Abstract
Heparan sulfate (HS) and chondroitin sulfate/dermatan sulfate (CS/DS) glycosaminoglycans (GAGs) participate in many important biological processes. Quantitative disaccharide analysis of HS and CS/DS is essential for the characterization of GAGs and enables modeling of the GAG domain structure. Methods involving enzymatic digestion and chemical depolymerization have been developed to determine the type and location of sulfation/acetylation modifications as well as uronic acid epimerization. Enzymatic digestion generates disaccharides with Δ-4,5-unsaturation at the non-reducing end. Chemical depolymerization with nitrous acid retains the uronic acid epimerization. This work shows the use of hydrophilic interaction liquid chromatography (HILIC)-MS for quantification of both enzyme-derived and nitrous acid depolymerization products for structural analysis of HS and CS/DS. This method enables biomedical researchers to determine complete disaccharide profiles on GAG samples using a single LC-MS platform.
Introduction
Proteoglycans (PGs) are key components of extracellular matrices in all eukaryotic cells. Members of this glycoprotein class contain one or more covalently attached polysaccharide chains, called glycosaminoglycans (GAGs).1 The major sulfated GAG chains of PGs, heparan sulfate (HS) and chondroitin sulfate/dermatan sulfate (CS/DS), are complex linear polysaccharides. These biopolymers are biosynthesized initially as repeating disaccharide units of β-D-glucuronic acid (GlcA) linked to either α-D-N-acetylglucosamine (GlcNAc) in HS or β-D-N-acetylgalactosamine (GalNAc) in CS/DS.2,3 The nascent chains are modified through a series of enzymatic reactions including N-deacetylation and N-sulfation (only in HS), epimerization of some GlcA to α-L-iduronic acid (IdoA), and sulfation to differing degrees at various hexuronic and hexosamine positions. In HS, sulfation occurs predominantly at C2 of IdoA and the N- and C6 positions of glucosamine (and rarely at C3).4 In CS/DS, sulfation can occur at C2 of IdoA as well as C4 or C6 of GalNAc.5 Because of these modifications, HS and CS/DS are comprised of disaccharide units with unique patterns of N-sulfation (only in HS), N-acetylation, O-sulfation, and HexA epimerization.
HS and CS/DS play critical roles in physiological and pathophysiological processes including homeostasis, cell migration and signaling, anticoagulation, inflammation, angiogenesis and cancer progression.6–9 These biological function are related to their structural diversity and ability to interact with cell surfaces and extracellular proteins, and structures of these GAGs are significant determinants of their binding interactions.10–12 The characterization of HS and CS/DS structures is, therefore, critical in elucidating these functions. However, characterization has been limited by the structural complexity and heterogeneity of these GAGs. Determination of disaccharide compositions of depolymerized GAGs is a key first step in the analysis of GAGs. GAG disaccharides are obtained by either enzymatic or chemical depolymerization. Polysaccharide lyase enzymatic digestion cleaves the hexosamine-hexuronic acid glycosidic bonds, resulting in the generation of unsaturated bonds between the C4 and C5 or hexuronic acid (termed Δ-4,5-unsaturated hexuronic acid).13 Chemical depolymerization involves mild hydrazinolysis to deacetylate N-acetylhexuronic acid residues, followed by deaminative cleavage using nitrous acid. This results in disaccharide units composed of uronic acid (GlcA or IdoA) and 2,5- anhydrohexose (aHex) bearing an aldehyde group.14,15 The advantage of this classic method for GAG depolymerization is that the original epimeric nature of the uronic acid is retained. However, information about the N-acetylation or N-sulfation in HS is lost because of the formation of the anyhydrohexose.
