Abstract
The diverse functions of Notch signalling imply that it must elicit context-specific programmes of gene expression. With the aim of investigating how Notch drives cells to differentiate, we have used a genome-wide approach to identify direct Notch targets in Drosophila haemocytes (blood cells), where Notch promotes crystal cell differentiation. Many of the identified Notch-regulated enhancers contain Runx and GATA motifs, and we demonstrate that binding of the Runx protein Lozenge (Lz) is required for enhancers to be competent to respond to Notch. Functional studies of targets, such as klumpfuss (ERG/WT1 family) and pebbled/hindsight (RREB1 homologue), show that Notch acts both to prevent the cells adopting alternate cell fates and to promote morphological characteristics associated with crystal cell differentiation. Inappropriate activity of Klumpfuss perturbs the differentiation programme, resulting in melanotic tumours. Thus, by acting as a master regulator, Lz directs Notch to activate selectively a combination of target genes that correctly locks cells into the differentiation programme.
Keywords: Lozenge/Runx, Notch, Chromatin immunoprecipitation, Haemocyte, Drosophila
INTRODUCTION
The Notch pathway operates during many different developmental decisions, as exemplified by haematopoiesis, where Notch regulates both the emergence of stem cells and the subsequent cell fate choices and differentiation (reviewed by Gering and Patient, 2010; Maillard et al., 2005; Pajcini et al., 2011; Radtke et al., 2010). With a simple transduction pathway, where receptor activation results in proteolytic release of the Notch intracellular domain (NICD), one of the primary outcomes of Notch activation is a change in transcription (reviewed by Bray, 2006; Kopan and Ilagan, 2009; Kovall, 2008; van Tetering and Vooijs, 2011). Recent studies have revealed large numbers of Notch-responsive genes in progenitor cells or in cancers (Hamidi et al., 2011; Krejcí et al., 2009; Li et al., 2012; Palomero et al., 2006; Wang et al., 2011) that appear to be involved in preventing differentiation and cross-regulation of other signalling pathways (Hamidi et al., 2011; Krejcí et al., 2009; Li et al., 2012). It is not yet clear whether a different spectrum of Notch targets is involved in promoting differentiation and, if so, what such targets tell us about the mechanisms involved.
To address these issues we have turned to a simple system, Drosophila blood cells (haemocytes), where Notch activity promotes crystal cell differentiation. The second wave of haemocyte development occurs in the lymph gland (Fig. 1A) (Crozatier and Meister, 2007; Evans and Banerjee, 2003; Jung et al., 2005) and primarily gives rise to two cell types: crystal cells and plasmatocytes (Fig. 1B). Previous studies have shown that Notch activity is required for expression of the Runx protein Lozenge (Lz) in haemocytes (Lebestky et al., 2003). As Lz is necessary for crystal cell development, this suggested a simple mechanism to explain how Notch directs crystal cell differentiation. However, there is as yet no evidence that lz is directly regulated by Notch pathway (Lebestky et al., 2003; Muratoglu et al., 2007). Furthermore, perturbations to Notch at late stages prevent crystal cell differentiation and compromise cell survival, suggesting that Notch activity may also be required in parallel to or subsequent to Lz, although no other downstream targets have been identified (Krzemien et al., 2010; Mukherjee et al., 2011). Likewise, although Notch1 appears to function upstream of Runx in several steps during mammalian haematopoiesis, no evidence that Runx genes are direct targets of Notch1 has emerged (e.g. Burns et al., 2009; Burns et al., 2005; Nottingham et al., 2007; Robert-Moreno et al., 2005). Indeed Runx factors are suggested to integrate with Notch1 activity in specifying the T-cell lineage (Guo et al., 2008) and Notch1 targets in leukaemic T-cell have signatures suggestive of co-regulation by Runx and Notch (Wang et al., 2011). Whether this reflects direct cooperation between Notch and Runx has not been established.
Fig. 1.
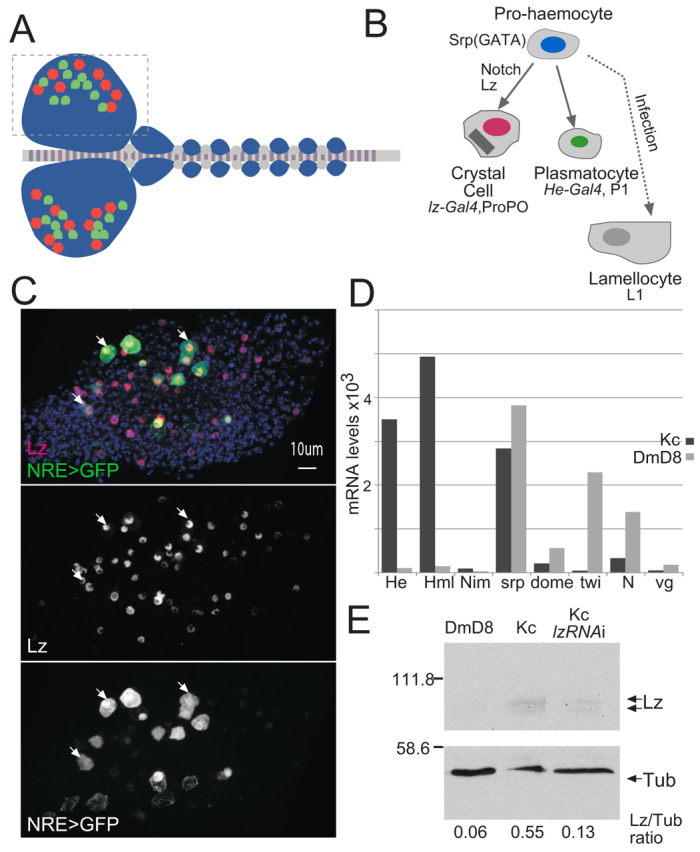
Haemocyte development: role of Notch and relationship to Kc167 cells. (A) Drosophila lymph gland where larval haemocyte development gives rise to crystal cells (red) and plasmatocytes (green). Boxed area indicates the region shown in confocal images in this and subsequent figures. (B) The haemocyte lineage, specific markers and relevant Gal4 drivers are indicated. Notch and Lozenge (Lz) are required for crystal cells (red nuclei) to develop from Serpent (Srp)-expressing pro-haemocytes. (C) Expression of a general Notch-responsive reporter, NRE-GFP (green), indicates that Notch is active in Lz-expressing (red) crystal cell precursors. Arrows indicate examples of cells co-expressing Lz and NRE-GFP. (D) ModEncode measurements of relative mRNA expression levels of the indicated genes (He, hemese; Hml, hemolectin; Nim, Nimrod; srp, serpent; dome, domeless; twi, twist; N, Notch; vg, vestigial) in Kc167 cells compared with DmD8 (muscle precursor related) (Cherbas et al., 2011); haemocyte-related genes Hemolectin (Hml) and Hemese (He) are specifically expressed in Kc cells. (E) Lz is present in Kc167 cells; western blot of total cell extracts from the indicated cells was probed with α-Lz and α-Tub. The quantified ratio of Lz/Tub is indicated for each lane (arbitrary units). Cells pre-treated with dsRNA to ablate Lz (Kc LzRNAi) have reduced protein. Scale bar: 10 μm.
Drosophila haemocytes therefore offer a simple model with which to investigate how Notch coordinates differentiation and the relationship it has with Lz/Runx in this process. To address these issues, we first identified direct transcriptional targets of Notch in haemocyte-related cells. Many of these direct Notch targets were associated with Lz/Runx-binding motifs and we demonstrate that Notch and Lz act in combination to regulate the enhancers. Furthermore, our analysis of target gene functions reveals that Notch simultaneously prevents cells adopting the alternate plasmatocyte fate, by upregulating klumpfuss (a ERG/WT1 family member) and promotes characteristics associated with differentiation, by upregulating pebbled/hindsight (a RREB1 homologue). Therefore, through these combined targets, Notch and Lz operate to tie cells into the differentiation programme, converting them from an unstable to a committed state.
MATERIALS AND METHODS
Genome-wide expression and ChIP analysis
Kc 167 cells, obtained from the Drosophila Genomics Resource Center (DGRC) were cultured in Schneider's Drosophila Medium (Invitrogen) supplemented with 5% foetal calf serum (Kc 167) and 1% penicillin-streptomycin at 25°C. Expression and ChIP-chip array experiments were performed as described previously (Krejcí et al., 2009). In brief, chromatin was prepared after 30 minutes of Notch activation by 2 mM EDTA and XChIP performed with α-Su(H) antibody (Krejcí et al., 2009). Precipitated genomic fragments and a fraction of the input DNA were amplified by ligation-mediated PCR, labelled with Cy3 or Cy5, mixed and hybridized to Nimblegen Drosophila 2.1 M tiling arrays, which have 50-75 bp probes distributed at 55 bp intervals across the Drosophila genome. In all cases, three independent biological replicates were analysed and data normalized, as described previously, before identification of statistically significant differences in expression levels (P≤0.05) or peaks of Su(H) occupancy. For the latter, Tamalpais (Bieda et al., 2006) was used to identify significant ChIP peaks (minimum cut-off of five adjacent probes, P<0.05) and after excluding atypical peaks clustered in a small region of Chromosome 2R, the analysis yielded a set of 185 peaks. A custom-written Perl script was used to determine genes in the vicinity of peaks. Peaks were assigned to genes only when they were within 10 kb of an upregulated gene on either strand. Results have been deposited in Gene Expression Omnibus with series Accession Numbers GSE43132 (ChIP) and GSE9964 (expression array).
