body wall muscle regeneration in the planarian Girardia tigrina. Dev. Genes Evol. 207, 306-316 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Kobayashi C., Umesono Y., Nakazawa M., Mineta K., Ikeo K., Gojobori T., Itoh M., Taira M., Sánchez Alvarado A., et al. (2002). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620-624 [DOI] [PubMed] [Google Scholar]
- Dubois F. (1949). ‘Contribution á l 'ètude de la migration des cellules de règènèration chez les Planaires dulcicoles’. Bull. Biol. Fr. Belg. 83, 213-283 [Google Scholar]
- Felix D. A., Aboobaker A. A. (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 6, e1000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E., Li B., Zewdu R., Wells V., Hebert J. M., Karner C., Anderson M. J., Williams T., Dixon J., Dixon M. J., et al. (2011). A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell 21, 627-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel D. J., Newmark P. A. (2009). Emerging patterns in planarian regeneration. Curr. Opin. Genet. Dev. 19, 412-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French C. R., Erickson T., Callander D., Berry K. M., Koss R., Hagey D. W., Stout J., Wuennenberg-Stapleton K., Ngai J., Moens C. B., et al. (2007). Pbx homeodomain proteins pattern both the zebrafish retina and tectum. BMC Dev. Biol. 7, 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviño M. A., Reddien P. W. (2011). A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr. Biol. 21, 294-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz O., Basler K. (2002). Wingful, an extracellular feedback inhibitor of Wingless. Genes Dev. 16, 1055-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giráldez A. J., Copley R. R., Cohen S. M. (2002). HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev. Cell 2, 667-676 [DOI] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C., Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Elliott S. A., Simakov O., Schmidt H. A., Holstein T. W., Sánchez Alvarado A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N., Agata K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371-380 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Motoishi M., Yazawa S., Itomi K., Tanegashima C., Nishimura O., Agata K., Tarui H. (2011). A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679-3688 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Saló E., Adell T. (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135, 1215-1221 [DOI] [PubMed] [Google Scholar]
- Kamps M. P., Murre C., Sun X. H., Baltimore D. (1990). A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell 60, 547-555 [DOI] [PubMed] [Google Scholar]
- Kato K., Orii H., Watanabe K., Agata K. (1999). The role of dorsoventral interaction in the onset of planarian regeneration. Development 126, 1031-1040 [DOI] [PubMed] [Google Scholar]
- Kurant E., Pai C. Y., Sharf R., Halachmi N., Sun Y. H., Salzberg A. (1998). Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125, 1037-1048 [DOI] [PubMed] [Google Scholar]
- Lapan S. W., Reddien P. W. (2011). dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genet. 7, e1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent A., Bihan R., Omilli F., Deschamps S., Pellerin I. (2008). PBX proteins: much more than Hox cofactors. Int. J. Dev. Biol. 52, 9-20 [DOI] [PubMed] [Google Scholar]
- Leyns L., Bouwmeester T., Kim S. H., Piccolo S., De Robertis E. M. (1997). Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 88, 747-756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Fire A. (2000). Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development 127, 5179-5190 [DOI] [PubMed] [Google Scholar]
- Moens C. B., Selleri L. (2006). Hox cofactors in vertebrate development. Dev. Biol. 291, 193-206 [DOI] [PubMed] [Google Scholar]
- Molina M. D., Saló E., Cebrià F. (2007). The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev. Biol. 311, 79-94 [DOI] [PubMed] [Google Scholar]
- Molina M. D., Saló E., Cebrià F. (2009). Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr. Patterns 9, 246-253 [DOI] [PubMed] [Google Scholar]
- Molina M. D., Neto A., Maeso I., Gómez-Skarmeta J. L., Saló E., Cebrià F. (2011). Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 21, 300-305 [DOI] [PubMed] [Google Scholar]
- Monica K., Galili N., Nourse J., Saltman D., Cleary M. L. (1991). PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol. Cell. Biol. 11, 6149-6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H. (1898). Experimental studies of the regeneration of Planaria maculata. Arch. Entw. Mech. Org. 7, 364-397 [Google Scholar]
- Morgan T. H. (1905). “Polarity” considered as a phenomenon of gradation of materials. J. Exp. Zool. 2, 495-506 [Google Scholar]
- Mukherjee K., Bürglin T. R. (2007). Comprehensive analysis of animal TALE homeobox genes: new conserved motifs and cases of accelerated evolution. J. Mol. Evol. 65, 137-153 [DOI] [PubMed] [Google Scholar]
- Murphy M. J., Polok B. K., Schorderet D. F., Cleary M. L. (2010). Essential role for Pbx1 in corneal morphogenesis. Invest. Ophthalmol. Vis. Sci. 51, 795-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P. A., Sánchez Alvarado A. (2000). Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142-153 [DOI] [PubMed] [Google Scholar]