Disaccharide analysis after depolymerization has been performed using gel or paper electrophoretic14,16,17, capillary electrophoretic18,19, or chromatographic20–22 analysis of the products. In recent years, liquid chromatography-mass spectrometry (LC-MS) has emerged as the tool of choice for GAG disaccharide analysis.23–28 The unique elution times of the disaccharides together with the m/z values provide sufficient information to assign each disaccharide. In addition, the use of tandem MS provides structural information on peak identities.29–32
We have used a size exclusion chromatography (SEC)-MS system for analysis of lyase enzyme generated GAG disaccharides.26,33–37 This method has the advantage that it is very robust due to the fact that the mobile phase does not contain ion pairs or other additives. In addition, it is applicable to analysis of GAGs purified from a variety of biological matrices. Although the sensitivity of the SEC-MS system is modest, no derivatization of the GAG disaccharides is necessary. Thus, the quantity of starting GAG necessary for successful disaccharide analysis using SEC-MS is only a few micrograms. This method uses tandem MS to differentiate sulfation positional isomers. SEC-MS is not suitable for the analysis of disaccharides generated using deaminative cleavage because the uronic acid epimers cannot be differentiated by tandem MS. We therefore applied graphitized carbon LC-MS for analysis of HS disaccharide compositions from deaminative cleavage.23 This work demonstrated that uronic acid epimerization positions and positional sulfation isomers could be determined using LC-MS. It is desirable that a single LC-MS platform be used for analysis of disaccharides generated by both enzymatic and deaminative cleavage. Unfortunately, recovery of disaccharides with more than two sulfate group has proven to be problematic using graphitized carbon chromatography. Such disaccharides are present in mammalian heparin and heparan sulfate as well as invertebrate chondroitin sulfate. Triply sulfated disaccharides are highly retained, the retention times variable, and the recovery poor. Thus, graphitized carbon chromatography does not provide robust performance for analysis of lyase enzyme generated GAG disaccharides.
Hydrophilic interaction chromatography (HILIC) is gaining importance in recent years for the separation of carbohydrates. HILIC is a variation of normal-phase chromatography wherein polar compounds are separated on polar stationary phases using water miscible organic solvents and water as the elutropic solvent.38 This technique offers the advantages of improved MS sensitivity because of the high organic content in the mobile phase, shortened sample preparation time with direct injection of organic-solvent extracts of biological samples and the potential for ultra-fast analysis because of low-column backpressure. However, the need for better understanding of the mechanism of retention of analytes with different mobile- and stationary-phase compositions and solutions to ion suppression are some of the challenges associated with this chromatography.
HILIC with online electrospray ionization (ESI) MS has been used for the analysis of released glycans39,40, glycopeptides41, and GAG oligosaccharides42–44. To date, there is only one report of GAG disaccharide analysis using HILIC.45 In this work, the analysis of HS, CS/DS, and hyaluronan enzyme-derived disaccharides was performed using glycoblotting-assisted sample preparation followed by zwitter-ionic-HILIC. Amide HILIC methods are very robust and applicable to all glycan classes. There is not a problem of recovery of highly sulfated GAGs using amide HILIC. The resolution using 3–5 micron amide-HILIC stationary phase is modest, however, and does not suffice for disaccharide analysis. In addition, unsulfated disaccharides co-elute with an artifact that is always present in 3–5 micron amide-HILIC elution profiles that interferes with MS-detection. For these reasons, 3–5 micron amide-HILIC is not suitable for disaccharide analysis. The recent availability of <2 micron HILIC stationary phases has the potential to improve chromatographic speed and resolution of GAG disaccharide analysis. In the present work, we demonstrate use of 1.9 micron HILIC-MS to analyze both enzyme-derived and HONO-derived HS and CS/DS disaccharides. This method offers speed and ease of use as no derivatization or ion-pairing is necessary. The HILIC stationary phase permits separation of the Δ-unsaturated disaccharides as well as the HexA-aHex residues which allowed quantification of all the HS and CS/DS depolymeirzation products using a single platform.
Experimental Section
Materials
HS (bovine kidney), chondroitin sulfate A (sturgeon notochord) and C (shark cartilage), dermatan sulfate (porcine intestinal mucosa), and chondroitinase ABC were purchased from Sigma-Aldrich (St. Louis, MO). Porcine intestinal mucosa HS was purchased from Celsus Laboratories (Cincinnati, OH). Heparin lyases I from Flavobacterium heparinum were purchased from Ibex (Montreal, QC) and heparin lyases I and III were the generous gifts of Prof. Jian Liu of the University of North Carolina, Chapel Hill. Heparan sulfate disaccharides were from Sigma and V-Labs (Covington, LA). K5 polysaccharide and epimerized N-sulfated K5 polysaccharide were from Iduron (Manchester, UK).