Motif and GO analysis
CONSENSUS Matrix-based motif discovery in RSAT was used to look for over-represented motifs (Thomas-Chollier et al., 2011). A position-weighted matrix (PWM) for Su(H), Srp and Lz was generated from a compilation of binding sites based on previously published data. Patser (Hertz and Stormo, 1999) was used to search for matches to PWM in the Drosophila genome, with a threshold of 5.5. Custom-written Perl scripts were used to calculate the distance between Su(H) PWM and other PWMs within 600 bp window of Su(H) ChIP peaks. Analysis of gene ontology enrichments was performed using the functional enrichment chart at the DAVID Bioinformatics Resource. Results were filtered for enrichment (over threefold) and for significance using modified Fisher's Exact Test (EASE) with Benjamini correction (http://david.abcc.ncifcrf.gov). Representative examples of Biological Functions (P≤0.005) are depicted to avoid repetitions between similar categories. All enriched molecular function categories with P≤0.1 are shown.
RNAi experiments and chromatin immunoprecipitation
For treating Kc cells with RNAi, double-stranded RNA duplexes corresponding to 400-800bp exonic regions were produced using T7 promoter-containing primers and subsequently transcribed with a MEGAscript T7 Kit (Ambion). Cells were treated with dsRNA for 72 hours before harvesting. Three biological replicates were performed in all experiments. RNA isolation, real-time PCR and ChIP experiments were performed as described (Krejcí and Bray, 2007). Antibodies for ChIP were goat α-Su(H) (Santa Cruz Biotechnology, sc15813) and mouse α-Lz (DSHB).
Fly strains and immunofluorescence staining of lymph glands
Alleles and fly stocks, as described in FlyBase (http://flybase.org/), were lz-Gal4 (Jackson Behan et al., 2005), He-Gal4 (Kurucz et al., 2003), pxn-Gal4 (Stramer et al., 2005), UAS-mamDN (Helms et al., 1999), UAS-Su(H)VP16 (Furriols and Bray, 2000), UAS-KluDN and UAS-kluEnR (Kaspar et al., 2008), UAS-lz (U. Banerjee), UAS-lzEnR (Wildonger et al., 2005), UAS-ush14A (Haenlin et al., 1997), UAS-ushN (Fossett et al., 2000), UAS-ush14A (Cubadda et al., 1997), UAS-hntRNAi (TRiP.JF03162), UAS-mycRNAi (TRiP.JF01761), UAS-whiteRNAi (TRiP.GL00094), UAS-CD8GFP, klu212lR51C and Klu-Gal4 (Klein and Campos-Ortega 1997), ush(–7462/-25)-lacZ (Muratoglu et al., 2007), and NRE-GRins (Housden et al., 2012).
Lymph glands were dissected from third instar larvae (cultured at 29°C from 48-72 hours AEL), fixed for 10 minutes in 4% formaldehyde in PBS. After three washes in PBS and three washes in PBT (PBS + 1% Triton X100), lymph glands were removed from contaminating tissue and mounted onto poly-lysine-coated slides. After blocking for 1 hour in PBTN (PBT + 4% horse serum) they were stained overnight with primary antibodies at 4°C. Primary antibodies were as follows: mouse anti-Hnt (1/20), mouse anti-Lz (1/20), mouse anti-Lam (all from DSHB), mouse anti-P1 (1/30; a gift from I. Ando, AFFILIATION), goat anti-GFP (1/600; Abcam, ab6673), mouse anti-Fib (1/500, Abcam, ab4566), rabbit anti-Ds-Red (1/25; Clontech, 632496) and rabbit anti-β-Galactosidase (1/5000; Cappel). After three 15-minutes washes in PBT, fluorescently conjugated secondary antibodies (Jackson Labs) were applied for 1.5 hours at room temperature followed by three 15-minute washes in PBT and one wash in PBS. Finally, the sample was mounted in Vectashield containing DAPI for imaging with Nikon D-Eclipse C1 or Leica SP2 confocal microscopes. For phenotypic experiments, cell and nuclear dimensions were measured in over 10 lymph glands using IMARIS. Typically, 300-1000 cells were scored for each genotype.
Construct design and mutagenesis
ChIP-enriched regions from klu, CG32369, rgr, peb and other putative targets were amplified from Drosophila genomic DNA, using primers containing restriction enzyme sequences (see supplementary material Table S2), and cloned into pGreenRabbit/pRedRabbit vectors (Housden et al., 2012) for in vivo reporter assays or pGL3min for luciferase assays. Site-directed mutagenesis was performed using a PCR-based approach with primers overlapping the Su(H)/Srp/Lz-binding site to be mutated with the sequences changed as follows: CGTGGGAA to CGTTGTTA for the Su(H) motif; GGATAAC to GTTCTAC for the Srp motif and AACCACA to AATGACC for the Lz motif. Luciferase assays were performed as described previously (Krejcí and Bray, 2007).
RESULTS
Notch responsive genes in Drosophila Kc cells
As Drosophila Kc cells exhibit characteristics of haemocytes (Echalier and Ohanessian, 1969; Lunstrum et al., 1988; Nelson et al., 1994; Schneider, 1972), express several haemocyte markers (including Hemese, Hemolectin and Serpent, Fig. 1D) (Cherbas et al., 2011; Jung et al., 2005) and contain detectable levels of Lz (Fig. 1E), they provide a suitable model for investigating the Notch response and its relationship with the Runx factor Lz. In addition, Kc cells differ substantially from the muscle-related DmD8 cells (Fig. 1D) (Cherbas et al., 2011), where Notch activity is involved in maintaining progenitor characteristics in collaboration with transcription factor Twist (Bernard et al., 2010), enabling us to discover how the Notch response differs between cell types.
To characterize Notch-responsive genes in Kc cells, we used a similar strategy to that used previously, activating Notch using a calcium chelator (Gupta-Rossi et al., 2001; Rand et al., 2000) and monitoring mRNA expression changes 30 minutes later (Krejcí et al., 2009; Krejcí and Bray, 2007). Probes prepared from control and Notch-activated mRNA populations were hybridized in pair-wise combinations to Drosophila transcriptome microarrays to identify genes that were significantly upregulated (P≤0.05). In parallel, we identified genomic regions that were occupied after Notch activation by Suppressor of Hairless [Su(H)], the core transcription factor in the Notch pathway (reviewed by Bray, 2006; Kopan and Ilagan, 2009; Kovall, 2008; van Tetering and Vooijs, 2011), using chromatin immunoprecipitation (ChIP) and hybridizing bound DNA to genomic tiling arrays. Integrating these data, the 185 Su(H) occupied regions, ‘peaks’, were assigned to genes if they were located within or close to genes that were upregulated (Fig. 2A). This identified 69 assigned peak genes (APGs) that fulfilled the criteria of Su(H) binding within 10 kb and upregulation with the Notch activation regime [supplementary material Table S1; ranked by AvgM, fold difference in expression (log2)]. Several APG were validated using quantitative PCR to confirm Su(H) binding in repeat ChIPs (e.g. Fig. 4C) and, using this method, we also detected Su(H) binding near one additional upregulated gene [pebbled (peb), also known as hindsight (hnt)]. As calcium chelation is a non-specific method of activating Notch, a subset, including peb/hnt, were further validated by testing their upregulation in cells transfected with NICD (supplementary material Fig. S1) (although this approach is hampered by the fact that transfection efficiency is less than 30% in these cells).
Fig. 2.
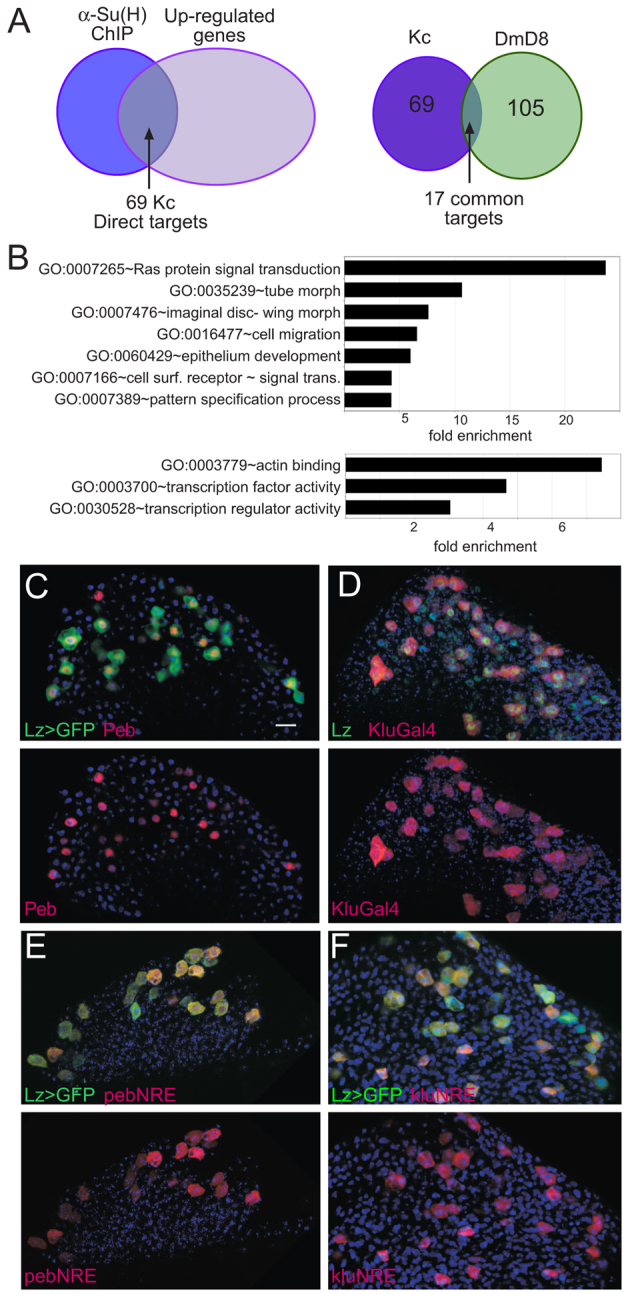
Identification of Notch targets in haemocytes. (A) Left: Venn diagram illustrating overlap between genes in proximity to Su(H)-bound regions [blue: anti-Su(H) ChIP, 185 bound regions] and upregulated after 30 minutes of Notch activation (mauve, 1302 upregulated transcripts) identifies 69 APGs that are putative direct Notch targets in Kc cells. Right: overlap between direct Notch targets identified in Kc cells (purple) and in DmD8 cells (green) is limited to 17 common genes. (B) Examples of over-represented GO (Biological Process, upper graph; Molecular Function, lower graph) categories in Kc direct Notch targets. (C-F) Expression of indicated targets and/or target enhancers in crystal cell lineage (marked by lz>GFP (C, E, F; green) and anti-Lz (D; green). See supplementary material Table S1 for a list of direct targets. Scale bar: 10 μm.
