Abstract
Environmental factors can stably perturb the epigenome of exposed individuals and even that of their offspring, but the pleiotropic effects of these factors have posed a challenge for understanding the determinants of mitotic or transgenerational inheritance of the epigenetic perturbation. To tackle this problem, we manipulated the epigenetic states of various target genes using a tetracycline-dependent transcription factor. Remarkably, transient manipulation at appropriate times during embryogenesis led to aberrant epigenetic modifications in the ensuing adults regardless of the modification patterns, target gene sequences or locations, and despite lineage-specific epigenetic programming that could reverse the epigenetic perturbation, thus revealing extraordinary malleability of the fetal epigenome, which has implications for ‘metastable epialleles’. However, strong transgenerational inheritance of these perturbations was observed only at transgenes integrated at the Col1a1 locus, where both activating and repressive chromatin modifications were heritable for multiple generations; such a locus is unprecedented. Thus, in our inducible animal models, mitotic inheritance of epigenetic perturbation seems critically dependent on the timing of the perturbation, whereas transgenerational inheritance additionally depends on the location of the perturbation. In contrast, other parameters examined, particularly the chromatin modification pattern and DNA sequence, appear irrelevant.
Keywords: Epigenetic inheritance, Mouse, Transgenerational, Intergenerational, Tetracycline, Inducible, Timing, Location
INTRODUCTION
Acute exposure to environmental factors such as stress, diet and toxicants can produce long-lasting biological effects on the affected individuals and even their offspring, which has tremendous clinical and evolutionary implications (Jirtle and Skinner, 2007; Rando and Verstrepen, 2007; Jablonka and Raz, 2009; Faulk and Dolinoy, 2011; Skinner, 2011; Daxinger and Whitelaw, 2012). Some of these effects are presumably mediated by aberrant epigenetic modifications (termed ‘epigenetic lesions’ hereafter) at target genes (Youngson and Whitelaw, 2008). Specifically, it is thought that although epigenetic lesions induced by environmental signals are often reversible, they can also be self-perpetuating mitotically and even meiotically, if the aberrant epigenetic modifications, once induced, can somehow serve as templates (or ‘cis epigenetic signals’) to attract chromatin-modifying enzymes for self-replication during cell division (Bonasio et al., 2010).
The stability and propagation of epigenetic lesions might be influenced by parameters such as the timing of lesion induction, lesion structure, target gene sequence and location, and developmental epigenetic reprogramming that may erase the lesion. These parameters have been difficult to dissect, in part because natural environmental factors are pleiotropic, affecting many genes and causing secondary confounding effects. Thus, if a gene is found, for example, to be heritably methylated following an acute exposure, it would be unclear whether the inheritance is due to self-perpetuation of the lesion (a cis-acting mechanism) or a consequence of the irreversible induction of a DNA methyltransferase (a trans-acting mechanism) (Bonasio et al., 2010). Further complicating the issue, environmental factors can mutate DNA (Guerrero-Bosagna et al., 2010).
The problems outlined above can be bypassed by directing epigenetic lesions to specific genes via recruitment of transcriptional regulators. If the target genes do not encode regulatory proteins controlling the inheritance of the epigenetic lesions, then the inheritance is likely to be mediated by cis-acting mechanisms. By necessity, the induction of epigenetic lesions in this setting is artificial, but the propagation of the lesions must depend on, and thus reflect, the physiological cis-acting mechanisms that mediate self-perpetuation of epigenetic states. This approach is the basis of the elegant ‘chromatin in vivo assay’ for assessing the dynamics and memory of heterochromatin in cell lines (Hathaway et al., 2012).
Here, we used reverse tetracycline-regulated transcription activator (rtTA) (Gossen et al., 1995; Schönig et al., 2010) to selectively alter the epigenetic states of its target genes in mice, and followed its consequences for generations. As the target genes had no apparent regulatory function, the epigenetic lesions were presumably propagated via cis-acting mechanisms. Using this system, we dissected various parameters that potentially influence epigenetic inheritance. The data reveal the surprising malleability of the fetal epigenome and identify a novel type of locus supporting transgenerational inheritance.
MATERIALS AND METHODS
Mice
CMV-GFP (C57B/6x129/sv) and CMV-CD4hTLR4 (C57/B6) mice have been described (Beard et al., 2006; Qureshi et al., 2006). The Cd4 minigene mice (C57B/6x129/sv) were created using the flip-in system (Beard et al., 2006).
Lymphocyte culture
Tail blood (10 μl) was collected using a pipet into 10 μl RPMI medium (Invitrogen) containing 50 ng/μl heparin. The cells were then washed with 200 μl PBS to remove serum, resuspended in 200 μl RPMI containing Dox (1 μg/ml; Sigma) and incubated at 37°C for the indicated times before red blood cell lysis and gene expression analysis by FACS or RT-PCR.
Fluorescence-activated cell sorting (FACS)
Cells were stained with anti-CD4 APC (Sungene, Tianjin, China), anti-CD8 PE-Cy7 and anti-B220 PE (BD Pharmingen) to resolve the lymphocytes into CD4, CD8 and B cells before the analysis of GFP expression in each cell type.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described (Yu et al., 2008; Lai et al., 2009) except that Dynal protein G magnetic beads (Invitrogen) instead of Sepharose beads were used to capture chromatin, which produced negligible nonspecific signals, and the DNA was quantified by real-time, multiplex PCR. The abundance of targets in the ChIP DNA, normalized to that in the input, was plotted relative to the abundance of similarly normalized control regions as fold enrichment over the control.
Embryonic stem (ES) cell culture
ES cells (C10-1) carrying the tetO-CMV promoter transgene integrated into the Col1a1 locus and a transgene ubiquitously expressing rtTA from the Rosa26 locus were obtained from the R. Jaenisch laboratory (MIT, Cambridge, MA, USA). ES cells were cultured and passaged every 2-3 days on mitomycin-treated mouse embryonic fibroblasts. Before analysis for GFP expression or chromatin alteration, ES cells in the culture were dissociated by trypsinization and contaminating feeder fibroblasts were depleted by gravity sedimentation.
DNA methylation of the CMV promoter
Genomic DNA (1 μg) isolated from lymph node cells and sperm was bisulfite-converted using the Epitech Bisulfite Kit (Qiagen). The resulting DNA was amplified using the PyroMark PCR Kit (Qiagen) with a primer pair targeting an 89 bp fragment spanning the transcription start site of the CMV promoter. An internal primer was then used to sequence the amplicon in the PyroMark Q24 system (Qiagen).
Sperm isolation
Sperm was isolated as described (Brykczynska et al., 2010), except that mice were first perfused with 30 ml PBS before dissecting the epididymis in order to eliminate contaminating blood cells.
RESULTS
A quantitative in vitro assay for GFP induction from the CMV-GFP transgene
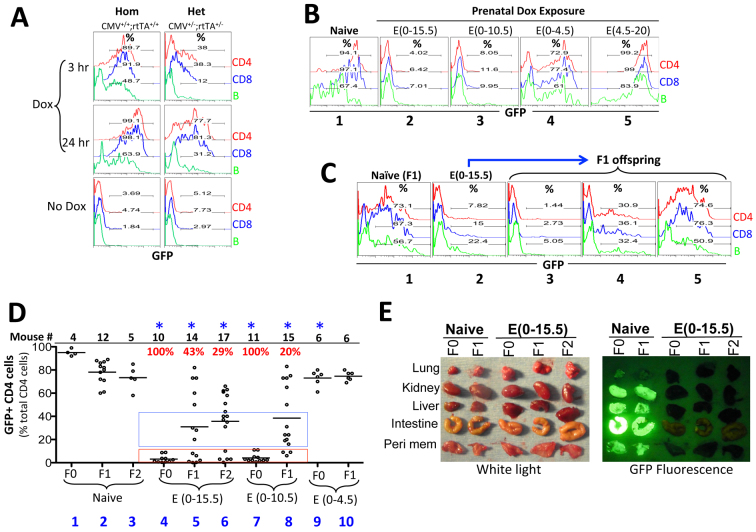
We analyzed CMV-GFP mice that ubiquitously express rtTA from the Rosa26 locus and also carry a doxycycline (Dox)-responsive GFP reporter transgene under the control of the human CMV minimal promoter inserted as a single-copy gene into the Col1a1 locus; Dox is known to induce widespread GFP expression in the adult mice (Beard et al., 2006). We first developed an in vitro assay to accurately quantify GFP induction using peripheral blood lymphocytes collected from adult CMV-GFP mice (see Materials and methods). Dox induced robust GFP expression in lymphocytes from homozygous (CMV+/+;rtTA+/+) mice in a time-dependent manner, especially in CD4 and CD8 cells, the majority of which expressed GFP within 3 hours of Dox stimulation (Fig. 1A, left). As expected, induction was less robust in lymphocytes from heterozygous CMV-GFP mice (CMV+/–;rtTA+/–; Fig. 1A, right). We then examined whether fetal Dox exposure could facilitate GFP induction in adults.