Enzymatic Digestion
HS from bovine kidney (HSBK) and porcine intestinal mucosa (HSPIM) as well as heparin were digested with 3.2 milliunits of heparin lyase I, 3.2 milliunits of heparin lyase II, and 1.6 milliunits of heparin lyase III in a final volume of 50 µL of 50 mMTris/HCl buffer, pH 7.45, in the presence of 5 mM CaCl2. Half of the total amount of the enzymes was added and the solutions were incubated at 37°C for 24 hrs. The rest of the enzymes were then added and incubation was continued for another 24 hrs.
CS samples were digested with 10 mU chondroitinase ABC in 40mM Tris, pH8.0, with 40mM sodium acetate and 0.01% BSA at 37 °C for 48 hours.
Hydrazinolysis and Nitrous Acid (HONO) Depolymerization
For the hydrazinolysis, polysaccharide (10–15 µg) was dissolved in 20 µL of a 70% hydrazine solution containing 1% (w/w) hydrazine sulfate. The mixture was heated at 97°C for 4 hours. After cooling the reaction in an ice-cold bath, the hydrazine was evaporated in a stream of air at room temperature and lyophilized to remove any residual hydrazine. After repeated evaporation to dryness, the sample was desalted using a PD-10 cartridge (GE Healthcare, Piscataway, NJ), and lyophilized.
To the dried deacetylated sample, 20 µL of cold HONO pH 1.5 reagent (0.5M H2SO4 and 0.5M Ba(NO2)2 with volume ratio of 1:1) was added. After 10 min, the pH of the solution was adjusted to 4 with 1M Na2CO3. 20 µL of HONO pH 4 reagent (5.5 M NaNO2 and 0.5M H2SO4 with 5:2 volume ratio) was added. After 15 min, the reaction was terminated by adjusting the pH to 8.5 using 1M Na2CO3. The HONO-treated cleavage products were reduced with 0.5 M NaBH4 for 8 h at 55 °C. After incubation, samples were acidified to ~pH 4 with acetic acid and then neutralized to pH 7 by addition of ammonia.
Liquid Chromatography/Mass Spectrometry
An amide HILIC prototype column was provided by Thermo Fisher Scientific (150 mm × 2.1 mm, 1.9 µm particle size). The samples were separated using a Waters Acquity Ultra Performance LC system. Solvent A was ammonium formate, pH 4.4, and solvent B was solvent A in 90% acetonitrile. The column temperature varied from room temperature to 60 °C and the flow rates used were 0.15 and 0.1 mL/min. Disaccharides eluting from the column were analyzed using an Applied Biosystems QSTAR Pulsar-I (Q-TOF) mass spectrometer operating in negative ion mode. The ionization of the LC flow was accomplished by a TurboIonSpray source with capillary voltage set at -3500, nebulizer gas at 60, curtain gas at 35, turbo gas at 30, temperature at 100, and the sprayer position optimized to give least sulfate loss for ΔHexA2S-GlcNS6S
The PGC column (Hypercarb, 150 mm × 4.6 mm, 5 µm particle size) was purchased from Thermo Electron Corporation (Waltham, MA). Solvent A was 0.1% formic acid adjusted to pH 5.5 by ammonia and solvent B was solvent A in 90% acetonitrile. The gradient used was 0–15% B in 4 min and 15% B over 50 min. The flow rate was 0.5 mL/min, split down before the mass spectrometer to approximately 100 PL/min. The following parameters were used for ionization of the LC flow: capillary voltage set at -3500, nebulizer gas at 60, curtain gas at 35, turbo gas at 30, and temperature at 100.
The SEC column (Superdex™ peptide PC 3.2/30) was purchased from GE Healthcare (Piscataway, NJ). The mobile phase, 12.5 mm formic acid, pH adjusted to 4.4 using ammonia, in 10% acetonitrile was delivered isocratically at 0.016 mL/min. The following parameters were used for ionization of the LC flow: capillary voltage set at -3500, nebulizer gas at 40, curtain gas at 25, and turbo gas at 0.