Fig. 4.
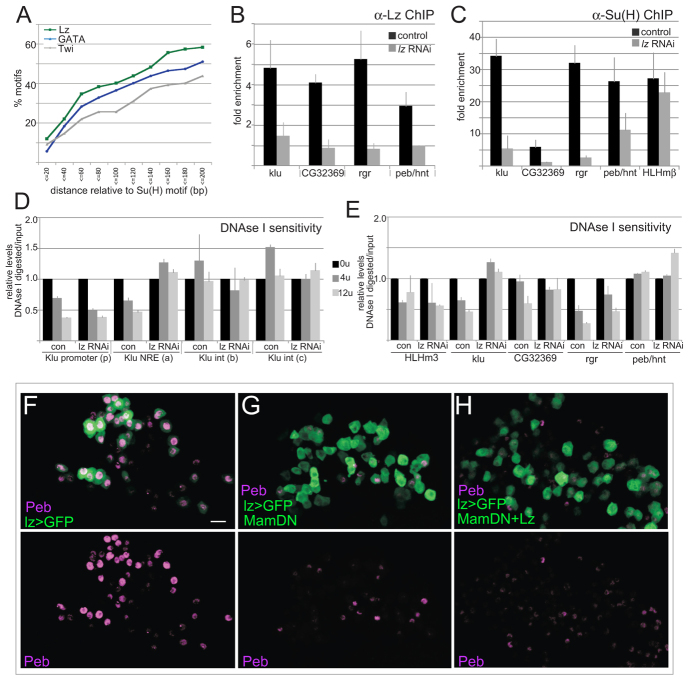
Lz influences Su(H) recruitment at Notch-responsive enhancers and is required with Notch in vivo. (A) Lz and GATA motifs are located in proximity to Su(H). Graph showing percentage of detected motifs located within the indicated distance (bp) of the Su(H) motif within Kc ChIP peaks (blue, GATA; green, Lz; grey, Twist). (B) Lz is present at NREs. Fold enrichment of the indicated NRE in α-Lz ChIP relative to the adjacent coding sequence (cds) in Kc cells (dark shading) and after lz RNAi (light shading). (C) Depletion of Lz compromises Su(H) binding. Fold enrichment of the indicated NRE in anti-Su(H) ChIP relative to the cds in Kc cells (dark shading) and after lz RNAi (light shading). (D,E) Effects of Lz depletion on DNase I sensitivity of the indicated regions in klu (D) and of NRE from other genes (E). Chromatin from control and Lz RNAi-treated cells was subjected to digestion with 0, 4 or 12 U of DNase I and the levels of intact DNA quantified by qPCR. Location of klu fragments are indicated in Fig. 3A. (F-H) Peb/Hnt expression in control glands (F, lz >GFP) and in glands where MamDN only (G) or Lz and MamDN (H) were expressed (lz>GFP indicates lz-Gal4 UAS-GFP). Scale bar: 10 μm. Data are mean±s.e.m.
A comparison with results from similar experiments in DmD8 cells demonstrated relatively little overlap of Su(H)-occupied sites or of upregulated genes. Thus, although the number of occupied sites in the two cell types was of similar magnitude, 260 in DmD8 and 185 in Kc cells, only 28 peaks were overlapping between the two. This suggests that the accessibility of sites to Su(H) is likely to be a major factor in determining the outcome from Notch activation. Most overlapping peaks were in proximity to upregulated genes, corresponding to 17 APGs that were common to both Kc and DmD8 cells (Fig. 2A). These included several of the E(spl) genes, myc and Notch (supplementary material Table S1). Such genes appear to be widespread targets of Notch in many different organisms, including humans, and are likely to identify mechanisms of fundamental importance to Notch signalling (Bray, 1997; Davis and Turner, 2001; Fischer and Gessler, 2007; Kageyama et al., 2007; Klinakis et al., 2006; Krejcí et al., 2009; Palomero et al., 2006; Satoh et al., 2004; Weng et al., 2006; Yashiro-Ohtani et al., 2009). Nevertheless, there were also substantial differences in the Notch-responsive genes in the two cell types, indicative of disparate functional outcomes from Notch activation. We note also that, as we were monitoring Su(H) occupancy and not recruitment of NICD to these targets (owing to NICD antibody not performing well in ChIP), we cannot formally rule out the possibility that expression from some of the Su(H)-bound upregulated genes may be independent of direct binding by NICD. Furthermore, 58% Su(H) peaks had no upregulated genes within 10 kb, and although some may be false positives, it suggests that some Su(H) sites may be associated with genes that require other co-factors or represent Notch-independent Su(H) targets.
To gain a global overview of the Su(H)-bound responsive genes in Kc cells, we analysed their functional characteristics using gene ontology (GO) annotations (http://david.abcc.ncifcrf.gov/) (Fig. 2B). Although the cohort of targets differed, several enriched Biological Process categories were similar to DmD8 cells, including cell surface receptor-linked signalling and imaginal disc morphogenesis. The cross-regulation of other signalling pathways, especially Ras signal transduction, therefore emerges as a common theme, with Gap1 and pointed being among the Kc-regulated genes. Pattern specification and cell migration were, however, more enriched in Kc than in DmD8 cells. Similarly, there was enrichment for the Molecular Function categories ‘transcription factors’ and ‘actin binding’. The former are candidates to coordinate the programme of differentiation in haemocytes, and many encode zinc-finger transcription factors (Interpro zinc-finger C2H2 type; fivefold enriched) such as klumpfuss (klu), pebbled (peb/hnt), regular (rgr) and Hnf4. We note, however, that lz was not among the genes bound by Su(H) or upregulated within 30 minutes, despite its expression in the lymph gland being dependent on Notch activity (Lebestky et al., 2003).
Kc Notch targets are expressed in differentiating crystal cells
None of the identified Kc Su(H) targets had been previously associated with haemocyte fates and several were as yet uncharacterized. To determine their relevance for blood cell differentiation in vivo, we investigated their expression in third instar lymph glands. First we used a Notch activity-sensing reporter (NRE-GFP; Housden et al., 2012) to confirm that Notch is specifically active in Lz-expressing crystal cell precursors (Fig. 1C), although we note that NRE-GFP expression was detected only in a subset of Lz+ cells, suggesting that there is a transient phase of Notch activity. Selecting two highly upregulated transcription factors, klu and peb/hnt, we compared their expression to the Lz crystal cell lineage marker (Fig. 2C,D) and found that both were expressed in the lymph gland in a pattern that overlapped with Lz. Indeed, peb/hnt protein was present in all Lz-expressing cells (detected with lz-Ga4 UAS-GFP), although there were also a few cells with Peb/Hnt only. Similarly, the majority of Klu-expressing cells (klu-Gal4 UAS-GFP) contained Lz. Thus, these Kc identified APG are expressed in a relevant lineage in vivo, where their expression appears to persist. For example, Peb/Hnt protein expression was detected in mature crystal cells, based on its colocalization with anti-ProPO (supplementary material Fig. S2) and with other markers in a recent study (Benmimoun et al., 2012). In addition, expression of peb/hnt was upregulated in NICD-expressing clones and was suppressed when Notch was ablated by RNAi (supplementary material Fig. S2).