Fig. 1.
Phenotypic analysis of prenatally exposed CMV-GFP transgenic mice. (A) An in vitro assay for GFP induction. Tail blood cells, collected from mice homozygous (Hom) or heterozygous (Het) for the CMV promoter and rtTA transgenes, were cultured in the presence of Dox (1 μg/ml) for 3 (top) or 24 (middle) hours. GFP expression in CD4 (red), CD8 (blue) and B (green) cells was then analyzed by FACS. As a control, cells were cultured for 24 hours without Dox (bottom panel). All mice were on a C57/B6x129sv background. (B) Effects of fetal Dox exposures on GFP induction in the ensuing adults. Homozygous males and females were mated, and plugged females were given Dox (1 mg/ml in drinking water) for the indicated times. For Dox exposure starting at E0, Dox water was given 1-2 days prior to mating. Blood was collected from the adult offspring and analyzed as in A. Naïve homozygous mice lacking prenatal Dox exposure were used as a control. All mice were on a C57/B6x129sv background. (C) Transgenerational inheritance of GFP repression via the female germline. Shown are the GFP induction patterns of a homozygous female (on a C57/B6x129sv background) prenatally exposed to Dox for 15 days (plot 2), her F1 offspring fathered by a CD1 male (plots 3-5), and a control F1 mouse produced by a naïve homozygous female (on a C57/B6x129sv background) mated with the same CD1 father (plot 1). (D) Summary of FACS data pooled from multiple experiments. F0 mice (on a C57/B6x129sv background) were homozygous males and females that were either naïve or pre-exposed to Dox. F1 mice were generated by mating F0 females with CD1 males, and F2 mice were derived by mating CD1 males with the F1 females with severe GFP repression. F0 mice were homozygous whereas F1 and F2 were heterozygous, which explained the strongest GFP induction in the F0 naïve mice. The plot displays the percentages of the CD4 cells expressing GFP following 24 hours of Dox stimulation; CD8 and B cells displayed a similar trend (not shown). The dots represent individual mice. The red and blue boxes highlight the mice with full and partial GFP repression, respectively. The sample sizes in each group and the percentage of mice with the severe phenotype are displayed at the top of the graph using black and red fonts, respectively. Asterisks indicate statistical significance (P=0.0001 to 0.003) when comparing all the mice in a particular Dox-exposed group with the naïve mice of the corresponding generation. Multiple independent litters were analyzed to avoid sampling errors. (E) GFP induction in various organs from adult mice following 3 weeks of Dox administration (2 mg/ml) via drinking water. The mice are as described in D. For the F1 and F2 mice from prenatally exposed mothers, only those with severe GFP repression are shown. Peri mem, peritoneal membrane.
Fetal rtTA activation silences the CMV promoter in the ensuing adults and subsequent generations
We exposed the homozygous embryos to Dox and analyzed GFP induction in the ensuing adults. Surprisingly, Dox exposure throughout gestation, i.e. from E0 to E20 [termed E(0-20)], prevented GFP induction (supplementary material Fig. S1A), as did exposure at E(0-15.5) and E(0-10.5), whereas E(0-4.5) exposure only mildly (but reproducibly, P=0.003) impaired GFP induction (Fig. 1B, plots 1-4; 1D, groups 1, 4, 7, 9). However, E(0-4.5) exposure was necessary for silencing, as E(4.5-20) exposure failed to silence GFP in lymphocytes and instead somewhat enhanced GFP induction (Fig. 1B, plot 5). Thus, rtTA activation during the first 10 days of embryogenesis was necessary and sufficient to fully induce mitotically stable GFP silencing.
To determine whether the GFP silencing in these prenatally exposed homozygous mice was heritable transgenerationally, we mated these mice (on C57/B6x129sv) with the outbred CD1 strain. For simplicity, we refer to the mice with prenatal exposures as F0 mice because they were the ‘founders’ in our system. We found F0 males unable to transmit the phenotype (supplementary material Fig. S1B; see also Fig. 9). By contrast, GFP silencing in F0 females with E(0-15.5) exposure was fully heritable to 43% (6/14) of the F1 pups (the remaining pups showed partial or no repression; Fig. 1C, plots 3-5 and 1D, group 5). The variation in GFP expressivity among the littermates might be purely stochastic in nature, as observed in genetically identical strains (Morgan et al., 1999; Kearns et al., 2000; Sutherland et al., 2000; Rakyan et al., 2002), or might reflect the segregation of a genetic modifier given that the pups were on a mixed (B6/129/CD1) background; preliminary data (not shown) suggest the former scenario.
Fig. 9.
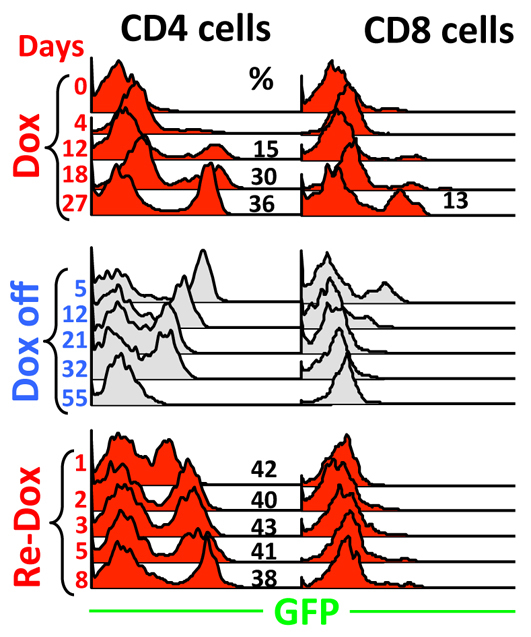
Effects of rtTA acting in adult naïve Cd4 minigene+/–; rtTA+/– mice. Mice were primed with Dox for 27 days (top), rested for 55 days (middle), and then re-challenged for 8 days (bottom). Blood was drawn at various times to monitor GFP expression in CD4 (left) and CD8 (right) cells.
Importantly, the F1 females with GFP silencing (but not those lacking GFP repression) could in turn produce some (29%) F2 offspring with a similar phenotype (Fig. 1D, group 6; data not shown). Therefore, mice with direct environmental exposure can transmit the environmental effect for at least two generations, thus satisfying the criterion for bona fide transgenerational inheritance (Skinner, 2008). GFP silencing was widespread (Fig. 1E). The GFP silencing in females with E(0-10.5) exposure was also heritable at least to the F1 offspring, albeit at reduced frequency (20%; Fig. 1D, group 8), whereas the effect of E(0-4.5) exposure was entirely non-heritable (Fig. 1D, compare group 10 with group 2). Finally, as expected, females with E(0-20) exposure could also transmit the phenotype to a fraction of pups (supplementary material Fig. S1A).