Results and Discussion
HS and CS GAGs are heterogeneous with respect to chain length, sulfation, and uronic acid epimerization. The chain structure depends on spatially and temporally regulation of expression in biological tissue. Disaccharide analysis is an essential first step in GAG analysis. The information produced enables modeling of the GAG domain structure.34,46,47 Such modeling requires compositional data from serial analysis of disaccharides produced by lyase enzymes. It also requires compositions of deaminative cleavage disaccharides. In previous studies, we have shown an SEC-MS method for the separation of enzyme-derived disaccharides as well as a PGCMS method for the analysis of nitrous acid-derived dp2s.23,26 Enzyme digestion entails some loss of information since the asymmetric configuration at C-5 of the HexA units is eliminated. In deaminative cleavage, the epimeric nature of the HexA residue is retained. However, information about the N-substituents of the GlcN units is lost due to the formation of the anhydromannose and anhydrotalose of HS and CS, respectively, after nitrous acid treatment. Enzyme digestion retains the N-substituents of the GlcN residues resulting in the generation of N-sulfated and N-sulfated disaccharides. We therefore investigated the use of a 1.9 micron stationary phase amide HILIC-MS method for the analysis of both enzyme-derived and deaminative cleavage dp2s. Described in this report is a sensitive and reliable method for complete disaccharide analysis of HS and CS GAGs using a single analytical platform.
Separation of Enzyme-Derived Disaccharides
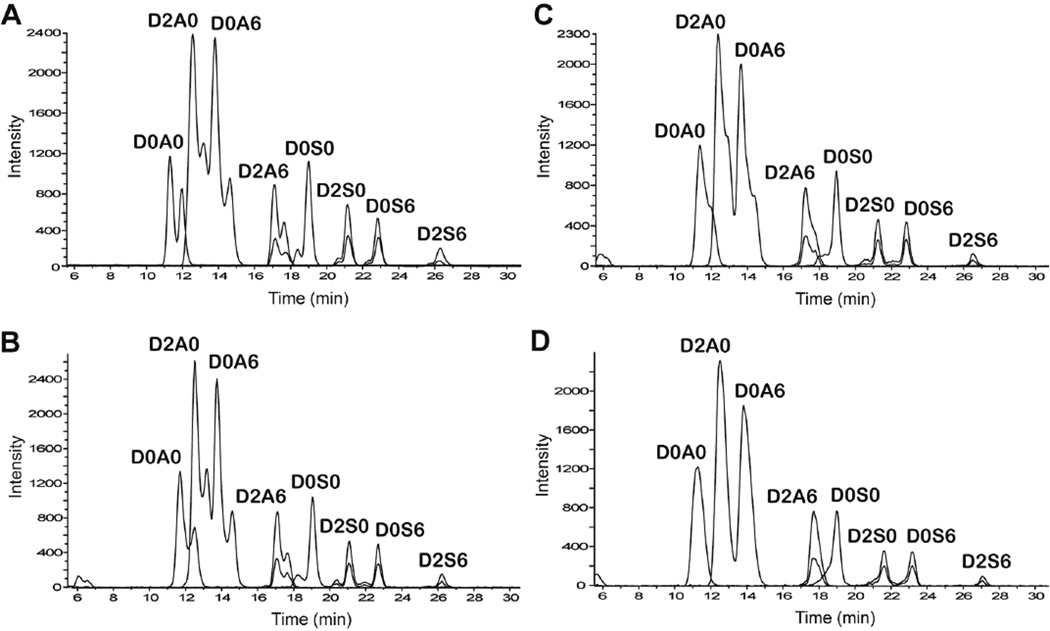
Optimization of the HILIC method for the analysis of enzyme-derived disaccharides was based on the chromatographic behavior of commercially purchased Δ-unsaturated disaccharide standards (see Supporting Information Figure S-1 for the disaccharide structures). The initial method used was based on the solvent composition used for HILIC-MS analysis of HS oligosaccharides (solvent A is 50 mM ammonium formate, pH 4.4 and solvent B is solvent A in 90% acetonitrile).48 The gradient used was 80%B for 4 min followed by 80–60% B in 25 min with a flow rate of 0.15 mL/min. Figure 1-A shows the extracted ion chromatograms of the eight disaccharide standards analyzed at room temperature. By gradient optimization it was possible to fully resolve the mixture of eight HS disaccharides with baseline separation. As a group, the N-acetylated dp2s eluted earlier than the N-sulfated disaccharides. Peaks corresponding to loss of sulfate were also observed for the di- and tri-sulfated disaccharides (overlaid on the chromatograms in Fig. 1). The reported abundances of the di- and tri-sulfated disaccharides include those of these sulfated loss ions. The dp2 standards, particularly the N-acetylated ones, showed split peaks corresponding to α and β