Abstract
Heterotrimeric guanine nucleotide–binding proteins (G proteins) are positioned at the top of many signal transduction pathways. The G protein α subunit is composed of two domains, one that resembles Ras and another that is composed entirely of α helices. Historically, most attention has focused on the Ras-like domain, but emerging evidence reveals that the helical domain is an active participant in G protein signaling.
Introduction
Most good stories have a strong central character, but they often work with a close confidant or collaborator. Sherlock Holmes relied on Doctor Watson, whose contributions might have escaped the attention of the casual observer. The same could be said of many proteins, protein subunits, and protein domains.
A trusty comrade is always of use; and a chronicler still more so.
—from The Adventures of Sherlock Holmes
Heterotrimeric guanine nucleotide–binding proteins (G proteins) play a central role in signal transduction. Various chemical and environmental signals bind to cell-surface receptors that activate G proteins, which are composed of an α subunit and an obligate βγdimer. The Gα subunits in particular are part of a large and diverse family whose members include the proto-oncogene Ras and the bacterial elongation factor Tu (EF-Tu). In general, the activity of these proteins is dictated by cycles of guanosine triphosphate (GTP) binding and hydrolysis (Fig. 1). Exchange of GTP for guanosine diphosphate (GDP) initiates G protein activation, whereas GTP hydrolysis terminates the signal. Both steps are tightly regulated by accessory proteins. Cell-surface receptors serve as guanine nucleotide exchange factors (GEFs). Conversely, regulator of G protein signaling (RGS) proteins act as guanosine triphosphatase (GTPase)–activating proteins (GAPs). At least one effector enzyme can likewise serve as a GAP (1, 2). Additional proteins can serve as guanine nucleotide dissociation inhibitors (GDIs).
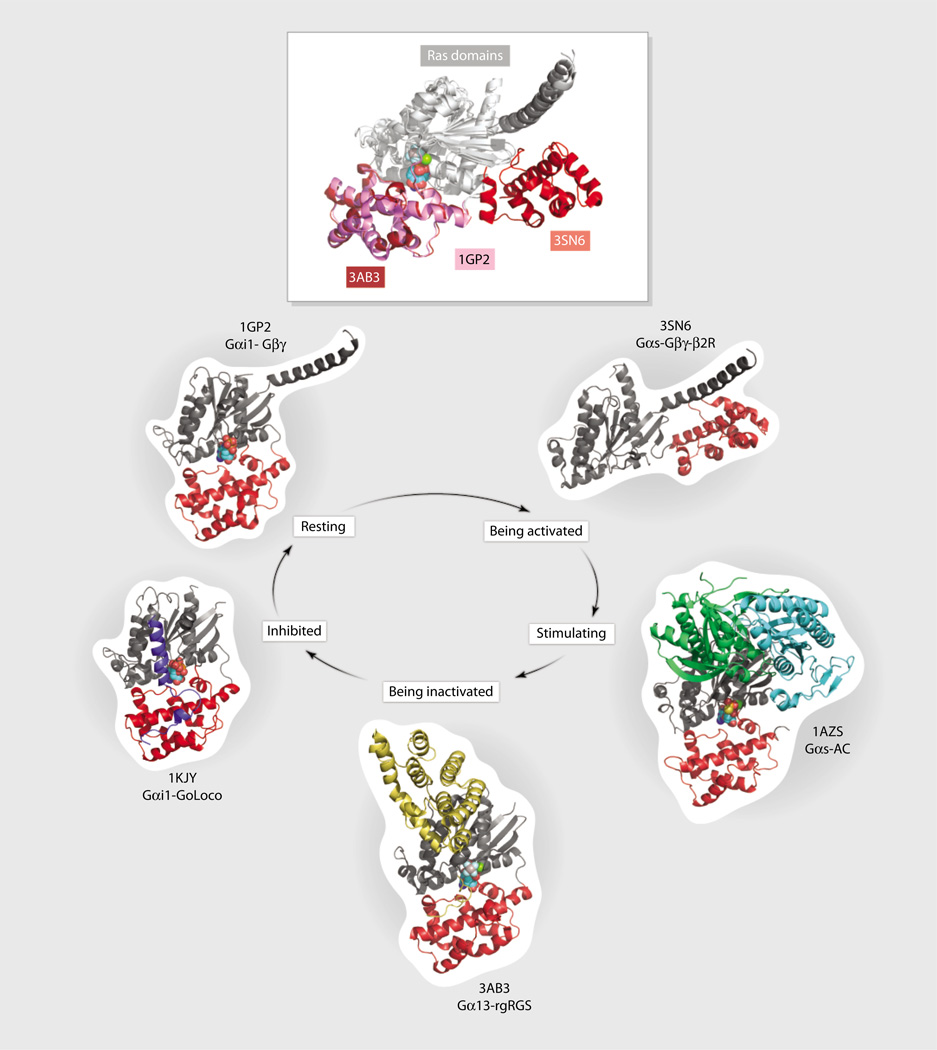
Fig. 1.