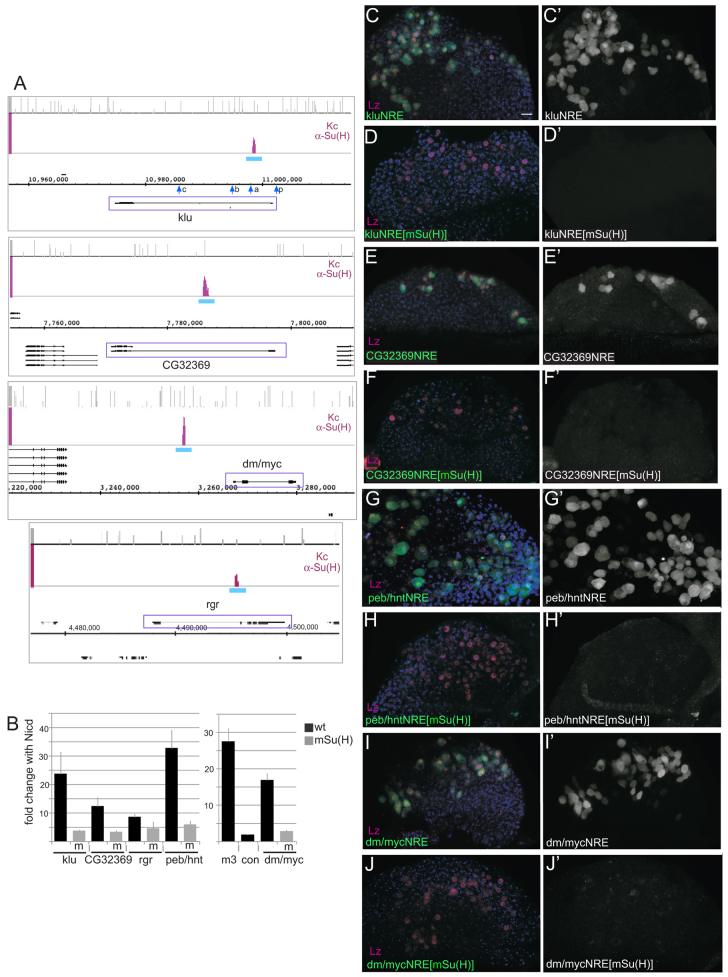
To further investigate whether the APGs were subject to Notch-mediated regulation in the lymph gland, reporter genes were generated containing putative Notch-responsive enhancers (NREs), as defined by Su(H) ChIP peaks, upstream of GFP or mCherry (Fig. 3A) (Housden et al., 2012). All exhibited expression in crystal cell precursors (Lz-expressing cells) (Fig. 3C,E,G,I), although levels of expression varied. For example, peb/hntNRE, kluNRE, mycNRE and CG32369NRE all directed expression at high levels in Lz+ cells (Fig. 2E,F; Fig. 3C,G,E,I). Other reporters, rgrNRE, Gap1NRE, CG6860NRE, CG11873NRE and hnf4NRE, were detected at low levels in Lz-expressing cells (supplementary material Fig. S3) but these levels could be augmented by expression of the constitutively active Su(H)VP16 (supplementary material Fig. S3). To confirm that the identified enhancers were regulated by Notch, we tested consequences of mutating all the Su(H) motifs in klu, CG32369, peb/hnt, myc and rgr NREs (Fig. 3D,F,H,J; supplementary material Fig. S3A). With kluNRE, CG32369NRE mycNRE and rgrNRE, the Su(H) motif mutations completely abolished expression in the lymph gland (Fig. 3D,F,J; supplementary material Fig. S3B). Effects on peb/hntNRE were intermediate (Fig. 3H), with some residual expression detected from the mutated NRE in a subset of glands (2/15). Moreover, luciferase assays revealed that mutated NREs had lost their response to NICD (Fig. 3B). These results confirmed the importance of Su(H) sites for expression in haemocytes, demonstrating that these targets are Notch responsive.
Fig. 3.
Notch-responsive enhancers direct expression in crystal cell lineage and require Su(H) motifs for activity. (A) Examples of genomic regions from representative Notch targets showing Su(H) ChIP-enriched regions in Kc cells (purple; fold enrichment relative to total input –0.1-2.5, log2 scale). Matches to the Su(H)-binding motif are indicated (bar height indicates affinity class of sites). Gene models are depicted in black. Blue bars represent the regions (NRE) that were cloned to test responsiveness in vivo (for distribution of motifs see Fig. 5). Blue arrows in klu indicate fragments analysed in DNase I sensitivity assays (see Fig. 4D). (B) Fold change in luciferase activity in the presence of the NICD from each unmutated NRE (wt) indicated and from NRE with mutated (m) Su(H) motifs. (C-J′) Expression from the indicated NRE, either unmutated (C,C′,E,E′,G,G′,I,I′) or where Su(H) motifs have been mutated [mSu(H)] (D,D′,F,F′,H,H′,J,J′). Levels of enhancer activity are detected by fluorescence (GFP or mCherry, green, C-J; single channel, white, C′-J′) in crystal cell precursors marked by expression of Lz (red, C-J). See supplementary material Fig. S1 for additional NRE expression. Scale bar: 10 μm.
Requirement for Lz/Runx
The differences between the Notch-responsive genes in Kc and DmD8 cells suggests that intrinsic factors alter which target enhancers can be regulated. One approach to identify such co-regulators is to look for motifs that are enriched within the regions bound by Su(H) in each cell type. Taking as our input the 300 bp flanking the mid-point of each Su(H) ChIP peak (a 600 bp window), we used CONSENSUS Matrix-based motif discovery in RSAT to look for over-represented motifs (Thomas-Chollier et al., 2011). This returned three assemblies. The first, CGTGGGAA, corresponds to Su(H) motif. The second, GATAAAGT, resembles a GATA-factor binding site, implicating Serpent (or one of the three other Drosophila GATA factors). The third, ACCATAGT, a variant on the established Runx motif suggesting that Lz could be a co-factor, was detected under a subset of conditions. We therefore used a position weight matrix to locate putative binding motifs for Lz/Runx and GATA, and analysed their relationship to Su(H) motifs in regions defined by Kc peaks, using for comparison the motif for Twist (Twi), a muscle-specific transcription factor. Approximately 40% of Su(H) motifs in Kc peaks were within 100 bp of Lz and 36% were within 100 bp of a GATA site, whereas fewer than 25% had a Twi site in similar proximity (under conditions where the total numbers of each motif in the genome were similar; Fig. 4A). Indeed, 49% of the assigned genes are associated with Kc peaks that contain GATA and Lz motifs, as well as Su(H). A further 17% are associated with peaks containing only Su(H) and Lz motifs, and 13% are associated with peaks containing only Su(H) and GATA motifs. By contrast, fewer than 39% of APGs in DmD8 muscle-related cells have GATA and Lz in proximity to Su(H), whereas 57% have a Twi motif. The highly conserved Lz/Runx and GATA factors may therefore provide the crucial specificity for Notch responses in Kc cells/haemocytes.
One hypothesis is that Lz (and GATA) are necessary to make the enhancers capable of responding to Su(H)/Notch. As we were interested whether targets required cooperation between Notch and Runx, we focused our analysis on Lz, for which there was a monoclonal antibody that was suitable for ChIP. First, we investigated whether Lz was present at candidate enhancers prior to Notch activation. Fragments that encompass Lz motifs in peak regions from klu, CG32369, rgr, peb/hnt were significantly enriched in the Lz ChIP, compared with control regions (Fig. 4B). We therefore asked whether reducing Lz, by treating cells with RNAi (Fig. 1E), would interfere with recruitment of Su(H) to target enhancers. Little residual Lz binding was detectable at target enhancers in Lz RNAi-treated cells (Fig. 4B). Su(H) binding at klu, CG32369, rgr, peb/hnt enhancers was similarly compromised in Lz-depleted cells (Fig. 4C), whereas it remained unchanged at E(spl)mβ, a region not linked to Lz sites (Fig. 4C). Conversely, there was no reduction in Lz binding in Su(H) RNAi-treated cells (supplementary material Fig. S4). Altogether, these results argue that Lz binding precedes recruitment of the Su(H)/Notch complex, and that it enhances Su(H) recruitment to target enhancers.
To investigate the mechanism of Su(H) recruitment by Lz, we first checked for direct interactions by co-immunoprecipitation, but were unable to detect any co-purification of Su(H) in Lz immunoprecipitates or vice versa (supplementary material Fig. S4). Second, we assessed whether Lz had an influence on chromatin accessibility, as measured by sensitivity to DNase I treatment. Focusing initially on klu, we tested the effects from two different concentrations of DNase I on digestion at different sites across the locus. This revealed that two sites were more sensitive than others, the promoter and the NRE (Fig. 4D). We subsequently tested whether this sensitivity was altered in Lz RNAi-treated cells and found that the NRE was less sensitive to DNase I under these conditions, suggesting that Lz alters the DNA accessibility (Fig. 4D). Finally, we tested whether other enhancers showed similar Lz-dependant DNase I accessibility. Although the effects were most robust with klu, both CG32369 and rgr also showed some decrease in accessibility following Lz RNAi (Fig. 4E). However, we were not able to detect similar sensitivity at the peb/hnt NRE (Fig. 4E). Taken together, the data indicate therefore that the effects of Lz on Su(H) recruitment are likely to be indirect and most likely involve a change in the chromatin accessibility at least at some loci, such as klu.