anomers. Although it is interesting that the anomeric forms of a disaccharide can be resolved by HILIC, this is usually not desirable. The use of higher temperature prevents peak broadening due to anomeric separation.49 In this case the anomeric peaks were observed to coalesce as the column temperature was increased from ambient to 60 °C (Figure 1B to 1C). The merging of the anomers into a single chromatographic peak was attained at 60 °C. Therefore, a column temperature of 60 °C was selected for further studies2. However, the peaks for the sulfated disaccharides (D2A6 and all N-sulfated disaccharides) are asymmetric because the anomers are not fully merged even at 60 °C.
Figure 1.
Effect of temperature on retention of heparin lyase-derived disaccharide standards. (A) Room temperature, (B) 40 °C, (C) 50 °C, (D) 60 °C. Extracted ion chromatograms of the dp2 standards are shown. The peaks overlaid on those of the di- and tri-sulfated dp2s correspond to sulfate loss. The disaccharide peaks are: D0A0, ΔHexA-GlcNAc; D2A0, ΔHexA2S-GlcNAc; D0A6, ΔHexA-GlcNAc6S; D2A6, ΔHexA2S-GlcNAc6S; D0S0, ΔHexA-GlcNS; D2S0, ΔHexA2S-GlcNS; D0S6, ΔHexA-GlcNS6S; D2S6, ΔHexA2S-GlcNS6S.
The presence of ammonium formate (50 mM) resulted in decrease in signal strength over time due to the build-up of salt on the mass spectrometer source optics. Hence, the elution behavior of the dp2 standards at 60 °C was compared at three different concentrations of ammonium formate in the aqueous phase (Supporting Information Figure S-2). At 50 mM ammonium formate (Figure S-2A), the gradient used was 80% B for 4 min followed by 80–60% B in 25 min. At 25 and 12.5 mM ammonium formate (Figure S-2B and S-1C), the gradient used was 85% B for 4 min followed by 85–65% B in 25 min. Figure S-2 shows that the order of elution of the dp2s was not altered by the mobile phase ionic strength. The separation of the dp2 standards was not affected by the change in salt concentration in the aqueous phase, except for D0A0 and D2A0, which were less resolved in 12.5 mM ammonium formate. Therefore, 12.5 mM ammonium formate was selected for further studies to prevent salt build-up in the mass spectrometer.
The effect of flow rate on the resolution was also evaluated (Figure S-3). It is clear the lower flow rate (0.10 mL/min) increased the retention of the disaccharides, especially the N-acetylated D0A0, D2A0 and D0A6. The order of elution, separation, and total analysis time of the dp2s was not altered by the decrease in flow rate. However, at 0.10 mL/min the total intensity of all the disaccharides increased by ~40%. The increase in signal is likely due to the increase in analyte concentration at the lower flow rate. Other benefits of using the lower flow rate are decreased pressure, reduced mobile phase and nebulization gas consumption. Hence, a flow rate of 0.10 mL/min was used for the analysis of HS and CS GAGs.