The role of the helical domain during the cycle of G protein activation and inactivation. Images of the indicated crystal structures at different stages of G protein activation. Shown are the G protein Ras-like domain (gray), the G protein helical domain (red), the Go-Loco peptide (blue), an adenylyl cyclase fragment (AC, green and cyan), and the RGS fragment from p115RhoGEF (rgRGS, yellow). For reference, the Ras-like domains were aligned and held to the same relative position in each image. In the resting state, the helical domain covers the bound guanine nucleotide. (Inset) Overlay of the helical domain from three different structures shows how it is displaced (relative to the fixed Ras-like domain) during the binding of receptor (red), Gβγ(pink), and RGS protein (burgundy).
Although G proteins are composed of three subunits, it is the α subunit that does much of the heavy lifting. Upon receptor activation, the α subunit undergoes profound conformational changes, liberates the G protein βγsubunits, activates downstream effectors, and binds to the RGS protein. The α subunit is in turn composed of two domains, one that resembles Ras and another that is composed entirely of α helices. For years the Ras-like domain has received almost all of the attention. Here, we focus on newly appreciated functions of the helical domain.
“Data! Data! Data!” he cried impatiently. “I can’t make bricks without clay.”
—from The Adventure of the Copper Beeches
Crystal Structure Data Reveal a Displacement of the G Protein Helical Domain
Three-dimensional (3D) structures of Gα, alone and in complex with Gβγor other binding partners, have provided critical insights into how these molecular “switches” operate. The first crystal structures of Gα revealed substantial changes in three segments known as Switch I, Switch II, and Switch III (3). All three switches lie within the Ras domain, although Switch I (which is also called linker 2) connects the Ras-like domain with the helical domain. These switch regions are primary contact sites for, and dictate the association with, the G βγ subunits (Switch II), RGS proteins, and effectors One exception to the rule is Gαt, for which the helical domain alone binds to an effector enzyme, cyclic guanosine monophosphate (cGMP) phosphodiesterase, and which does so with equivalent apparent affinity and subtype selectivity to that of the full-length Gα protein (4, 5).
All of the available crystal structures indicate that nucleotide binding occurs primarily within the Ras-like domain. Bound GTP or GDP is sequestered between the Ras-like domain and the helical domain; thus, exchange of one nucleotide for the other must depend on the separation of the two domains (Fig. 1). Indeed, early studies revealed that each domain can exist separately and, when recombined, the free exchange of GDP and GTP is slowed (6). Paradoxically, the rate of dissociation of GDP from native Gαi and G αs is greater than that from Ras, which possesses no helical domain. The rapid nucleotide exchange rate of G αrelative to that of Ras is likely a result of the low affinity of Gα-GDP for Mg2+ (7). On the other hand, the helical domain may help to stabilize nucleotide binding in the absence of coordination of Mg2+ (8).
Further findings uncovered additional functions for the helical domain. One insight comes from the determination of the crystal structure of the G protein αsubunit from the plant Arabidopsis thaliana (AtGPA1). The plant G protein represents an interesting counterexample to G proteins in animals, because Arabidopsis appears to lack a canonical G protein–coupled receptor (GPCR). Instead, AtGPA1 activates itself at a rate that is two to three orders of magnitude faster than those of its animal counterparts (9). Detailed structural, biophysical, and molecular dynamics analyses indicate that the helical domain in AtGPA1 is more disordered and dynamic than those in other known G proteins. Could the flexibility within the helical domain account for its spontaneous activation property? Indeed, domain-swapping experiments confirmed that it is the helical domain of AtGPA1 that accounts for its higher basal activity (10).