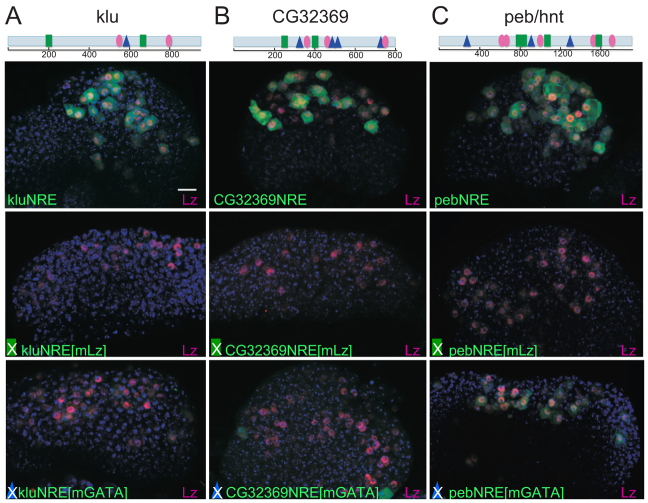
To address whether Notch is required in combination with Lz, we compared the consequences on Peb/Hnt expression of perturbing Notch function in Lz-expressing cells using a dominant-negative form of Mam (MamDN) with the effect of LzEnR, a constitutive repressor form of Lz (Wildonger et al., 2005). The MamDN peptide occupies the Mam-binding groove on the Su(H)-NICD complex, blocking the functional Mam protein, an essential co-activator of Notch, from binding. Expression of MamDN alone or of LzEnR was sufficient to severely reduce levels of Peb/Hnt expression (e.g. Fig. 4F,G). Only 56.5% of Lz>GFP-expressing cells stained positive for Peb/Hnt in the presence of MamDN, and only 66.6% in the presence of LzEnR, compared with 97.2% in control glands (supplementary material Fig. S5). Importantly, co-expression of wild-type Lz with MamDN was unable to rescue the Peb/Hnt expression (Fig. 4H). Thus, reduced Notch activity cannot be compensated for by increased Lz. This implies that the combination of Notch activity and Lz is required for Peb/Hnt expression and establishes that the phenotype caused by MamDN cannot be attributed to failure in Lz. To confirm the relevance of Lz/Runx in vivo, we tested the consequences of mutating the identified Lz motifs on expression from the klu, peb/hnt and CG32369 NREs (Fig. 5). Expression from all three NREs was abolished when Lz/Runx sites were eliminated (kluNRE[mLz], CG32369NRE[mLz], pebNRE[mLz]; Fig. 5). Altogether, these data indicate that Lz confers specificity on the Notch response in differentiating haemocytes. We note that klu is also regulated by Lz in another context, through a different enhancer (Wildonger et al., 2005).
Fig. 5.
Notch-responsive enhancers require Lz and Srp sites for full activity. (A-C) Consequences on indicated NRE activity of mutating Lz (mLz) or GATA (mGATA) motifs. Expression of the resulting GFP reporters (green) in crystal cell lineage (α-Lz, purple). Diagrams above depict the location of the Lz-(green rectangles) and GATA (blue triangles)-binding motifs within each enhancer relative to Su(H) motifs (pink ovals). Scale bar: 10 μm.
Finally we also tested the relevance of GATA motifs for expression from klu, peb/hnt and CG32369 NREs. Expression was reduced by mutations disrupting the GATA motifs (Fig. 5), but with variable effects: CG32369NRE[mGATA] lost all detectable expression, kluNRE[mGATA] exhibited low level expression in few cells and peb/hntNRE[mGATA] had moderate levels of activity. Nevertheless, the results make it likely that a GATA factor, possibly Srp, also cooperates with Notch on these enhancers.
Inhibition of alternate plasmatocyte fates by the Notch target klu
The identified Kc Notch targets are expressed in response to Notch and Lz in the crystal cell lineage. So far, relatively little is known about events downstream of Notch activation in these cells, although recent studies have demonstrated that Notch is required not only for crystal cell specification, but also for their expansion and maintenance (Mukherjee et al., 2011). Target gene functions should therefore reveal how Notch implements its role in promoting the specific differentiation programme.
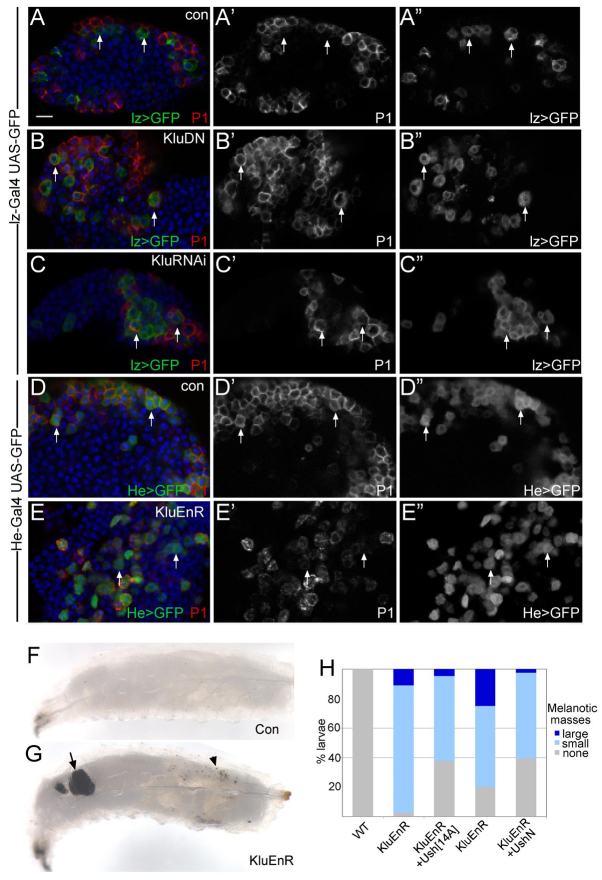
klu is one of the genes that is most highly upregulated in Kc cells and encodes a zinc-finger protein related to the Wilms Tumor 1 (WT-1)/Early growth response (EGR) gene family, which regulate differentiation in haematopoetic lineages (Alberta et al., 2003; Friedman, 2007; Georgescu et al., 2008; Klein and Campos-Ortega, 1997) and which exhibit altered activity in acute myeloid and lymphoid leukaemias (see Huff, 2011; Tosello et al., 2009; Yang et al., 2007). We investigated whether klu has a similar crucial role in haemocytes by expressing a dominant-negative form of the Klu protein (KluDN; Kaspar et al., 2008; Klein and Campos-Ortega, 1997) in the developing crystal cells with lz-Gal4. A characteristic of lz-Gal4-expressing cells is that they are devoid of P1, a marker for the alternate plasmatocyte lineage (Fig. 6A) (Krzemien et al., 2010; Minakhina and Steward, 2010). Strikingly, in the presence of KluDN, many lz-Gal4-expressing cells were found to express P1 (Fig. 6B), although they retained Lz (supplementary material Fig. S6). Similarly, we observed P1 in Lz-expressing cells in lymph glands from klu mutant larvae (kluG4/klu212lR51C; supplementary material Fig. S7) and when klu was ablated using short hairpin RNAi (Fig. 6C). These results suggest that Klu is important for repressing the alternate plasmatocyte fate in crystal cell precursors.
Fig. 6.
Regulation of klu by Notch blocks plasmatocyte fates. (A-A″) Expression of the plasmatocye marker P1 (red, A; white, A′) does not overlap with Lz (lz>GFP, green in A; white in A″) in wild-type glands (arrows). (B-C″) P1 is detected in Lz cells that express KluDN (B-B″, e.g. arrows) or kluRNAi (C-C″, arrows), labelled as in A. (D-D″) Expression of P1 is detected in all He-Gal4-expressing cells (He-Gal4 UAS-nlsGFP, He>GFP) in control glands (arrows). (E-E″) Expression of KluEnR compromises P1 expression in many of the He>GFP cells (arrows). (F,G) Expression of KluEnR leads to melanotic masses, (F) wild-type larvae, (G) KluEnR-expressing larvae with both large (arrows) and small (arrowhead) small masses. (H) Larvae were scored for melanotic masses (large, larvae had at least one large mass; small, larvae had only small dots of melanin). Co-expression of Ush reduces the percentage of KluEnR larvae that exhibit melanotic masses. Over 80 larvae were scored for each genotype. See supplementary material Fig. S6 for Lz expression in KluEnR. Scale bar: 10 μm.
Studies of Klu function in PNS development demonstrated that a constitutive repressor form of Klu, KluEnR, behaved as a strong gain of function for Klu when overexpressed (Kaspar et al., 2008). We therefore tested the consequences of KluEnR expression using He-Gal4, which drives expression in P1-expressing cells (Fig. 6D). Many of the KluEnR-expressing cells had reduced P1 expression (Fig. 6E, arrows) and those with significant residual P1 expression exhibited altered morphology (Fig. 6E). Furthermore, many larvae showed striking accumulations of melanotic cells, resembling melanotic tumours (Fig. 6F-H). However, the KluEnR expression was not sufficient to fully convert the cells to crystal cells, as there was little increase in the number of He>GFP cells that contained Peb/Hnt or Lz (supplementary material Fig. S6). Thus, KluEnR suppresses plasmatocyte characteristics but is not sufficient to fully direct the cells into crystal cell lineage.
One possible mechanism through which Klu might prevent plasmatocyte differentiation is by repressing ush, the Drosophila homologue of FOG1, because a decrease in ush expression is thought to be important for crystal cell maturation in the embryo (Fossett et al., 2003; Fossett et al., 2001; Waltzer et al., 2003). We therefore investigated whether, by restoring Ush in the presence of KluEnR, we could suppress the melanotic cell masses. The number of larvae with melanotic masses was significantly reduced in combination with ush (Fig. 6H), supporting the hypothesis that one mechanism through which Klu prevents plasmatocyte fates is by antagonizing ush. Although expression of ush (ush-lacZ) was widespread and heterogeneous in the lymph gland (supplementary material Fig. S7), the majority of Lz-positive cells had little or no ush-lacZ expression, consistent with the diminished expression in crystal cells observed in the embryo (supplementary material Fig. S7). Broad (pxn-Gal4) expression of KluDN alone (supplementary material Fig. S7) or MamDN alone (supplementary material Fig. S7) had little effect on ush expression in Lz-positive cells, although there was more variability in levels in the former. However, combined overexpression of kluDN and mamDN together resulted in a significant increase in the number of Lz-expressing cells exhibiting strong ush-lacZ (30% of Lz-expressing cells; supplementary material Fig. S7). This suggests that Ush may be one factor involved in the switch downstream of Klu, but suggests that its regulation may require additional inputs from Notch.