We conclude that fetal Dox exposures can produce long-lasting effects on the CMV promoter, with a brief (4.5 day) exposure minimally sufficient for inducing the silencing in the ensuing adults, whereas longer exposures (10-15 days) are necessary for transgenerational inheritance of this phenotype.
Epigenetic marks associated with GFP silencing: CpG methylation and loss of H3K4me2 at the CMV promoter
We first analyzed DNA methylation at the CMV promoter. In lymphocytes from naïve mice, the promoter showed minimal (<11.4%) methylation (Fig. 2A, groups 1-2). E(0-15.5) exposure dramatically increased the methylation to ∼90% (Fig. 2A, group 3), and this pattern was inherited by the F1 offspring with severe GFP repression (group 4, mice in the box) but not by the littermates with normal GFP induction (groups 4, the mouse outside the box). The methylation pattern seemed to fade somewhat in the F2 generation, where the levels were reduced to below 70% in three of the four F2 pups analyzed (Fig. 2A, group 5). E(0-10.5) exposure similarly caused CpG hypermethylation, but the level of methylation and its transgenerational inheritability appeared lower than those caused by E(0-15.5) exposure (Fig. 2A, groups 6 and 7 versus 3 and 4).
Fig. 2.
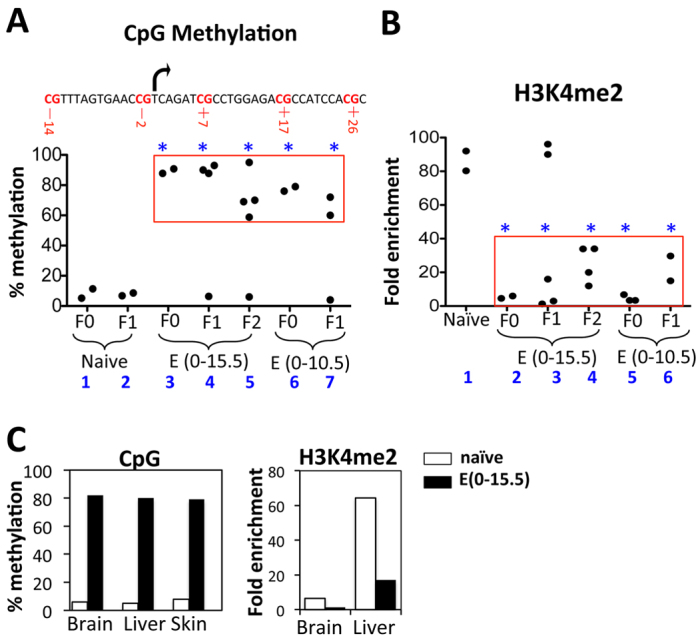
Epigenetic lesions at the CMV promoter in adult tissues. (A,B) Epigenetic lesions in lymphocytes. (A) Five CpGs at the promoter (top) were analyzed. Their methylation levels were similar (supplementary material Fig. S2) and averaged (bottom). The dots represent individual mice and the red box encloses mice with severe GFP repression, whereas the remaining mice in the plot showed no phenotype. Asterisks indicate significant differences between the mice in the red box and naïve mice (P=0.005 to 0.01). (B) H3K4me2 levels at the CMV promoter relative to an internal control at the Col1a1 locus ∼11 kb downstream. PCR was performed in triplicate and the values averaged. The mice and red box are as in A. The naïve mice were F0. Asterisks indicate significant differences between the mice in the red box and naïve mice (P=0.0004 to 0.02). (C) Epigenetic lesions in other tissues. The tissues were pooled from three naïve (white bars) and three prenatally exposed (black bars) adults before parallel analysis of CpG methylation (left) and H3K4me2 (right), except that ChIP analysis of skin was not performed for technical reasons. The experiment was performed once.
We next determined whether the phenotype is associated with any histone modifications. H3K9ac, H3K27me3 and H3K9me3 were hardly detectable at the CMV promoter in naïve mice, and remained so following fetal Dox exposure (not shown). By contrast, H3K4me2, a marker of poised promoters, was highly enriched (80-fold over an internal control) in naïve mice and severely depleted (to only ∼5-fold over the control) in mice with E(0-15.5) exposure (Fig. 2B, groups 1 and 2). The effect was fully heritable to the F1 offspring showing severe GFP repression (Fig. 2B, group 3), but seemed to fade in the F2 mice, where the H3K4me2 level was partially restored (to ∼35-fold over the control) in two of the four mice analyzed (Fig. 2B, group 4; also recall that CpGs were partially demethylated in three of them); interestingly, all four mice retained the ability to severely repress GFP, presumably because the epigenetic lesion, although partially repaired, remained sufficient to block GFP induction under our assay conditions. H3K4me2 was also severely depleted in mice with E(0-10.5) exposure, but was already partially restored in the F1 generation (Fig. 2B, groups 5 and 6).
Thus, fetal Dox exposures can cause CpG methylation and depletion of H3K4me2 at the CMV promoter in lymphocytes of the ensuing adults and their offspring. Similar lesions were detected in other tissues in the prenatally exposed adults (Fig. 2C), consistent with widespread GFP silencing (Fig. 1E). Of note, the lesions in different tissues seemed divergent, with H3K4me2 much higher in the liver than in the brain (Fig. 2C), which presumably reflects the effect of lineage-specific developmental reprogramming.
Dox induces a similar epigenetic lesion in ES cells
rtTA is a well-known activator in adult mice. How could rtTA, acting in early embryos, paradoxically cause repressive lesions in the adults? Perhaps early fetal cells are unique, enabling rtTA to silence the CMV promoter, which is then propagated to adults. We used ES cells to test this hypothesis; ES cells are derived from E3.5 embryos, around the period when rtTA action is minimally sufficient and absolutely essential for the silencing. In the ES cell line carrying the CMV-GFP transgene and expressing rtTA, from which line the CMV-GFP mice were derived, Dox robustly induced GFP within 2 days (Fig. 3A), as reported (Beard et al., 2006). Remarkably, longer stimulation led to progressive GFP repression (Fig. 3A), which was not an artifact of cell death (because the cells remained healthy; Fig. 3B) and occurred at the level of transcription (Fig. 3C) concomitant with CpG hypermethylation and H3K4me2 depletion (Fig. 3D,E). By contrast, prolonged Dox treatment (2 months) of adult CMV-GFP mice failed to silence GFP in any tissue examined (supplementary material Fig. S3). These data confirm the uniqueness of ES cells and suggest that Dox can induce a similar lesion in early embryos, which was then propagated to the adults, although some secondary modifications can apparently occur during the propagation as a result of lineage-specific reprogramming as discussed above.
Fig. 3.
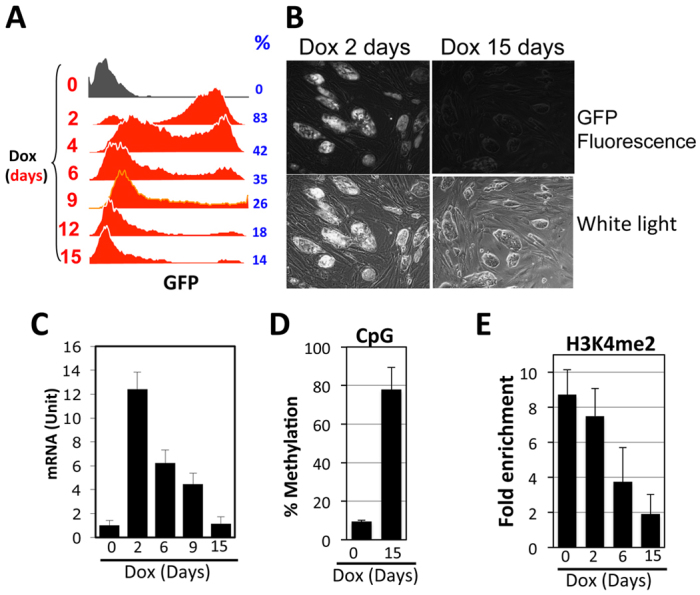
Effects of rtTA on the CMV-GFP transgene in ES cells. Cells were cultured in the presence of Dox (1 μg/ml) for various days before analysis. (A) FACS analysis of GFP expression. Dox stimulation times and percentage of GFP-expressing cells are shown at left and right, respectively. (B) Micrographs of ES cells following 2 (left) and 15 (right) days of Dox exposure. (C) GFP mRNA levels, normalized to β-actin mRNA. (D,E) CpG methylation and H3K4me2 at the CMV promoter, as described in Fig. 2. The values were averaged from two and three independent experiments for D and E, respectively. Error bars indicate variations between the independent trials.