A major breakthrough came in the form of a crystal structure of the animal G protein Gs in complex with an agonist-occupied β2 adrenergic receptor (Fig. 1) (11). That work confirmed interactions that were long suspected but never demonstrated directly. Specifically, it revealed that direct interactions occurred between the receptor and the N- and C-terminal segments of Gαs but that there were no direct contacts between the receptor and the Gβγsubunits. Within Gα, the largest conformational change caused by the receptor was in the Ras-like domain and in particular the α5 helix, which was rotated and displaced toward the core of the receptor. Previous studies with various approaches had predicted an important role for the α5 helix in the activation of G proteins by receptors (12, 13).
Whereas previous structures of Gα were solved in the presence of GDP, GTP analogs, or the transition state analog GDP-AlF4−, attempts to isolate Gα in the absence of nucleotide proved difficult (14). The receptor-bound complex, in contrast, is nucleotide-free. That structure—caught in the act of activation—reveals a massive displacement of the helical domain away from the Ras-like domain (Fig. 1). Thus, upon receptor binding, the two domains dissociate, exposing the nucleotide-binding pocket. It remains uncertain whether domain dissociation is the cause or consequence of nucleotide release. Nevertheless, the observed changes reveal a plausible mechanism for nucleotide exchange, whereby the two domains open in a clamshell motion to liberate GDP and accommodate GTP. This mechanism has since been corroborated with various complementary biophysical methods.
We must look for consistency. Where there is a want of it we must suspect deception.
—from The Problem of Thor Bridge
Experimental Validation of New Structural Models
A crystal structure is a model (and a static model at that), and any model requires experimental validation. In the case of the receptor–G protein complex, corroboration was swift and persuasive. Even before seeing the receptor and G protein complex, Van Eps et al. had obtained evidence in support of key features of the structure. The authors employed double electron-electron resonance spectroscopy to measure the change in distance between chemical probes positioned on the Ras-like and helical domains of G αi. With this method, they observed a large (up to 20 Å) increase in the distance between domains, compatible with the release of nucleotide after receptor activation (15). In addition, they observed broad distance distributions for several labeling pairs, indicating the existence of multiple conformations in dynamic equilibrium. The distances observed in the nucleotide-free state corresponded to minor populations observed for the nucleotide-bound state. Thus, receptor activation appears to shift the equilibrium from a closed state to an open state(s).
A second strategy entailed measurements of deuterium exchange. By changing the solvent from H2O to D2O, deuterons are incorporated into the amide positions accessible to the solvent. The pattern of deuterium labeling provides information about protein tertiary structure. With this method, it was established that receptor binding promotes an increase in solvent accessibility within the nucleotide-binding site of Gα and throughout the interface between the helical and Ras-like domains. In comparison, there is little exchange within the core of the helical domain itself, which suggests that it remains mostly intact and folded (16). Thus, the deuterium-exchange studies confirmed that the interface between the Ras-like and helical domains becomes exposed after receptor activation.
Finally, a third study used single-particle EM (EM) to visualize the complex of receptor and G protein. With this approach, it was demonstrated that the helical domain occupies various positions relative to the Ras-like domain, receptor, and Gβγ. The helical domain was visible in only about a third of the particles (17), however, and in those instances the position of the helical domain with respect to the Ras domain was similar to that seen in the structures of Gαs-GTPγS and Gαi-GDP/Gβγ(18, 19), but entirely different from the receptor-Gαs crystal complex. These results indicated that the helical domain undergoes a transition from a conformationally stabilized (inactivated) state to a more flexible (activated) state that is invisible to EM.
Thus, a consistent picture emerges from structural and biophysical analysis. These investigations showed that the Ras-like and helical domains undergo a domain separation compatible with the release of nucleotide. But is this motion recapitulated in cells? Resonance energy transfer analysis has been used to monitor the dissociation of G protein domains and subunits in response to receptor activation. Early studies reported a loss of resonance energy transfer between probes in the Gα subunit and probes in either Gβ (20) or Gγ (21). Given the placement of the probes at sites within the helical domain of Gα, and with the benefit of new structural information, it is possible that the loss of interaction was actually due to the movement of the helical domain away from the Ras-like domain and Gβγ (22).