Other Notch targets implement cellular programmes associated with crystal cell fates
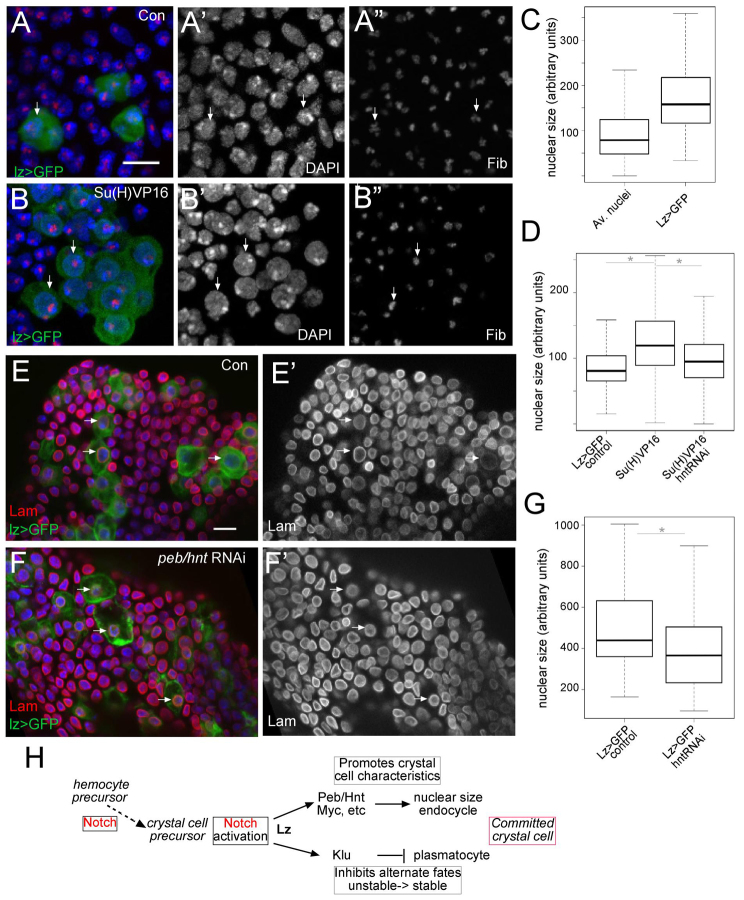
One characteristic of crystal cells is that they undergo DNA replication but do not appear to be mitotically active, suggesting that they are in endocycle (Krzemien et al., 2010). In the follicular epithelium, peb/hnt is required downstream of Notch for implementing the switch to endoreplication (Sun and Deng, 2007; Sun et al., 2008). We therefore considered whether it might function similarly in the lymph gland. Endocycling cells possess an increased DNA content relative to neighbouring mitotic cells, evident in the DAPI staining. We first investigated whether Notch activity affected the DNA content in lymph-gland nuclei, by expressing Su(H)VP16 in Lz-expressing cells (lz>GFP). This had the striking consequence of increasing the dimensions of the nucleus (based on DAPI staining and nuclear Lamin; Fig. 7A,B,E) and also enlarged the patch of Fibrillarin (Fig. 7A″,B″), a nucleolar marker. We noted that there was also considerable heterogeneity among the lz>GFP population even in the absence of additional Su(H)VP16 (Fig. 7C), suggesting that nuclear enlargement is a feature of their differentiation. To investigate whether peb/hnt has a role in this nuclear enlargement, we ablated peb/hnt, by expressing peb/hnt-RNAi, in the presence of the Su(H)VP16. This resulted in a significant reduction in the nuclear dimensions compared with expression of Su(H)VP16 alone (Fig. 7D; supplementary material Fig. S8). Thus, peb/hnt is in part responsible for the effects of Su(H)VP16 on nuclear size. Then, we examined the consequences of perturbing peb/hnt on nuclear dimensions in the largest lz>GFP-expressing cells (Fig. 7E,F). Knockdown of peb/hnt led to a significant reduction in the nuclear diameters in these cells, consistent with a role in promoting nuclear enlargement associated with endoreplication (Fig. 7G).
Fig. 7.
Notch regulates nuclear size via Hnt. (A-B″) Crystal cell lineage marked by lz>GFP (green, e.g. arrows) in control (A) and Su(H)VP16-expressing (B) lymph gland stained to detect DNA (DAPI, blue, single channel A′,B′) and nucleolus (Fibrillarin, red, single channel A″,B″). Expression of Su(H)VP16 results in enlarged nuclei (compare arrows). (C) Size of nuclei based on DAPI staining in lz>GFP-expressing lymph glands, GFP-expressing nuclei (crystal cell lineage) are larger than average (P=3.331e-14, 15 LG, 75 Lz+ cells and 1833 non Lz+ cells). Box and whisker plot. Horizontal line indicates median, box indicates interquartile range (IQR), and whiskers indicate maximum and minimun within 1.5 IQR. (D) The measurable increase in nuclei (DAPI) size obtained by expressing Su(H)VP16 (with lz-Gal4) is suppressed by co-expressing RNAi targeting peb/hnt or myc. RNAi targeting white was used as control (nuclear size in C,D was calculated by measuring the diameter/volume of DAPI staining; 15 LG, 732 cells for Lz>+; 11 LG, 1325 cells for LzG4>Su(H)VP16+whiteRNAi; 17 LG, 527 cells for LzG4>Su(H)VP16+hntRNAi). Asterisk indicates that results were significantly different, P<2.2e-16, using used two-sample Kolmogorov-Smirnov test. Numbers are arbitrary units that differ between experiments owing to the method used to obtain images. Box and whisker plot as in C. (E-F″) Nuclear diameters (nuclear Lamin, red E,F; single channel E′,F′) in large lz>GFP crystal cells from control (E, whiteRNAi; F, peb/hntRNAi). (G) Knockdown of peb/hnt leads to a reduction in nuclear diameter. Asterisk indicates that results were significantly different, P=0.004799, using used two-sample Kolmogorov-Smirnov test (11 LG per genotype, 60 Lz+ cells for controls and 47 Lz+ cells for hntNRAi). Box and whisker plot as in C. (H) The proposed role of Notch acting in combination with Lz in the crystal cell lineage. Notch activity is also required at earlier stages in haemocyte development, where the target genes are likely to differ, as it will operate in a different context. Scale bar: 10 μm.
Myc (also known as dimutive) is another of the Kc Notch targets, which is implicated in cell growth and regulates polyploidy in some tissues (Maines et al., 2004; Pierce et al., 2004). Indeed when overexpressed in Lz>GFP cells, Myc caused an increase in cell, nuclear and nucleolus size (supplementary material Fig. S8). When we assessed the consequences of myc-RNAi on the Su(H)VP16 phenotype, there was also a reduction in nuclear size (supplementary material Fig. S8). Therefore, it appears that myc may also be important in implementing the Notch-dependent changes in nuclear size/ploidy that appear intrinsic to the crystal cell differentiation programme, although quantitative measurements of DNA synthesis in individual lz>GFP cells will be needed to confirm its role.
DISCUSSION
As signalling pathways, such as Notch, are used iteratively during development, one of the challenges is to understand their context-specific effects. Taking a genome-wide approach, we have elucidated the transcriptional outputs of Notch in Drosophila Kc cells, a model for haemocyte development, and shown that, in vivo, the targets are involved in locking cells into a specific fate, by shutting down the alternatives and by promoting specific characteristics of the differentiated cells (Fig. 7H). The context specificity for this programme is provided by the Runx factor Lz, probably acting in combination with GATA factors, as mutation of Lz or GATA sites eliminates expression from the Notch-regulated enhancers. Furthermore, ablation of Lz compromises the recruitment of Su(H) to these targets enhancers. Thus, Lz appears to act as a lineage master regulator for Notch in the manner proposed for cell-specific factors acting with BMP and Wnt signals in regeneration of haematopoietic lineages (Trompouki et al., 2011) and with TGFβ in differentiation (Mullen et al., 2011).
Our analysis of haemocyte Su(H) targets has thus uncovered a cooperative activity between Lz and Notch. In most previous studies, Notch and Runx have been shown to act in a hierarchical manner, with Notch functioning upstream of Runx/Lz (Burns et al., 2009; Burns et al., 2005; Lebestky et al., 2003; Nottingham et al., 2007; Robert-Moreno et al., 2005). However, a similar cooperative mechanism may operate at late stages in thymocyte development, where Runx1 confers the capability to promote T-ell fate in response to Notch activity (Guo et al., 2008). Analysis of Notch-regulated targets in T-ALL cells also uncovered a signature that was suggestive of Notch and Runx co-regulation in these malignant cells (Wang et al., 2011). Although Lz/Runx expression is itself dependent on Notch activity, there is as yet no evidence to suggest direct regulation in other systems and we have not detected Lz as a Su(H)-bound target in our experiments, making it plausible that the regulation by Notch is indirect. Nevertheless, the observation that Lz/Runx is required first downstream of Notch and second in combination with Notch at target enhancers suggests that a feed-forward ratchet mechanism may contribute to cell fate specification both in Drosophila crystal cells and mammalian lineages. This also highlights the fact that Notch is needed at several different stages in such lineages, and that the specific targets regulated are likely to differ depending on the stage.