Fetal Dox exposure leads to silencing of a randomly integrated CMV promoter in adult mice
To further explore the basis of the unusual behavior of the CMV-GFP transgene integrated at the Col1a1 locus, we analyzed another transgene bearing the rtTA-regulated CMV minimal promoter (Qureshi et al., 2006). This transgene (CMV-CD4hTLR4) is integrated into an unknown region, and the sequence outside the tetO-CMV minimal promoter is completely different from that of the CMV-GFP transgene (Fig. 4A).
Fig. 4.
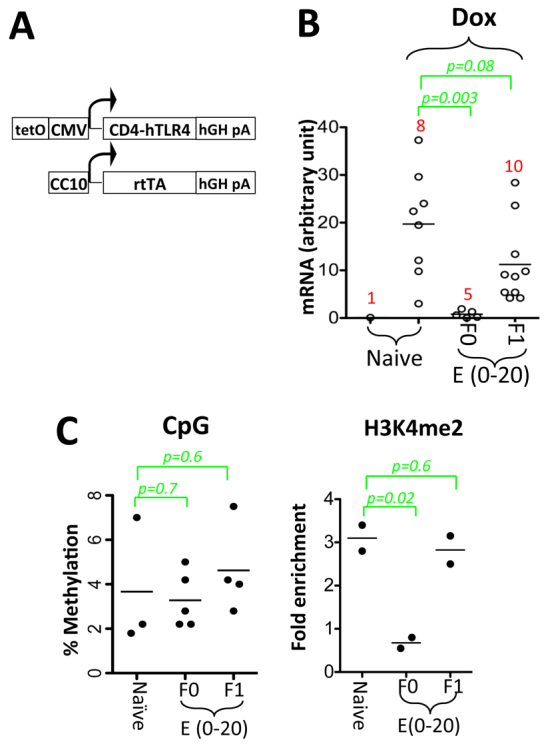
Fetal Dox exposure silences a randomly integrated CMV promoter in the ensuing adults, but the effect is non-heritable. (A) The CMV-CD4hTLR4 transgene consists of a CMV promoter transgene (top) and a co-integrated transgene (bottom), the former differing from the CMV-GFP transgene (Fig. 6A) in that it expresses a fusion protein consisting of the extracellular domain of mouse CD4 and the intracellular domain of human TLR4 (instead of GFP), carries a poly(A) signal from the human growth hormone gene [instead of rabbit β-globin poly(A)] and is integrated as a concatemer. The co-integrated transgene carries the CC10 (Scgb1a1) promoter and express rtTA specifically in the lung. This CMV-CD4hTLR4 transgene is therefore distinct from the CMV-GFP transgene in terms of integration site and sequence, except that it also carries the tetO-CMV minimal promoter. (B) Tail blood cells (CMV-CD4hTLR4+/–;Rosa26-rtTA+/–) were cultured in the presence or absence of Dox for 24 hours before CD4hTLR4 mRNA was quantified using β-actin as a loading control. Three prenatally exposed females were mated with CD1 males to produce the F1 offspring. (C) CpG methylation (left) and H3K4me2 (right) at the CMV promoter in lymphocytes. Experiments were performed as in Fig. 2 (note the differences in the y-axis scales and in the basal H3K4me2 levels with Fig. 2).
To determine the effect of rtTA on the CMV-CD4hTLR4 transgene, we introduced the Rosa26-rtTA allele (on C57/B6x129sv) into the CMV-CD4hTLR4 mice (on C57/B6). The resulting mice were therefore on a genetic background that is highly similar to that of the CMV-GFP mice. In the lymphocytes of these mice, Dox could induce CD4hTLR4 mRNA (increased from 0.1 to 20±10 units), which was prevented by prenatal Dox exposure, although the silencing was hardly heritable (Fig. 4B). Consistent with this, the lesion at the CMV promoter induced by fetal Dox exposure was structurally distinct from that at the CMV-GFP transgene, showing no CpG hypermethylation (Fig. 4C, left) and only a moderate (3.5-fold) decrease in H3K4me2 (as opposed to 16-fold in the CMV-GFP mice; compare Fig. 4C, right, with Fig. 2B).
Thus, rtTA induced mitotically heritable silencing at both the CMV-GFP and the CMV-CD4hTLR4 transgenes, even though the two transgenes differ in sequence, location and lesion structure, suggesting that these parameters are irrelevant to mitotic inheritance. However, the silencing was transgenerationally heritable at the CMV-GFP but not the CMV-CD4hTLR4 transgene. Which of these parameters is responsible for this discrepancy? The following experiment suggested that the Col1a1 locus, where the CMV-GFP transgene is integrated, is the decisive factor.
The Col1a1 locus also supports strong transgenerational inheritance of an activating epigenetic lesion: analysis of the Cd4 minigene
The Col1a1 locus is euchromatic, but supports the inheritance of GFP silencing at the CMV-GFP transgene. We suspect that the locus might be able to insulate the repressive lesion from reprogramming enzymes, thus allowing its transgenerational inheritance; this would predict that the locus can also preserve activating lesions, i.e. those that are marked with activating chromatin modifications and function to facilitate transgene expression.
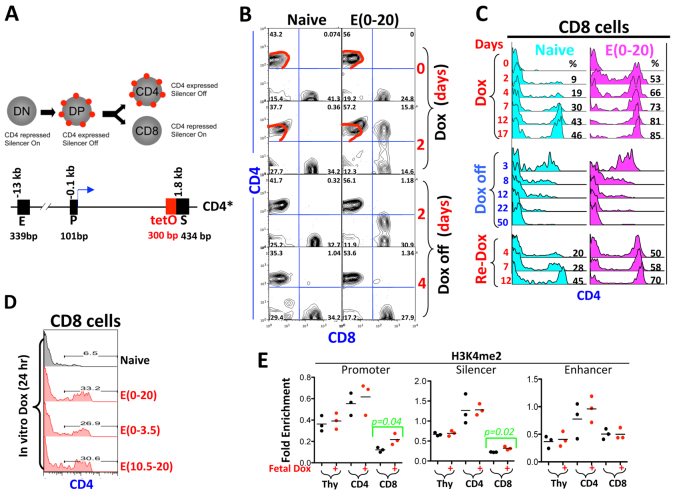
To test this, we replaced the CMV-GFP transgene with the Cd4 minigene, which contains regulatory elements of the murine Cd4 gene, namely the Cd4 silencer, enhancer and promoter (Fig. 5) (Ellmeier et al., 1999). In adult mice heterozygous for the Cd4 minigene and rtTA (Cd4 minigene+/–;rtTA+/–, on the C57B6x129sv background, termed ‘Cd4 minigene mice’ hereafter), GFP was not expressed in CD4 or CD8 cells, whereas Dox administration via drinking water led to slow, variegated GFP induction, with ∼30% of CD4 and ∼20% of CD8 cells fully expressing GFP, whereas the remainder completely lacked GFP expression after 2 weeks of Dox stimulation (Fig. 6A, column 1). Such a ‘digital’ transcription response, which is reminiscent of position-effect variegation mediated by heterochromatin (Hendrich and Willard, 1995; Girton and Johansen, 2008), indicates that Dox controls the probability of transcription in individual cells, presumably by regulating chromatin accessibility at the Cd4 promoter (Festenstein and Kioussis, 2000; Sen and Grosschedl, 2010).