The position of the helical domain in the crystal structure is likely to represent just one of several possible conformations available to the protein. Moreover, Switch domains I and II are inherently flexible and, in some cases, could not be modeled because of a lack of electron density. In studies of the “cytosolic” GEF, Ric-8A, the nucleotide-free G αi displayed virtually no thermal transition upon heating, which suggests the loss of stable tertiary interactions in the nucleotide-free state (23). Similarly, we have speculated that intrinsic flexibility accounts for the ability of the plant G protein to “self-activate,” whereas flexibility in the animal G proteins is regulated by the agonist-bound receptor (10).
Aside from G proteins, there is growing evidence that a substantial proportion of cellular proteins contain segments that are flexible or unstructured in solution. Such “intrinsically disordered proteins” carry out important biological functions that may require the absence of a specific 3D structure (24). Disorder could confer function by increasing the surface area available to binding partners, including enzymes that impart a posttranslational modification. For example, it has been demonstrated that intrinsically disordered proteins are considerably more likely than ordered regions to be phosphorylated in vivo (25).
Singularity is almost invariably a clue.
—from The Boscombe Valley Mystery
Posttranslational Modifications That Target the Helical Domain
Although rare, there are examples of posttranslational modifications within the G protein helical domain. One early paper reported the phosphorylation of a site within the helical domain of Gα2 from Dictyostelium discoideum (Ser113, between the αB and αC helices) (26). Mammalian Src phosphorylates Gαt-GDP in vitro, and the site of phosphorylation was later mapped to the helical domain (Tyr114, between αD and αE) (27). Phosphorylation by Src is likely to be functionally important, given that phosphorylation improves receptor-catalyzed activation of the G protein (26). It remains to be determined if and how these phosphorylation events affect G protein signaling in cells (28).
Much work has centered on the phosphorylation and ubiquitination of Gpa1, a G protein α subunit from the yeast Saccharomyces cerevisiae. Gpa1 is a particularly interesting example because it undergoes monoubiquitination as well as the more common polyubiquitination (that is, the formation of ubiquitin-ubiquitin conjugates). Work spanning the past decade has revealed that mono- and polyubiquitination are mediated by distinct enzymes (29, 30), that these modifications have distinct consequences for Gpa1 trafficking and function (31), and that polyubiquitination is dynamically regulated through phosphorylation (32). All of these modifications occur within the helical domain of Gpa1, and in particular within a unique 109-residue insert close to the αB helix (32, 33). Thus, the helical domain of Gpa1 binds to a number of modifying proteins, including the protein kinase(s) and at least two ubiquitin ligases (29, 30, 32).
Skill is fine, and genius is splendid, but the right contacts are more valuable than either.
—Arthur Conan Doyle, Sr.
Contact Sites in the Helical Domain Contribute to Protein-Binding Specificity
All G protein subtypes exhibit a high degree of sequence similarity. Sequence conservation is greatest within the Ras-like domain, particularly the switch regions, and weakest in the helical domain. This distinction has led to speculation that the helical domain contributes to subtype-selective interactions with at least some binding partners. Here, we highlight examples of contacts with the helical domain that have been documented through change-of-function mutations or crystal structures of G proteins and their binding partners.
Crystal structures of various Gαand RGS pairs have revealed helical domain contacts, including complexes of Gαo with RGS16, G αi1 with RGS4, and G αi3 with RGS8 and RGS10 (34, 35). In each case, the contact sites are few in number (two to three amino acid residues in each protein) but heterogeneous in sequence and thus likely to contribute to Gα -binding selectivity. In support of this idea, domain-swapping experiments revealed that it is the helical domain of Gαt that confers binding selectivity to RGS9 (36). Another prominent example is the receptor kinase GRK2, which has an RGS domain that binds to selected Gα protein subtypes, although it lacks appreciable GAP activity (37). GRK2 binds well to Gα16, as well as to chimeric proteins that contain the helical domain of Gα16 and the Ras-like domain of Gαq, but not to the reciprocal chimeras (38). Through site-directed mutagenesis studies, the helical domain interactions were subsequently mapped to a higher resolution (39).