Among the newly identified Notch-regulated genes, many are transcription factors (Klu, Peb/Hnt, Myc, Hnf4, Rgr, p53) whose expression and functions illustrate their importance for the crystal cell differentiation programme. Strikingly, our analysis and manipulation of Klu, a zinc-finger protein related to the ERG/WT1 family (Klein and Campos-Ortega, 1997), revealed that it is necessary to inhibit alternate cell fates in the Lz-expressing cells. In some respects, this is surprising as lineage-tracing experiments indicated that most, if not all, Lz-expressing cells were fated to become crystal cells in healthy animals owing to signalling events at much earlier stages (Krzemien et al., 2010; Minakhina and Steward, 2010). Why is Klu required? One possibility is that Lz+ cells are in a transitional state prior to Notch activation/Klu expression, retaining the potential to adopt alternate fates depending on environmental challenges. For example, hypoxia appears to be important for crystal cell differentiation to be maintained (Mukherjee et al., 2011) and wasp infection can reroute differentiation to lamellocyte fates (e.g. Krzemień et al., 2007). Notch-induced upregulation of Klu may be important to lock cells into the crystal cell programme, by inhibiting activity of alternate lineage regulators. A similar role has been proposed for the mammalian homologues EGR1/2, which are part of an antagonistic regulatory circuit that maintains lineage fidelity, downstream of ‘pioneer’ transcription factors, by repressing alternate fate choices (Laslo et al., 2006).
Other targets appear to be actively involved in promoting the crystal cell differentiation programme. In particular, the transcription factor Peb/Hnt is important for the change in nuclear size/DNA content that probably reflects endoreplication in crystal cells (Krzemien et al., 2010), a process that may also require Myc. As both Peb/Hnt and Myc are involved in regulating the switch from mitotic to endocycle downstream of Notch in the Drosophila follicle cells (Maines et al., 2004; Shcherbata et al., 2004; Sun and Deng, 2007; Sun et al., 2008), this may be a conserved cassette that is deployed at multiple stages in development. Elsewhere, Myc has been shown to regulate polyploidy and differentiation in several cell types, including megakaryocytes and in T-ALL cells (e.g. Munoz-Alonso et al., 2012; Palomero et al., 2006; Pierce et al., 2004; Zanet et al., 2005). Other targets include Hnf4, which was previously implicated in causing haemocyte expansion downstream of AML (Runx)-ETO (Sinenko et al., 2010), and several genes involved in cell motility (e.g. CLIP-120), as well as other conserved genes with unknown functions, such as CG32369 (LONRF1-3).
A comparison with Notch-regulated genes in a different cell type (DmD8 cells) demonstrates that differences in Su(H) binding, elicited by cooperating transcription factors such as Lz, are likely to be the major factor in determining the outcome from Notch activation. Nevertheless, despite these differences, a cohort of genes was upregulated in both cell types. Besides the well characterized E(spl) genes, other common targets included myc and Notch itself. myc has emerged as a frequent and important target of Notch in several tissues, as well as in cancers (Klinakis et al., 2006; Palomero et al., 2006; Song and Lu, 2011; Weng et al., 2006). Positive feedback on Notch expression has also been observed in T-cell precursors (Yashiro-Ohtani et al., 2009), as well as in C. elegans (Christensen et al., 1996), and is likely to be important in maintaining Notch receptor levels in signalling cells as the process of activation results in destruction of the receptor (Kopan and Ilagan, 2009). Another common target in Kc and DmD8 cells encodes Rgl, a member of the RalGEF family that is suggested to couple Ras to Ral signalling (Ferro and Trabalzini, 2010). The shared and related targets from the Kc and DmD8 cells may thus identify core elements in the Notch response that are relevant in many different contexts.
Despite these similarities, it is clear that there is a context-specific response to Notch activation in Drosophila haemocytes that ensures proper cell fate specification and stabilization in the crystal cell lineage (Fig. 7H). Cooperating with the lineage-determining factor Lz, Notch activation antagonizes alternate cell fates, by eliciting expression of Klu, and promotes key aspects of the differentiation programme, through other targets. Lz may therefore be crucial in providing a transcriptional ‘priming’, converting cells into a transitional state that can then be canalized by the Notch activation.
Supplementary Material
Acknowledgments
We are very grateful to Bettina Fischer, Steve Russell and FlyChip for their help with the genome-wide arrays, and to Boris Adryan and Robert Stojnic for advice over motif analysis. We thank I. Ando, Utpal Banerjee, Nancy Fosset,Pascal Heitzler, Paul Martin, Thomas Klein, Barry Yebovnik and other members of the fly community for sharing fly stocks and antibodies, as well as the Bloomington Stock Center and the Developmental Hybridoma Bank.
Footnotes
Funding
This work was supported by Medical Research Council programme grant to S.J.B. [G0800034], S.C. was funded by a Biotechnology and Biological Sciences Research Council studentship, J.L. by China Scholarship Council Cambridge Scholarship and I.R. by Genetics Society summer studentship. Research in A.K.'s lab is supported by Grantová agentura České republiky [P305/11/0126].
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.086785/-/DC1
References
- Alberta J. A., Springett G. M., Rayburn H., Natoli T. A., Loring J., Kreidberg J. A., Housman D. (2003). Role of the WT1 tumor suppressor in murine hematopoiesis. Blood 101, 2570-2574 [DOI] [PubMed] [Google Scholar]
- Benmimoun B., Polesello C., Waltzer L., Haenlin M. (2012). Dual role for Insulin/TOR signaling in the control of hematopoietic progenitor maintenance in Drosophila. Development 139, 1713-1717 [DOI] [PubMed] [Google Scholar]
- Bernard F., Krejci A., Housden B., Adryan B., Bray S. J. (2010). Specificity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development 137, 2633-2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. (2006). Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16, 595-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J. (1997). Expression and function of Enhancer of split bHLH proteins during Drosophila neurogenesis. Perspect. Dev. Neurobiol. 4, 313-323 [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689 [DOI] [PubMed] [Google Scholar]
- Burns C. E., Traver D., Mayhall E., Shepard J. L., Zon L. I. (2005). Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331-2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. E., Galloway J. L., Smith A. C., Keefe M. D., Cashman T. J., Paik E. J., Mayhall E. A., Amsterdam A. H., Zon L. I. (2009). A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood 113, 5776-5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L., Willingham A., Zhang D., Yang L., Zou Y., Eads B. D., Carlson J. W., Landolin J. M., Kapranov P., Dumais J., et al. (2011). The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 21, 301-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S., Kodoyianni V., Bosenberg M., Friedman L., Kimble J. (1996). lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122, 1373-1383 [DOI] [PubMed] [Google Scholar]
- Crozatier M., Meister M. (2007). Drosophila haematopoiesis. Cell. Microbiol. 9, 1117-1126 [DOI] [PubMed] [Google Scholar]
- Cubadda Y., Heitzler P., Ray R. P., Bourouis M., Ramain P., Gelbart W., Simpson P., Haenlin M. (1997). u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 11, 3083-3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Turner D. L. (2001). Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20, 8342-8357 [DOI] [PubMed] [Google Scholar]
- Echalier G., Ohanessian A. (1969). [Isolation, in tissue culture, of Drosophila melangaster cell lines]. C.R. Hebd. Seances Acad. Sci., Ser. D, Sci. Nat. 268, 1771-1773 [PubMed] [Google Scholar]
- Evans C. J., Banerjee U. (2003). Transcriptional regulation of hematopoiesis in Drosophila. Blood Cells Mol. Dis. 30, 223-228 [DOI] [PubMed] [Google Scholar]
- Ferro E., Trabalzini L. (2010). RalGDS family members couple Ras to Ral signalling and that’s not all. Cell. Signal. 22, 1804-1810 [DOI] [PubMed] [Google Scholar]
- Fischer A., Gessler M. (2007). Delta-Notch – and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 35, 4583-4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N., Zhang Q., Gajewski K., Choi C. Y., Kim Y., Schulz R. A. (2000). The multitype zinc-finger protein U-shaped functions in heart cell specification in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 97, 7348-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N., Tevosian S. G., Gajewski K., Zhang Q., Orkin S. H., Schulz R. A. (2001). The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc. Natl. Acad. Sci. USA 98, 7342-7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N., Hyman K., Gajewski K., Orkin S. H., Schulz R. A. (2003). Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc. Natl. Acad. Sci. USA 100, 11451-11456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. D. (2007). Transcriptional control of granulocyte and monocyte development. Oncogene 26, 6816-6828 [DOI] [PubMed] [Google Scholar]
- Furriols M., Bray S. J. (2000). Dissecting the mechanisms of suppressor of hairless function. Dev. Biol. 227, 520-532 [DOI] [PubMed] [Google Scholar]
- Georgescu C., Longabaugh W. J., Scripture-Adams D. D., David-Fung E. S., Yui M. A., Zarnegar M. A., Bolouri H., Rothenberg E. V. (2008). A gene regulatory network armature for T lymphocyte specification. Proc. Natl. Acad. Sci. USA 105, 20100-20105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M., Patient R. (2010). Notch signalling and haematopoietic stem cell formation during embryogenesis. J. Cell. Physiol. 222, 11-16 [DOI] [PubMed] [Google Scholar]
- Guo Y., Maillard I., Chakraborti S., Rothenberg E. V., Speck N. A. (2008). Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T-cell specification. Blood 112, 480-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001). Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276, 34371-34378 [DOI] [PubMed] [Google Scholar]
- Haenlin M., Cubadda Y., Blondeau F., Heitzler P., Lutz Y., Simpson P., Ramain P. (1997). Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11, 3096-3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi H., Gustafason D., Pellegrini M., Gasson J. (2011). Identification of novel targets of CSL-dependent Notch signaling in hematopoiesis. PLoS ONE 6, e20022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms W., Lee H., Ammerman M., Parks A. L., Muskavitch M. A., Yedvobnick B. (1999). Engineered truncations in the Drosophila mastermind protein disrupt Notch pathway function. Dev. Biol. 215, 358-374 [DOI] [PubMed] [Google Scholar]