- Nogi T., Watanabe K. (2001). Position-specific and non-colinear expression of the planarian posterior (Abdominal-B-like) gene. Dev. Growth Differ. 43, 177-184 [DOI] [PubMed] [Google Scholar]
- Nourse J., Mellentin J. D., Galili N., Wilkinson J., Stanbridge E., Smith S. D., Cleary M. L. (1990). Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 60, 535-545 [DOI] [PubMed] [Google Scholar]
- Orii H., Watanabe K. (2007). Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev. Growth Differ. 49, 345-349 [DOI] [PubMed] [Google Scholar]
- Orii H., Kato K., Kiyokazu A., Watanabe K. (1998). Molecular cloning of bone morphogenetic protein (BMP) gene from the planarian Dugesia japonica. Zoolog. Sci. 15, 871-877 [Google Scholar]
- Orii H., Kato K., Umesono Y., Sakurai T., Agata K., Watanabe K. (1999). The planarian HOM/HOX homeobox genes (Plox) expressed along the anteroposterior axis. Dev. Biol. 210, 456-468 [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Newmark P. A., Sánchez Alvarado A. (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 226, 326-333 [DOI] [PubMed] [Google Scholar]
- Pai C. Y., Kuo T. S., Jaw T. J., Kurant E., Chen C. T., Bessarab D. A., Salzberg A., Sun Y. H. (1998). The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12, 435-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E., Sánchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Wieschaus E. (1990). Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 4, 1209-1223 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327-330 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2009a). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056-1068 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2009b). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. USA 106, 17061-17066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöpperl H., Rikhof H., Chang H., Haffter P., Kimmel C. B., Moens C. B. (2000). lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol. Cell 6, 255-267 [DOI] [PubMed] [Google Scholar]
- Rauskolb C., Wieschaus E. (1994). Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 13, 3561-3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C., Peifer M., Wieschaus E. (1993). extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell 74, 1101-1112 [DOI] [PubMed] [Google Scholar]
- Rauskolb C., Smith K. M., Peifer M., Wieschaus E. (1995). extradenticle determines segmental identities throughout Drosophila development. Development 121, 3663-3673 [DOI] [PubMed] [Google Scholar]
- Reddien P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005a). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005b). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Kicza A. M., Sánchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043-4051 [DOI] [PubMed] [Google Scholar]
- Rink J. C., Gurley K. A., Elliott S. A., Sánchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406-1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R., Fibla J., Bueno D., Sumoy L., Soriano M. A., Baguñà J. (1991). Monoclonal antibodies as markers of specific cell types and regional antigens in the freshwater planarian Dugesia (G) tigrina. Hydrobiologia 227, 73-79 [Google Scholar]
- Sánchez Alvarado A. (2007). Stem cells and the Planarian Schmidtea mediterranea. C. R. Biol. 330, 498-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M., Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665 [DOI] [PubMed] [Google Scholar]
- Scimone M. L., Srivastava M., Bell G. W., Reddien P. W. (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387-4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemer G., Podbilewicz B. (2002). LIN-39/Hox triggers cell division and represses EFF-1/fusogen-dependent vulval cell fusion. Genes Dev. 16, 3136-3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Rouhana L., Agata K. (2010). Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev. Growth Differ. 52, 27-41 [DOI] [PubMed] [Google Scholar]
- Takács-Vellai K., Vellai T., Chen E. B., Zhang Y., Guerry F., Stern M. J., Müller F. (2007). Transcriptional control of Notch signaling by a HOX and a PBX/EXD protein during vulval development in C. elegans. Dev. Biol. 302, 661-669 [DOI] [PubMed] [Google Scholar]
- Van Auken K., Weaver D., Robertson B., Sundaram M., Saldi T., Edgar L., Elling U., Lee M., Boese Q., Wood W. B. (2002). Roles of the Homothorax/Meis/Prep homolog UNC-62 and the Exd/Pbx homologs CEH-20 and CEH-40 in C. elegans embryogenesis. Development 129, 5255-5268 [DOI] [PubMed] [Google Scholar]
- Vlachakis N., Ellstrom D. R., Sagerström C. G. (2000). A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev. Dyn. 217, 109-119 [DOI] [PubMed] [Google Scholar]
- Wagner K., Mincheva A., Korn B., Lichter P., Pöpperl H. (2001). Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech. Dev. 103, 127-131 [DOI] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E., Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A., Moens C. B. (2002). Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 3, 723-733 [DOI] [PubMed] [Google Scholar]
- Wenemoser D., Reddien P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas R. M., Cebrià F., Guo T., Feng J., Newmark P. A. (2010). The use of lectins as markers for differentiated secretory cells in planarians. Dev. Dyn. 239, 2888-2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.