Different binary gradients were tested to optimize the separation as well the solubility of the disaccharides. The best results were obtained using 85% B for 4 min followed by gradient elution from 85% to 60% B (solvent A is 12.5 mM ammonium formate, pH 4.4, and solvent B is 25 mM ammonium formate in 90% acetonitrile) in 25 min at 60 °C and a flow rate of 0.10 mL/min. Mass spectra for each disaccharide standard are shown in Supporting Figure S-4. A peak was observed for each disaccharide at m/z 378.1, 458.1, 538.0, 416.1, 496.0, and 576.0. Low abundance peaks corresponding to loss of sulfate were also observed in the mass spectra of the di- and tri-sulfated disaccharides. Background ions from the liquid chromatography mobile phase are also present. The mass spectra reflect the different ionization efficiencies for each of the disaccharides. N-Sulfated disaccharides eluted from the HILIC column at a higher level of ammonium formate than the N-acetylated ones, and the decreased desolvation that accompanies the higher salt levels results in decreased sensitivity for MS detection. Significant ion suppression occurred for the latter eluting disaccharides, especially the N-sulfated structures. Thus, MS peak areas do not directly correspond to the actual proportions of each disaccharide within a sample, unlike with UV detection. In order to calculate appropriate correction factors for disaccharides, HS dp2 standards were analyzed by HILIC-MS (4 replicates of each) and the corresponding UV absorbance and MS peak areas were compared. The integrated peak area of each disaccharide from the UV chromatogram was correlated to the integrated peak area in the HILIC-MS extracted ion chromatogram according to the following equation:
| (Equation 1) |
where ε is the molar absorptivity and q is the ionization efficiency correction factor. The q value for each disaccharide was normalized to that of the D0A0 dp2 standard and is shown in Table 1. To be able to determine the relative amounts of the different disaccharide species using the HILIC-MS chromatograms, their respective MS peak areas should be multiplied by their q values to compensate for the differences in the ionization efficiency between the disaccharides.
Table 1.
Ionization efficiency correction factors for the HS disaccharides.
| dp2 | q |
|---|---|
| D0A0 | 1.00 |
| D2A0 | 0.72 |
| D0A6 | 1.02 |
| D2A6 | 1.16 |