- Hertz G. Z., Stormo G. D. (1999). Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563-577 [DOI] [PubMed] [Google Scholar]
- Housden B. E., Millen K., Bray S. J. (2012). Drosophila reporter vectors compatible with PhiC31 integrase transgenesis techniques and their use to generate new notch reporter fly lines. G3 (Bethesda), 2, 79-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V. (2011). Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat. Rev. Cancer 11, 111-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson Behan K., Fair J., Singh S., Bogwitz M., Perry T., Grubor V., Cunningham F., Nichols C. D., Cheung T. L., Batterham P., et al. (2005). Alternative splicing removes an Ets interaction domain from Lozenge during Drosophila eye development. Dev. Genes Evol. 215, 423-435 [DOI] [PubMed] [Google Scholar]
- Jung S. H., Evans C. J., Uemura C., Banerjee U. (2005). The Drosophila lymph gland as a developmental model of hematopoiesis. Development 132, 2521-2533 [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243-1251 [DOI] [PubMed] [Google Scholar]
- Kaspar M., Schneider M., Chia W., Klein T. (2008). Klumpfuss is involved in the determination of sensory organ precursors in Drosophila. Dev. Biol. 324, 177-191 [DOI] [PubMed] [Google Scholar]
- Klein T., Campos-Ortega J. A. (1997). klumpfuss, a Drosophila gene encoding a member of the EGR family of transcription factors, is involved in bristle and leg development. Development 124, 3123-3134 [DOI] [PubMed] [Google Scholar]
- Klinakis A., Szabolcs M., Politi K., Kiaris H., Artavanis-Tsakonas S., Efstratiadis A. (2006). Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 103, 9262-9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M. X. (2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R. A. (2008). More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27, 5099-5109 [DOI] [PubMed] [Google Scholar]
- Krejcí A., Bray S. (2007). Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21, 1322-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A., Bernard F., Housden B. E., Collins S., Bray S. J. (2009). Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2, ra1 [DOI] [PubMed] [Google Scholar]
- Krzemień J., Dubois L., Makki R., Meister M., Vincent A., Crozatier M. (2007). Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature 446, 325-328 [DOI] [PubMed] [Google Scholar]
- Krzemien J., Oyallon J., Crozatier M., Vincent A. (2010). Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev. Biol. 346, 310-319 [DOI] [PubMed] [Google Scholar]
- Kurucz E., Zettervall C. J., Sinka R., Vilmos P., Pivarcsi A., Ekengren S., Hegedüs Z., Ando I., Hultmark D. (2003). Hemese, a hemocyte-specific transmembrane protein, affects the cellular immune response in Drosophila. Proc. Natl. Acad. Sci. USA 100, 2622-2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P., Spooner C. J., Warmflash A., Lancki D. W., Lee H. J., Sciammas R., Gantner B. N., Dinner A. R., Singh H. (2006). Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126, 755-766 [DOI] [PubMed] [Google Scholar]
- Lebestky T., Jung S. H., Banerjee U. (2003). A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev. 17, 348-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hibbs M. A., Gard A. L., Shylo N. A., Yun K. (2012). Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells 30, 741-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunstrum G. P., Bächinger H. P., Fessler L. I., Duncan K. G., Nelson R. E., Fessler J. H. (1988). Drosophila basement membrane procollagen IV. I. Protein characterization and distribution. J. Biol. Chem. 263, 18318-18327 [PubMed] [Google Scholar]
- Maillard I., Fang T., Pear W. S. (2005). Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23, 945-974 [DOI] [PubMed] [Google Scholar]
- Maines J. Z., Stevens L. M., Tong X., Stein D. (2004). Drosophila dMyc is required for ovary cell growth and endoreplication. Development 131, 775-786 [DOI] [PubMed] [Google Scholar]
- Minakhina S., Steward R. (2010). Hematopoietic stem cells in Drosophila. Development 137, 27-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee T., Kim W. S., Mandal L., Banerjee U. (2011). Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science 332, 1210-1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A. C., Orlando D. A., Newman J. J., Lovén J., Kumar R. M., Bilodeau S., Reddy J., Guenther M. G., DeKoter R. P., Young R. A. (2011). Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565-576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Alonso M. J., Ceballos L., Bretones G., Frade P., León J., Gandarillas A. (2012). MYC accelerates p21CIP-induced megakaryocytic differentiation involving early mitosis arrest in leukemia cells. J. Cell. Physiol. 227, 2069-2078 [DOI] [PubMed] [Google Scholar]
- Muratoglu S., Hough B., Mon S. T., Fossett N. (2007). The GATA factor Serpent cross-regulates lozenge and u-shaped expression during Drosophila blood cell development. Dev. Biol. 311, 636-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R. E., Fessler L. I., Takagi Y., Blumberg B., Keene D. R., Olson P. F., Parker C. G., Fessler J. H. (1994). Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 13, 3438-3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham W. T., Jarratt A., Burgess M., Speck C. L., Cheng J. F., Prabhakar S., Rubin E. M., Li P. S., Sloane-Stanley J., Kong-A-San J., et al. (2007). Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110, 4188-4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajcini K. V., Speck N. A., Pear W. S. (2011). Notch signaling in mammalian hematopoietic stem cells. Leukemia 25, 1525-1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Lim W. K., Odom D. T., Sulis M. L., Real P. J., Margolin A., Barnes K. C., O’Neil J., Neuberg D., Weng A. P., et al. (2006). NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 103, 18261-18266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Yost C., Britton J. S., Loo L. W., Flynn E. M., Edgar B. A., Eisenman R. N. (2004). dMyc is required for larval growth and endoreplication in Drosophila. Development 131, 2317-2327 [DOI] [PubMed] [Google Scholar]
- Radtke F., Fasnacht N., Macdonald H. R. (2010). Notch signaling in the immune system. Immunity 32, 14-27 [DOI] [PubMed] [Google Scholar]
- Rand M. D., Grimm L. M., Artavanis-Tsakonas S., Patriub V., Blacklow S. C., Sklar J., Aster J. C. (2000). Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 20, 1825-1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., de la Pompa J. L., Bigas A. (2005). RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development 132, 1117-1126 [DOI] [PubMed] [Google Scholar]
- Satoh Y., Matsumura I., Tanaka H., Ezoe S., Sugahara H., Mizuki M., Shibayama H., Ishiko E., Ishiko J., Nakajima K., et al. (2004). Roles for c-Myc in self-renewal of hematopoietic stem cells. J. Biol. Chem. 279, 24986-24993 [DOI] [PubMed] [Google Scholar]
- Schneider I. (1972). Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353-365 [PubMed] [Google Scholar]
- Shcherbata H. R., Althauser C., Findley S. D., Ruohola-Baker H. (2004). The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development 131, 3169-3181 [DOI] [PubMed] [Google Scholar]
- Sinenko S. A., Hung T., Moroz T., Tran Q. M., Sidhu S., Cheney M. D., Speck N. A., Banerjee U. (2010). Genetic manipulation of AML1-ETO-induced expansion of hematopoietic precursors in a Drosophila model. Blood 116, 4612-4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Lu B. (2011). Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev. 25, 2644-2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino R. P., Tokusumi T., Schulz R. A. (2007). The Friend of GATA protein U-shaped functions as a hematopoietic tumor suppressor in Drosophila. Dev. Biol. 311, 311-323 [DOI] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M. J., Redd M. J., Jacinto A., Parkhurst S. M., Martin P. (2005). Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168, 567-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng W. M. (2007). Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12, 431-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Smith L., Armento A., Deng W. M. (2008). Regulation of the endocycle/gene amplification switch by Notch and ecdysone signaling. J. Cell Biol. 182, 885-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Defrance M., Medina-Rivera A., Sand O., Herrmann C., Thieffry D., van Helden J. (2011). RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 39, W86-W91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosello V., Mansour M. R., Barnes K., Paganin M., Sulis M. L., Jenkinson S., Allen C. G., Gale R. E., Linch D. C., Palomero T., et al. (2009). WT1 mutations in T-ALL. Blood 114, 1038-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E., Bowman T. V., Lawton L. N., Fan Z. P., Wu D. C., DiBiase A., Martin C. S., Cech J. N., Sessa A. K., Leblanc J. L., et al. (2011). Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G., Vooijs M. (2011). Proteolytic cleavage of Notch: “HIT and RUN”. Curr. Mol. Med. 11, 255-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L., Ferjoux G., Bataillé L., Haenlin M. (2003). Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 22, 6516-6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zou J., Zhao B., Johannsen E., Ashworth T., Wong H., Pear W. S., Schug J., Blacklow S. C., Arnett K. L., et al. (2011). Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc. Natl. Acad. Sci. USA 108, 14908-14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A. P., Millholland J. M., Yashiro-Ohtani Y., Arcangeli M. L., Lau A., Wai C., Del Bianco C., Rodriguez C. G., Sai H., Tobias J., et al. (2006). c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096-2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildonger J., Sosinsky A., Honig B., Mann R. S. (2005). Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 19, 1034-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Han Y., Suarez Saiz F., Minden M. D. (2007). A tumor suppressor and oncogene: the WT1 story. Leukemia 21, 868-876 [DOI] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y., He Y., Ohtani T., Jones M. E., Shestova O., Xu L., Fang T. C., Chiang M. Y., Intlekofer A. M., Blacklow S. C., et al. (2009). Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 23, 1665-1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Riva L., Xie H., Schindler Y., Moran T. B., Cheng Y., Yu D., Hardison R., Weiss M. J., Orkin S. H., et al. (2009). Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36, 682-695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Pibre S., Jacquet C., Ramirez A., de Alborán I. M., Gandarillas A. (2005). Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J. Cell Sci. 118, 1693-1704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.