Particularly extensive contacts with the helical domain have been documented for two of the more unusual RGS proteins, RGS14 and p115RhoGEF (Fig. 1). RGS14 is one of a handful of RGS family members that contain a GoLoco motif, in addition to a canonical RGS core domain. Whereas RGS proteins promote GTP hydrolysis, GoLoco proteins interact specifically with GDP-bound Gα proteins and act to slow nucleotide exchange. Thus, RGS14 functions as both a GAP and a GDI. Crystallographic analysis revealed extensive contacts between the GoLoco fragment of RGS14 and portions of Gαi1-GDP, including the αA and αB helices of the helical domain, the bound nucleotide, and Switch I of the Ras-like domain (40). The helical domain interactions appear to alter the conformation of Gα, causing the αB-αC loop to become displaced from the Ras-like domain. The contact sites within the helical domain also appear to be important for Gα binding specificity. RGS14 is a GDI for Gαi1 but not Gαo; however, it is evident from the crystal structure that the contact sites within the Ras-like domain are conserved in both Gα subtypes. Whereas a chimera consisting of the Ras-like domain of Gαo and the helical domain of Gαi1 was responsive to the GoLoco peptide, the reciprocal chimera was not (40).
An especially striking example of a helical domain–binding protein comes from a crystal structure of the RGS domain of p115RhoGEF in complex with Gα13 or a hybrid Gα assembled from Gα13 and Gαi3 (41, 42). p115RhoGEF is a GAP for Gα13 and also serves as a GEF for Rho family GTPases; activation of Gα13 stimulates Rho GEF activity (43, 44). Thus, the protein is both an inhibitor and an effector for Gα, directly coupling activation of Gα13 to the activation of Rho. The GAP domain of p115RhoGEF has only weak sequence similarity to other RGS proteins, however, and the structure of the p115RhoGEF-Gα complex indicated some major differences compared with those of other RGS-Gα complexes. Specifically, the p115RhoGEFGα structure revealed regions N-terminal to the RGS core domain that form extensive contacts with the helical domain. Moreover, whereas the structures of Gα13 and Gαi are largely superimposable, the helical domain contact sites are distinct. These contacts are evidently important for GAP activity, as was demonstrated by domain-swapping experiments (41, 42).
Thus, it appears that the helical domain may confer binding selectivity for a number of RGS proteins and their homologs. There is also limited evidence that the helical domain is an active participant in signal transmission. One of the original crystal structures of Gαi bound to GDP and Gβγ revealed several differences when compared with that of Gαi-GTPγS (45). Aside from the usual changes in Switch I, II, and III, a fourth segment within the helical domain was unwound by approximately one turn, thereby extending the αB-αC loop. The authors dubbed the αB-α C loop “Switch IV.” These observations raise the possibility of a nucleotide-dependent conformational change in the helical domain. The crystal structures of RGS14 GoLoco and p115Rho-GEF also revealed modest conformational changes within the helical domain of their cognate Gα, particularly within the αB and αC helices. Moreover, molecular dynamics simulations have pointed to unexpected differences in inter- and intradomain motion when comparing the animal and plant G proteins, Gαi and AtGPA1. Subsequent experimental analysis of chimeric proteins established that the αA helix is almost entirely responsible for the >100-fold faster rate of nucleotide exchange exhibited by AtGPA1 (46). Thus, changes in the helical domain, even changes that are far from the nucleotide-binding pocket, can have profound effects on the nucleotide-binding function of the G protein.
You know my methods, Watson. There was not one of them which I did not apply to the inquiry. And it ended by my discovering traces, but very different ones from those which I had expected.
—from The Crooked Man
Opportunities for New Pharmacology
What do these observations mean for pharmacology? Roughly half of all drugs work, either directly or indirectly, on GPCRs. These include receptors for opioids, angiotensin II, epinephrine, histamine, and serotonin. The crystal structure of a receptor in complex with a G protein will no doubt provide new opportunities for receptor pharmacology. Likewise, the Gα helical domain has features that could be useful in drug interactions, including potential binding pockets and a conformational flexibility that could accommodate the binding of small molecules, features that are not afforded the “small” G proteins. With recent advances, it is evident that emerging opportunities are not limited to receptors but also include the G protein and the growing number of G protein–binding proteins that rely on the helical domain for their activity.