Fig. 5.
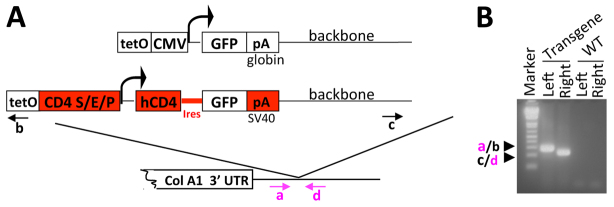
Generation and phenotype of the Cd4 minigene transgenic line. (A) Comparison of the CMV-GFP transgene and the Cd4 minigene. Regions unique to the Cd4 minigene are highlighted in red, including the Cd4 silencer (S), enhancer (E) and promoter (P). To further diversify the two transgene sequences and therefore to help isolate the role of the Col1a1 locus in transgene behavior, we inserted the cDNA encoding human CD4 upstream of GFP and replaced the β-globin poly(A) with an SV40 poly(A) sequence. The arrows indicate the PCR primers a/b and c/d used for screening ES cells for integration of the left and right ends of the flip-in vector, respectively. (B) PCR analysis of a correctly targeted ES clone using primers a/b and c/d. DNA from the parental KH2 ES cells was used as a wild-type control (WT).
Fig. 6.
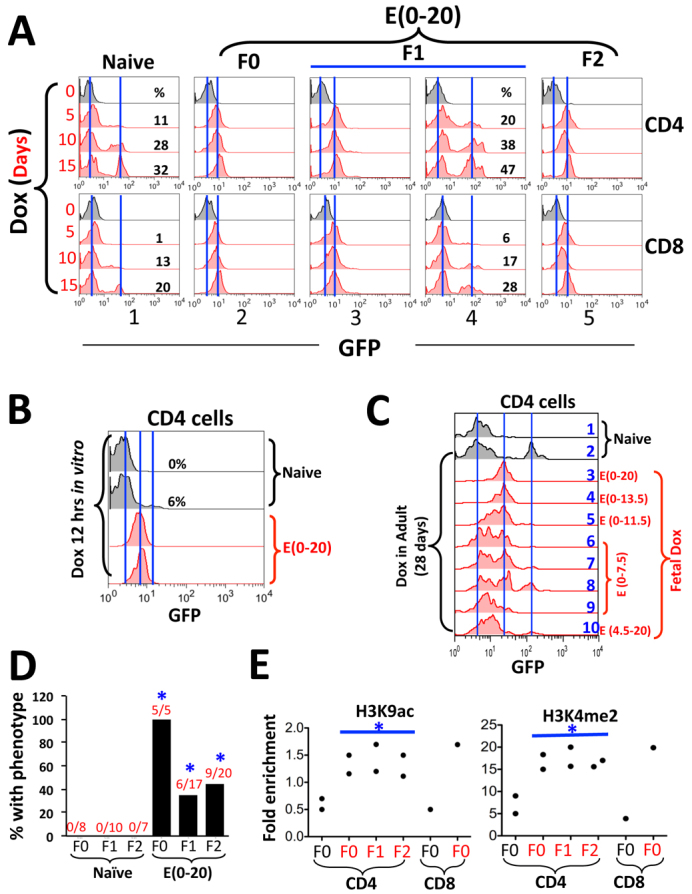
Phenotype of the Cd4 minigene;rtTA mice. (A) Effects of fetal Dox exposure on Cd4 minigene+/–;rtTA+/– mice. F0 females, exposed to Dox at E(0-20) (column 2), were mated with CD1 males to derive F1 pups (columns 3 and 4). F1 females that had inherited the phenotype (column 3) were again mated with CD1 males to produce F2 mice (column 5). Dox was given via drinking water, and tail blood was drawn at various times to monitor GFP induction in CD4 (top) and CD8 (bottom) cells. The left and right vertical lines mark the peaks of basal and induced GFP fluorescence, respectively. (B) GFP induction following 12 hours of Dox stimulation in vitro. CD4 cells from two naïve and two pre-exposed Cd4 minigene+/–;rtTA+/– mice were compared. (C) Developmental windows of rtTA action. Embryos (Cd4 minigene+/+;rtTA+/+) were exposed to Dox for various times. The ensuing adults were re-exposed to Dox for 28 days before analysis of peripheral blood CD4 cells. The left, middle and vertical lines mark the peaks of basal GFP fluorescence, the GFP fluorescence in mice with E(0-20) exposure and that in naïve mice, respectively. (D) Summary of the effects of E(0-20) exposure. The bars show the percentages of mice that displayed the phenotype (rapid, uniform but low-level GFP induction). The numerators and the denominators in the fractions above the bars indicate the numbers of the mice displaying the phenotype and the total numbers of mice analyzed, respectively. Control mice (naïve) were produced in the same way except that the F0 mice were not prenatally exposed. Asterisks indicate statistical significance (P=0.002 to 0.04) in the differences between the Dox-exposed mice and the naïve mice of the corresponding generations. (E) Histone modifications in Cd4 minigene+/–;rtTA+/– mice with E(0-20) exposure (F0) and in their offspring that had inherited the phenotype (red dots), normalized to an internal control ∼11 kb downstream of the transgenic Cd4 promoter. Naïve F0 mice were used as control (Ctr). The ChIP PCR primers target the transgenic Cd4 promoter, but the signals presumably also reflect histone modifications at the adjacent enhancer and silencer. Asterisks indicate statistical significance (P=0.02 to 0.04) in the differences between each generation of pre-exposed and naïve mice.
Remarkably, prenatal E(0-20) Dox exposure greatly facilitated GFP induction in both CD4 and CD8 cells: the induction occurred rapidly and uniformly, such that all cells expressed GFP and the plateau of expression was reached within 5 days of Dox stimulation, although the GFP expression level (∼10 fluorescence units) was ∼5-fold lower than in the control mice (∼50 fluorescence units; Fig. 6A, column 1 versus 2). An in vitro culture assay indicated that, in CD4 cells from prenatally exposed mice, GFP induction almost plateaued within 12 hours of Dox stimulation (Fig. 6B). Thus, fetal Dox exposure increased the kinetics and probability of GFP induction while reducing its expression level. Of note, because all cells now uniformly express GFP upon Dox stimulation, fetal Dox exposure might have converted the digital transcription response into an ‘analog’ response, in which transcription rate instead of probability is subject to regulation (Sen and Grosschedl, 2010).
We also defined the minimal window of prenatal Dox exposure necessary to induce long-lasting effects. Mice were exposed for various times during fetal development, and the ensuing adults were challenged with Dox for 28 days before the analysis of GFP induction in the peripheral blood. In naïve mice lacking fetal Dox exposure, Dox induced GFP in a fraction of CD4 cells (Fig. 6C, row 2). E(0-20) exposure led to lower but uniform GFP expression in all CD4 cells, as expected, which was fully recapitulated by E(0-13.5) but not other exposures including E(4.5-20) (Fig. 6C, rows 3-10), in general agreement with the scenario at the CMV-GFP transgene.
To determine the transgenerational inheritability of the phenotype, we crossed the mice with E(0-20) exposure to CD1 mice. As in the CMV-GFP mice, the phenotype was fully transmittable via females (but not males) to sizable fractions of their offspring for at least two generations: to 35% (6/17) of the F1 and 45% (9/20) of the F2 mice (Fig. 6A, columns 3-5; data summarized in 6D). The GFP induction pattern in the remaining littermates was comparable to that in naïve mice (e.g. Fig. 6A, column 1 versus 4), indicating that the phenotype was inherited largely in an all-or-none manner.
To characterize the epigenetic lesion, we examined H3K9ac and H3K4me2, two of the best-defined marks for active chromatin. Both marks were present at low levels at the Cd4 minigene in CD4 and CD8 cells in naïve mice, but were enriched in the prenatally exposed F0 mice and in their offspring that had inherited the phenotype (Fig. 6E). By contrast, CpG methylation at the Cd4 minigene was unaffected by prenatal Dox (supplementary material Fig. S4).
We conclude that fetal rtTA activation induces an activating epigenetic lesion at the Cd4 minigene that is heritable via the female germline, just as in the case of the repressive lesion at the CMV promoter, indicating that the Col1a1 locus can indeed protect both repressive and activating lesions.
The Col1a1 locus is susceptible to reprogramming in the male germline
To further characterize the Col1a1 locus, we explored why the phenotype in F0 males is not transgenerationally heritable. We examined CpG methylation at the CMV-GFP transgene in sperm, using lymphocytes from the same males as control. Sperm and lymphocytes both showed little methylation in naïve mice (Fig. 7, groups 1-2). Fetal Dox exposure led to high-level (∼90%) methylation in lymphocytes as described before (group 3). Interestingly, partial methylation (∼30%) was detectable in sperm (group 4), which was transmitted to the lymphocytes in one of the three F1 males analyzed (group 5, box) but not to the sperm in the same male (group 6). The data demonstrate that the male germline can erase pre-existing methylation at the Col1a1 locus, suggesting that Dox might have induced CpG methylation in the male primordial germ cells in the F0 mice, which was then progressively and completely erased, perhaps during spermatogenesis, over two successive generations. We also examined H3K4me2 and found it undetectable even in sperm from naïve males (not shown).
Fig. 7.
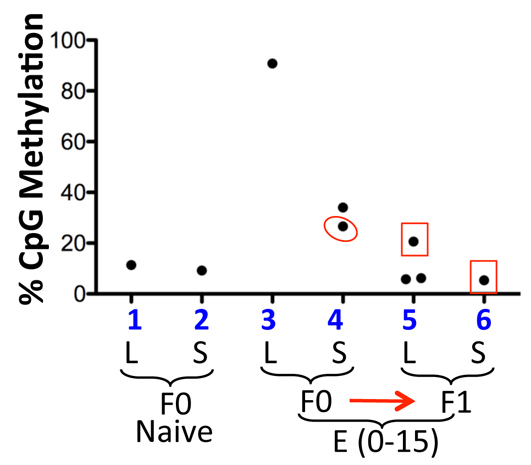
CMV promoter CpG methylation in lymphocytes and sperm from CMV-GFP mice. Sperm (S) from two pre-exposed F0 males was analyzed. One of the males (circled) was mated with a CD1 female to produce three F1 males, all of which were analyzed for lymphocyte (L) methylation and one for sperm methylation as well (box). Naïve CMV-GFP mice (F0) were used as a control.
Fetal Dox exposure impairs lineage-specific epigenetic modifications at the endogenous Cd4 gene
The experiments described so far were performed on transgenes, which differ from endogenous genes in many ways. In particular, endogenous genes often undergo lineage-specific chromatin modifications that can potentially prevent or erase the fetal epigenetic lesions. This and other issues unique to endogenous genes cannot be addressed using our transgenes, including the Cd4 minigene; although the native Cd4 locus undergoes multiple rounds of epigenetic modification during T-cell development (Fig. 8A), the Cd4 minigene failed to recapitulate this programming, given the comparable GFP expression pattern and epigenetic states of the Cd4 minigene in CD4 and CD8 cells (Fig. 6A,E).
Fig. 8.
Epigenetic lesion at the endogenous Cd4 locus in Cd4*/*;rtTA+/+ mice. (A) (Top) CD4 expression (red dot) during T-cell development. (Bottom) The murine Cd4 locus. CD4 is expressed in DP cells but shut off during DP CD8 development (by the action of the Cd4 silencer). The position of the Cd4 promoter (P), enhancer (E) and silencer (S) relative to the transcription start site (+1, blue arrow) and their sizes are indicated above and below the DNA, respectively. The tetO sequence (red box) was inserted upstream of the silencer to create the Cd4* allele. DN, double negative for CD4 and CD8; DP, double positive for CD4 and CD8. (B) CD4 expression in naïve (left) and pre-exposed (right) mice following 2 days of Dox administration in vivo. Note that Dox superactivated CD4 expression in CD4 cells (red contours) and that CD4 induction in T cells was reversible upon Dox withdrawal (Dox off). (C) Kinetics of CD4 induction in CD8 cells in vivo following the first (top) and second (bottom) rounds of Dox administration. (D) CD4 induction in CD8 cells after 24 hours of Dox stimulation in vitro. Cells were from mice with various prenatal exposures. (E) H3K4me2 at the Cd4 regulatory elements in naïve (black dots) and prenatally exposed (red dots) mice, normalized to β-actin.
To address the effects of fetal Dox exposure on endogenous genes, we used rtTA to manipulate the native Cd4 gene. CD4 expression is driven by the Cd4 promoter and enhancer, but repressed by the Cd4 silencer (Fig. 8A) (Ellmeier et al., 1999). We inserted the tetO sequence upstream of the Cd4 silencer, which should allow it to influence not only the Cd4 silencer by proximity, but also the Cd4 promoter/enhancer via chromatin looping (Jiang and Peterlin, 2008). Mice bearing the modified Cd4 locus (Cd4*) were produced and crossed with mice carrying the Rosa26-rtTA allele to generate Cd4*/*;rtTA+/+ mice. T-cell development was normal in these mice (Wan et al., 2013).
Intrathymic T-cell development starts with double-negative (DN) cells lacking CD4 or CD8 expression and proceeds via intermediate CD4+ CD8+ double-positive (DP) cells to produce mature T cells with mutually exclusive expression of CD4 or CD8 (Singer and Bosselut, 2004) (Fig. 8A). This developmental process begins in late embryos and continues into adulthood, and most of the peripheral CD4 and CD8 cells are generated postnatally from the thymus. We found that prenatal rtTA action interferes with CD4 regulation during postnatal T-cell development. Specifically, although fetal Dox exposure did not overtly alter CD4 expression in the ensuing adults (Fig. 8B, top right), it undermined the stability of CD4 repression, such that a brief (2 day) Dox stimulation, which had little effect on naïve mice, sufficed to disrupt CD4 silencing in ∼50% of CD8 cells in prenatally exposed mice (Fig. 8B, Dox day 2).
A detailed kinetic analysis indicates that, in naïve mice, CD4 induction in CD8 cells was slow and inefficient, with only 2% and 46% of CD8 cells expressing CD4 following 2 and 17 days of Dox stimulation, respectively (Fig. 8C, top left). By contrast, the induction was rapid and efficient in mice with fetal Dox exposure, reaching 53% and 85% on days 2 and 17, respectively (Fig. 8C, top right). Surprisingly, brief exposures at E(0-3.5) or E(10.5-20) were largely sufficient to recapitulate the effect of E(0-20) exposure (Fig. 8D), indicating that the Cd4 locus is highly susceptible to perturbation. Despite this susceptibility, even E(0-20) exposure induced only a weak transgenerational effect, reinforcing the distinction in the determinants of mitotic versus transgenerational inheritance (data not shown).
The phenotype in CD8 cells was associated with a defect in H3K4me2 reprogramming: in naïve mice, H3K4me2 was present at the Cd4 locus in DP cells and erased at the Cd4 promoter and silencer during DP→CD8 development (but persists in CD4 cells; Fig. 8F, black dots). Fetal Dox exposure partially countered this lineage-specific depletion; the effect was subtle (<2-fold) but specific, being absent at the Cd4 enhancer in CD8 cells and at any Cd4 regulatory elements in DP and CD4 cells (Fig. 8F, red dots). However, although Cd4 promoter DNA is reported to be highly methylated in T cells (Zou et al., 2001), we found little (<10%) CpG methylation at the promoter (or silencer) even in naïve mice, suggesting that DNA methylation is not involved in the epigenetic lesion (supplementary material Fig. S5).