Acknowledgments
We thank A. Jones, D. Isom, and J. Hurst for insightful comments on the manuscript.
Funding: This work was supported by a grant from the National Institute of General Medical Sciences (R01GM080739) to H.G.D. J.C.J. was supported by a Ruth L. Kirschstein postdoctoral award.
Footnotes
Author contributions
J.C.J. wrote the manuscript and prepared the figure; H.G.D. wrote the manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Biddlecome GH, Berstein G, Ross EM. Regulation of phospholipase C-β1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. J. Biol. Chem. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [published erratum appears in J. Biol. Chem. 271, 33705 (1996)]. [DOI] [PubMed] [Google Scholar]
- 2.Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, Harden TK. Kinetic scaffolding mediated by a phospholipase C-β and Gq signaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Clark WA, Sharma P, Northup JK. Mechanism of allosteric regulation of the rod cGMP phosphodiesterase activity by the helical domain of transducin α subunit. J. Biol. Chem. 1998;273:34284–34292. doi: 10.1074/jbc.273.51.34284. [DOI] [PubMed] [Google Scholar]
- 5.Liu W, Northup JK. The helical domain of a G protein α subunit is a regulator of its effector. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12878–12883. doi: 10.1073/pnas.95.22.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a Gα subunit. Science. 1993;262:1895–1901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 7.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. Effects of Mg2+ and the β γ-subunit complex on the interactions of guanine nucleotides with G proteins. J. Biol. Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 8.Coleman DE, Sprang SR. Crystal structures of the G protein Gi α 1 complexed with GDP and Mg2+: A crystallographic titration experiment. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 9.Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones JC, Duffy JW, Machius M, Temple BR, Dohlman HG, Jones AM. The crystal structure of a self-activating G protein α subunit reveals its distinct mechanism of signal initiation. Sci. Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conklin BR, Bourne HR. Structural elements of G alpha subunits that interact with G β γ, receptors, and effectors. Cell. 1993;73:631–641. doi: 10.1016/0092-8674(93)90245-l. [DOI] [PubMed] [Google Scholar]
- 13.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson KM, Higashijima T, Smigel MD, Gilman AG. The influence of bound GDP on the kinetics of guanine nucleotide binding to G proteins. J. Biol. Chem. 1986;261:7393–7399. [PubMed] [Google Scholar]
- 15.Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL. Interaction of a G protein with an activated receptor opens the interdomain interface in the α subunit. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Jr, Sunahara RK. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Structural flexibility of the G α s α-helical domain in the β2-adrenoceptor Gs complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunahara RK, Tesmer JJ, Gilman AG, Sprang SR. Crystal structure of the adenylyl cyclase activator Gsα. Science. 1997;278:1943–1947. doi: 10.1126/science.278.5345.1943. [DOI] [PubMed] [Google Scholar]
- 19.Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer Gi β 1 α 1 γ 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 20.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 21.Yi TM, Kitano H, Simon MI. A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10764–10769. doi: 10.1073/pnas.1834247100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galés C, Van Durm JJ, Schaak S, Pontier S, Percherancier Y, Audet M, Paris H, Bouvier M. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat. Struct. Mol. Biol. 2006;13:778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- 23.Thomas CJ, Briknarová K, Hilmer JK, Movahed N, Bothner B, Sumida JP, Tall GG, Sprang SR. The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS ONE. 2011;6:e23197. doi: 10.1371/journal.pone.0023197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hausdorff WP, Pitcher JA, Luttrell DK, Linder ME, Kurose H, Parsons SJ, Caron MG, Lefkowitz RJ. Tyrosine phosphorylation of G protein α subunits by pp60c-src. Proc. Natl. Acad. Sci. U.S.A. 1992;89:5720–5724. doi: 10.1073/pnas.89.13.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell MW, Desai N, Guo XX, Ghalayini AJ. Tyrosine phosphorylation of the α subunit of transducin and its association with Src in photo-receptor rod outer segments. J. Neurochem. 2000;75:2006–2019. doi: 10.1046/j.1471-4159.2000.0752006.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen MY, Devreotes PN, Gundersen RE. Serine 113 is the site of receptor-mediated phosphorylation of the Dictyostelium G protein α-subunit G α 2. J. Biol. Chem. 1994;269:20925–20930. [PubMed] [Google Scholar]