The adult epigenome is relatively resilient
Given that rtTA is routinely used to achieve reversible gene regulation in adult mice and tumor lines (Schönig et al., 2010), it is unexpected that fetal rtTA activation could so readily produce long-lasting effects at each of the four target genes we examined. This discrepancy might reflect the uniqueness of our target genes and/or of the fetal epigenome, which can be addressed by comparing target gene responses to rtTA acting in the fetus versus adult. However, this comparison is difficult for the CMV promoter, which displays opposite responses to rtTA (i.e. repression versus activation) when stimulated in fetus versus adult. Therefore, we focused on the Cd4 minigene and the endogenous Cd4 gene.
In naïve adult Cd4 minigene mice, GFP induction was slow and variegated in CD4 cells, being hardly detectable after 4 days of Dox exposure and reaching 36% only on day 27 (Fig. 9, top left; see also Fig. 6A, column 1), which was reversed slowly (within 55 days) but completely following Dox withdrawal (Fig. 9, middle left). Interestingly, upon re-exposure, it took only 1 day to induce GFP in a comparable fraction (42%) of CD4 cells (Fig. 9, bottom left). Thus, the initial Dox exposure in adult mice stably primed GFP for rapid induction in response to the second exposure. However, the effect of adult exposure was much less pronounced compared with fetal Dox exposure: whereas fetal Dox exposure increased the kinetics and probability of GFP induction but reduced the GFP expression level, adult Dox exposure affected the kinetics but not the probability of GFP induction, nor did it change the GFP expression level. Furthermore, the adult lesion was not transgenerationally heritable (not shown). Finally, rtTA acting in adults failed to stably affect CD8 cells in that the initial Dox exposure was unable to facilitate the second round of GFP induction (Fig. 9, right), which stands in contrast to the ability of fetal Dox exposure to affect GFP induction in adult CD8 cells (Fig. 6A, bottom). These data indicate that adults are relatively resilient epigenetically.
The scenario at the endogenous Cd4 locus supports this conclusion. In the experiment shown in Fig. 8C, after 17 days of Dox exposure, Dox was withdrawn for 50 days to allow restoration of CD4 repression in CD8 cells. We found that the CD4 induction kinetics upon Dox re-stimulation (Re-Dox) was comparable to that in the first round, indicating that the first round of Dox failed to induce a stable epigenetic lesion (Fig. 8C, bottom).
We conclude that the ability of fetal rtTA activation to induce epigenetic lesions reflects the extraordinary malleability of the fetal epigenome, a characteristic that is not shared by the adult epigenome.
DISCUSSION
We have analyzed four Dox-regulated genes to assess five parameters that potentially affect the inheritance of epigenetic perturbations caused by environmental factors: the timing and location of the perturbation, the chromatin modification pattern of the epigenetic lesion, the DNA sequence involved, and the lineage-specific epigenetic reprogramming subsequent to the epigenetic perturbation. It seems that in our system it is the timing and location but not the other parameters that are the decisive factors of epigenetic inheritance. Our data reveal the extraordinary malleability of the fetal epigenome and the existence of a novel type of locus supporting transgenerational epigenetic inheritance.
Timing of perturbation: the surprising malleability of the fetal epigenome
Whereas the adult epigenome was relatively resilient to rtTA-mediated perturbation, rtTA acting at the appropriate times during fetal development irreversibly altered the function of each of the four target genes in the ensuing adults, regardless of gene sequence, gene location or the nature of the epigenetic lesion, and despite antagonizing lineage-specific epigenetic reprogramming. Consistent with this extraordinary malleability of the fetal epigenome, it has been shown that a transcriptional repressor acting during the first few days of mouse development can irreversibly silence distinct transgenes in somatic cells (Wiznerowicz et al., 2007).
Early embryos have hyperdynamic chromatin that is undergoing global epigenetic reprogramming (Morgan et al., 2005; Meshorer et al., 2006). We propose that this feature makes the fetal chromatin highly vulnerable to environmental disruption, and that the effects tend to be mitotically heritable. Of particular interest, the first 4.5 days of embryogenesis were crucial for rtTA to induce full-blown phenotypes. Dramatic reprogramming is known to occur within this period, including the demethylation and remethylation of non-imprinted genes, where the resulting methylation patterns tend to self-perpetuate in somatic lineages thereafter. rtTA-induced disruption of such reprogramming can conceivably have profound effects, introducing an ‘original sin’ that may influence, shape and even dictate the effects of rtTA acting later (e.g. at E4.5-20) during embryogenesis. In this scenario, the ‘full-blown’ phenotype seen in mice with E(0-20) exposure was perhaps established in a stepwise manner, with E(0-4.5) exposure laying the foundation and the subsequent E(4.5-20) exposure enhancing or modifying the effect of the earlier exposure. This model might explain, for example, why in CMV-GFP mice E(0-20) as well as E(0-4.5) exposures caused silencing but E(4.5-20) exposure instead caused mild activation. Of note, although the E(0-4.5) exposure sufficed to affect the ensuing adults, the effect was not heritable transgenerationally, in contrast to the effect of E(0-20) exposure. Since CpG methylation is normally erased in the germline starting at E11.5 (Morgan et al., 2005), the germline lesions established by rtTA in E4.5 fetal cells might be similarly erased in the germline unless protected or consolidated by the continuous action of rtTA.
The malleability of the fetal epigenome has an important implication. Specifically, it is well known that only some genes can undergo heritable epigenetic changes following transient prenatal environmental exposure. These environment-sensitive genes, including the ‘metastable epialleles’, are of obvious clinical importance and are being actively sought (Rakyan et al., 2002; Waterland et al., 2006; Waterland et al., 2010; Weinhouse et al., 2011). It is intuitive to assume that these genes are endowed with the unique ability to propagate environmentally induced epigenetic alterations. Our data, however, raise the possibility that the apparent uniqueness of these genes might simply reflect the target selectivity of environmental factors, based on our finding that epigenetic changes are prone to self-perpetuation independently of the nature of the target genes if induced at the appropriate time during fetal development.
Paradoxically, although Dox-regulated activators are often used to control gene expression in mice (Schönig et al., 2010), the epigenetic phenomena that we describe have never been reported. However, the previous studies were not designed to address the long-term effects of fetal exposure, and to our knowledge there is no existing evidence to show that epigenetic manipulation of early embryos does not affect the adults.
Location of perturbation: novelty of the Col1a1 locus
Although fetal Dox exposures readily caused long-lasting effects in the ensuing adults, strong transgenerational inheritance of epigenetic lesions (via eggs) was observed only at the transgenes integrated at the Col1a1 locus. The physical structure of the heritable lesions in the eggs remains to be determined, although for the CMV-GFP transgene it might involve CpG methylation, given that fetal Dox exposure caused CpG methylation in various cell types including sperm. Despite this uncertainty, it is clear that the Col1a1 locus can support the inheritance of not only repressive but also activating lesions. This indicates that the locus acts ‘neutrally’, perhaps by insulating the environmentally induced epigenetic lesions from reprogramming enzymes, thereby preserving the lesions regardless of their structure, rather than by spontaneously silencing or activating integrated transgenes. This mode of action is novel. So far, only four transgenic mouse lines are known to show transgenerational epigenetic inheritance, all involving heritable silencing of integrated transgenes that is presumably due to the heterochromatic nature of the loci (Hadchouel et al., 1987; Allen et al., 1990; Kearns et al., 2000; Sutherland et al., 2000). Transgenerational inheritance has also been described at the Avy allele, the expression of which is influenced by the cryptic promoter of an inserted retrotransposon (Morgan et al., 1999); this promoter is constitutively active but undergoes stochastic and partially heritable silencing (Dolinoy et al., 2010), indicating that heritable silencing underlies the epigenetic phenomenon at the Avy allele.
rtTA-induced silencing of the CMV promoter
rtTA acting during embryogenesis led to transgenerationally heritable activation and silencing at the Cd4 minigene and CMV promoter, respectively. The former is reminiscent of the fact that environmental factors acting on Drosophila embryos can cause transgenerationally heritable gene activation (Cavalli and Paro, 1998; Cavalli and Paro, 1999; Seong et al., 2011), but the latter scenario appears to be novel. The mechanism of activation turned silencing is unclear, but might involve induction of non-coding RNA, disruption of higher order chromatin structure, and/or activation of a negative-feedback loop. ES cells provide a tractable system for addressing this fascinating problem.
Supplementary Material
Acknowledgments
We thank Dr R. Jaenisch for the CMV-GFP mice and related plasmids, Drs S. T. Qureshi and P. J. Lee for the CMV-CD4hTLR4 mice, and Dr D. J. Pan for comments.
Footnotes
Funding
Supported by the National Institutes of Health [1R56AI074916-01A2 and 1R21AI094000-01 to T.C.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
T.C. conceived the project and wrote the manuscript. M.W. and M.Y. created Cd4* and Cd4 minigene mice, respectively. M.W., H.G., J.W., H.H., J.Z., R.K.K., M.Y., R.K., B.H.C. and E.D. analyzed these and other mice described in the paper.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.088229/-/DC1
References
- Allen N. D., Norris M. L., Surani M. A. (1990). Epigenetic control of transgene expression and imprinting by genotype-specific modifiers. Cell 61, 853-861 [DOI] [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. (2006). Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23-28 [DOI] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D. (2010). Molecular signals of epigenetic states. Science 330, 612-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U., Hisano M., Erkek S., Ramos L., Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., Peters A. H. F. M. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679-687 [DOI] [PubMed] [Google Scholar]
- Cavalli G., Paro R. (1998). The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93, 505-518 [DOI] [PubMed] [Google Scholar]
- Cavalli G., Paro R. (1999). Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286, 955-958 [DOI] [PubMed] [Google Scholar]
- Daxinger L., Whitelaw E. (2012). Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 13, 153-162 [DOI] [PubMed] [Google Scholar]
- Dolinoy D. C., Weinhouse C., Jones T. R., Rozek L. S., Jirtle R. L. (2010). Variable histone modifications at the A(vy) metastable epiallele. Epigenetics 5, 637-644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W., Sawada S., Littman D. R. (1999). The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17, 523-554 [DOI] [PubMed] [Google Scholar]
- Faulk C., Dolinoy D. C. (2011). Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6, 791-797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R., Kioussis D. (2000). Locus control regions and epigenetic chromatin modifiers. Curr. Opin. Genet. Dev. 10, 199-203 [DOI] [PubMed] [Google Scholar]
- Girton J. R., Johansen K. M. (2008). Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv. Genet. 61, 1-43 [DOI] [PubMed] [Google Scholar]
- Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. (1995). Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766-1769 [DOI] [PubMed] [Google Scholar]
- Guerrero-Bosagna C., Settles M., Lucker B., Skinner M. K. (2010). Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE 5, e13100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadchouel M., Farza H., Simon D., Tiollais P., Pourcel C. (1987). Maternal inhibition of hepatitis B surface antigen gene expression in transgenic mice correlates with de novo methylation. Nature 329, 454-456 [DOI] [PubMed] [Google Scholar]
- Hathaway N. A., Bell O., Hodges C., Miller E. L., Neel D. S., Crabtree G. R. (2012). Dynamics and memory of heterochromatin in living cells. Cell 149, 1447-1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B. D., Willard H. F. (1995). Epigenetic regulation of gene expression: the effect of altered chromatin structure from yeast to mammals. Hum. Mol. Genet. 4, 1765-1777 [DOI] [PubMed] [Google Scholar]
- Jablonka E., Raz G. (2009). Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131-176 [DOI] [PubMed] [Google Scholar]
- Jiang H., Peterlin B. M. (2008). Differential chromatin looping regulates CD4 expression in immature thymocytes. Mol. Cell. Biol. 28, 907-912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle R. L., Skinner M. K. (2007). Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns M., Preis J., McDonald M., Morris C., Whitelaw E. (2000). Complex patterns of inheritance of an imprinted murine transgene suggest incomplete germline erasure. Nucleic Acids Res. 28, 3301-3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D., Wan M., Wu J., Preston-Hurlburt P., Kushwaha R., Grundström T., Imbalzano A. N., Chi T. (2009). Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc. Natl. Acad. Sci. USA 106, 1169-1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. (2006). Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. D., Sutherland H. G., Martin D. I., Whitelaw E. (1999). Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314-318 [DOI] [PubMed] [Google Scholar]
- Morgan H. D., Santos F., Green K., Dean W., Reik W. (2005). Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14, R47-R58 [DOI] [PubMed] [Google Scholar]
- Qureshi S. T., Zhang X., Aberg E., Bousette N., Giaid A., Shan P., Medzhitov R. M., Lee P. J. (2006). Inducible activation of TLR4 confers resistance to hyperoxia-induced pulmonary apoptosis. J. Immunol. 176, 4950-4958 [DOI] [PubMed] [Google Scholar]
- Rakyan V. K., Blewitt M. E., Druker R., Preis J. I., Whitelaw E. (2002). Metastable epialleles in mammals. Trends Genet. 18, 348-351 [DOI] [PubMed] [Google Scholar]
- Rando O. J., Verstrepen K. J. (2007). Timescales of genetic and epigenetic inheritance. Cell 128, 655-668 [DOI] [PubMed] [Google Scholar]
- Schönig K., Bujard H., Gossen M. (2010). The power of reversibility regulating gene activities via tetracycline-controlled transcription. Methods Enzymol. 477, 429-453 [DOI] [PubMed] [Google Scholar]
- Sen R., Grosschedl R. (2010). Memories of lost enhancers. Genes Dev. 24, 973-979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong K. H., Li D., Shimizu H., Nakamura R., Ishii S. (2011). Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145, 1049-1061 [DOI] [PubMed] [Google Scholar]
- Singer A., Bosselut R. (2004). CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83, 91-131 [DOI] [PubMed] [Google Scholar]
- Skinner M. K. (2008). What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 25, 2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. K. (2011). Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics 6, 838-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H. G., Kearns M., Morgan H. D., Headley A. P., Morris C., Martin D. I., Whitelaw E. (2000). Reactivation of heritably silenced gene expression in mice. Mamm. Genome 11, 347-355 [DOI] [PubMed] [Google Scholar]
- Wan M., Kaundal R., Huang H., Zhao J., Yang X., Chaiyachati B., Chi T. (2013). A general approach for controlling gene expression and probing regulatory mechanisms: Application to the Cd4 locus. J. Immunol. 190, 737-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R. A., Dolinoy D. C., Lin J. R., Smith C. A., Shi X., Tahiliani K. G. (2006). Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis 44, 401-406 [DOI] [PubMed] [Google Scholar]
- Waterland R. A., Kellermayer R., Laritsky E., Rayco-Solon P., Harris R. A., Travisano M., Zhang W., Torskaya M. S., Zhang J., Shen L., et al. (2010). Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet. 6, e1001252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse C., Anderson O. S., Jones T. R., Kim J., Liberman S. A., Nahar M. S., Rozek L. S., Jirtle R. L., Dolinoy D. C. (2011). An expression microarray approach for the identification of metastable epialleles in the mouse genome. Epigenetics 6, 1105-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M., Jakobsson J., Szulc J., Liao S., Quazzola A., Beermann F., Aebischer P., Trono D. (2007). The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 282, 34535-34541 [DOI] [PubMed] [Google Scholar]
- Youngson N. A., Whitelaw E. (2008). Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 9, 233-257 [DOI] [PubMed] [Google Scholar]
- Yu M., Wan M., Zhang J., Wu J., Khatri R., Chi T. (2008). Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc. Natl. Acad. Sci. USA 105, 3873-3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y. R., Sunshine M. J., Taniuchi I., Hatam F., Killeen N., Littman D. R. (2001). Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 29, 332-336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.