- 29.Torres MP, Lee MJ, Ding F, Purbeck C, Kuhlman B, Dokholyan NV, Dohlman HG. G protein mono-ubiquitination by the Rsp5 ubiquitin ligase. J. Biol. Chem. 2009;284:8940–8950. doi: 10.1074/jbc.M809058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappell SD, Baker R, Skowyra D, Dohlman HG. Systematic analysis of essential genes reveals important regulators of G protein signaling. Mol. Cell. 2010;38:746–757. doi: 10.1016/j.molcel.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Marotti LA, Jr, Lee MJ, Dohlman HG. Differential regulation of G protein α subunit trafficking by mono- and polyubiquitination. J. Biol. Chem. 2005;280:284–291. doi: 10.1074/jbc.M411624200. [DOI] [PubMed] [Google Scholar]
- 32.Torres MP, Clement ST, Cappell SD, Dohlman HG. Cell cycle-dependent phosphorylation and ubiquitination of a G protein α subunit. J. Biol. Chem. 2011;286:20208–20216. doi: 10.1074/jbc.M111.239343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marotti LA, Jr, Newitt R, Wang Y, Aebersold R, Dohlman HG. Direct identification of a G protein ubiquitination site by mass spectrometry. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 34.Slep KC, Kercher MA, Wieland T, Chen CK, Simon MI, Sigler PB. Molecular architecture of Gαo and the structural basis for RGS16-mediated deactivation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6243–6248. doi: 10.1073/pnas.0801569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundström M, Doyle DA, Siderovski DP. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein α-subunits. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skiba NP, Yang CS, Huang T, Bae H, Hamm HE. The α-helical domain of Gαt determines specific interaction with regulator of G protein signaling 9. J. Biol. Chem. 1999;274:8770–8778. doi: 10.1074/jbc.274.13.8770. [DOI] [PubMed] [Google Scholar]
- 37.Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. Selective regulation of Gα(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J. Biol. Chem. 1999;274:34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- 38.Day PW, Carman CV, Sterne-Marr R, Benovic JL, Wedegaertner PB. Differential interaction of GRK2 with members of the Gαq family. Biochemistry. 2003;42:9176–9184. doi: 10.1021/bi034442+. [DOI] [PubMed] [Google Scholar]
- 39.Day PW, Tesmer JJ, Sterne-Marr R, Freeman LC, Benovic JL, Wedegaertner PB. Characterization of the GRK2 binding site of Gαq. J. Biol. Chem. 2004;279:53643–53652. doi: 10.1074/jbc.M401438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Structural determinants for GoLo-co-induced inhibition of nucleotide release by Gα subunits. Nature. 2002;416:878–881. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- 41.Hajicek N, Kukimoto-Niino M, Mishima-Tsumagari C, Chow CR, Shirouzu M, Terada T, Patel M, Yokoyama S, Kozasa T. Identification of critical residues in Gα13 for stimulation of p115RhoGEF activity and the structure of the Gα13-p115RhoGEF regulator of G protein signaling homology (RH) domain complex. J. Biol. Chem. 2011;286:20625–20636. doi: 10.1074/jbc.M110.201392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z, Singer WD, Sternweis PC, Sprang SR. Structure of the p115RhoGEF rgRGS domain-Gα13/i1 chimera complex suggests convergent evolution of a GTPase activator. Nat. Struct. Mol. Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 43.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, Bollag G, Sternweis PC. p115 RhoGEF, a GTPase activating protein for Gα12 and Gα13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 44.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Gα13. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 45.Mixon MB, Lee E, Coleman DE, Berghuis AM, Gilman AG, Sprang SR. Tertiary and quaternary structural changes in Giα1 induced by GTP hydrolysis. Science. 1995;270:954–960. doi: 10.1126/science.270.5238.954. [see comments]. [DOI] [PubMed] [Google Scholar]
- 46.Jones JC, Jones AM, Temple BR, Dohlman HG. Differences in intradomain and interdomain motion confer distinct activation properties to structurally similar Gα proteins. Proc. Natl. Acad. Sci. U.S.A. 2012;109:7275–7279. doi: 10.1073/pnas.1202943109. [DOI] [PMC free article] [PubMed] [Google Scholar]