| D0S0 | 0.91 |
| D2S0 | 1.90 |
| D0S6 | 1.62 |
| D2S6 | 2.98 |
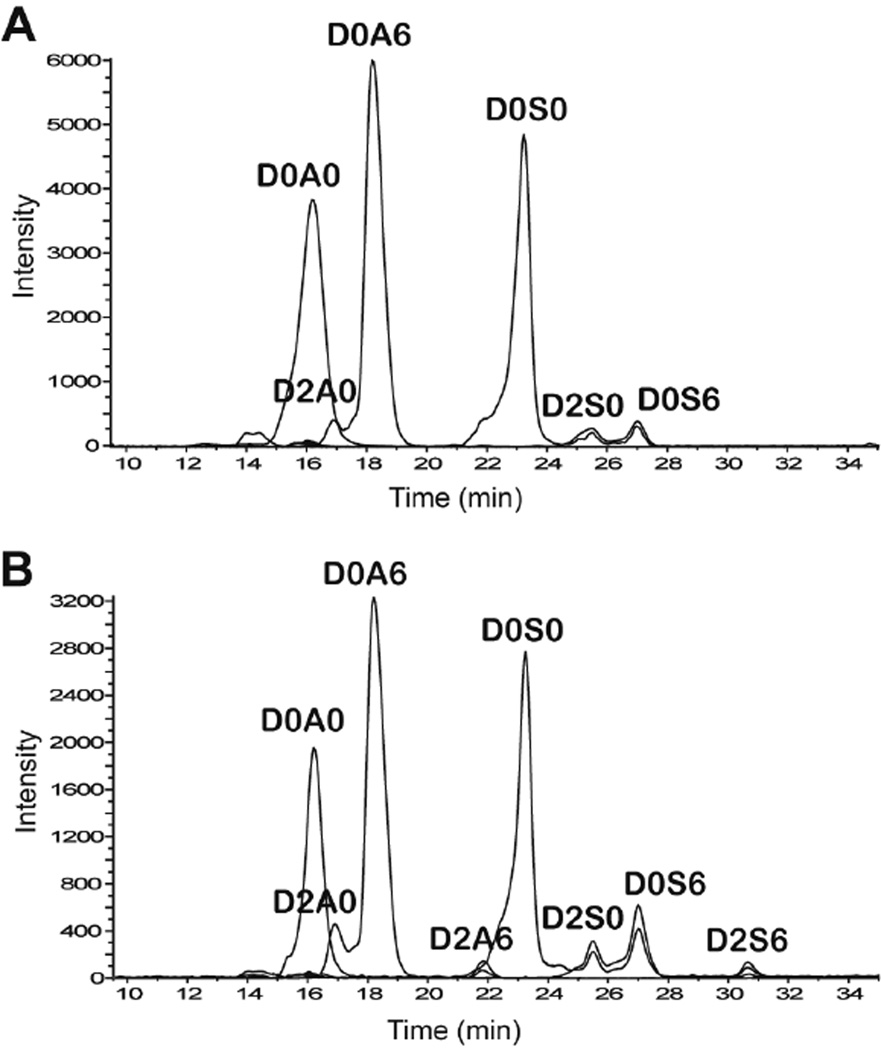
Samples of HS from bovine kidney (HSBK) and porcine intestinal mucosa (HSPIM) were treated with a mixture of heparin lyases I, II and III, resulting in complete depolymerization of the HS chains to disaccharides. The separation of these 4,5-Δ-unsaturated disaccharides using HILIC is shown in Figure 2. HILIC-MS gives baseline separation of the major isomeric ΔHexA2S-GlcNAc and ΔHexA-GlcNAc6S as well as ΔHexA2S-GlcNS and ΔHexA-GlcNS6S disaccharides. The relative quantities of the chromatographically separated disaccharides extracted from the HILIC-MS data are summarized in Table 2. As expected, HSPIM yielded more N-sulfated disaccharides. In contrast, HSBK generated more abundant non-sulfated dp2s. Also, the D2A6 disaccharide is barely discernible in HSBK. The EICs of the saturated disaccharides from the heparin lyase digests are shown in Figure 3. This figure demonstrates the ability to detect disaccharides that derive from the non-reducing ends of the parent polysaccharide mixture. This information is valuable because the ration of Δ-unsaturated to saturated disaccharide abundances determines the average polysaccharide chain length. The composition of the saturated disaccharides also provides information on the types of disaccharide units on the non-reducing ends relative to those found in the interior of the polysaccharide chains.
Figure 2.
Extracted ion chromatograms of unsaturated heparin lyase-derived HS disaccharides analyzed by HILC-MS from (A) HSBK and (B) HSPIM. The overlaid traces show abundances of sulfate loss peaks for D2S6, D0S6 and D2S0.
Table 2.
Comparison of the relative abundances of disaccharides from HSBK and HSPIM after heparin lyase digestion analyzed using SEC-MS and HILIC-MS
| HSBK | HSPIM | ||||||
|---|---|---|---|---|---|---|---|
| SEC-MS | HILIC-MS | SEC-MS | HILIC-MS | ||||
| D0A0 | 22 ± 1 | D0A0 | 25 ± 2 | D0A0 | 9.1 ± 0.6 | D0A0 | 11.2 ± 0.9 |
| D2A0/ D0A6 |
38 ± 2 | D2A0 | 1.6 ± 0.2 | D2A0/ D0A6 |
25 ± 1 | D2A0 | 2.3 ± 0.1 |
| D0A6 | 35 ± 2 | D0A6 | 22 ± 1 | ||||
| D2A6 | 0.10 ± 0.01 | D2A6 | 0.08 ± 0.01 | D2A6 | 1.1 ± 0.1 | D2A6 | 1.1 ± 0.2 |
| D0S0 | 13 ± 1 | D0S0 | 17 ± 2 | D0S0 | 9.9 ± 0.8 | D0S0 | 13 ± 1 |
| D2S0/ D0S6 |
17 ± 1 | D2S0 | 8.3 ± 0.4 | D2S0/ D0S6 |
27 ± 1 | D2S0 | 10.4 ± 0.6 |
| D0S6 | 7.8 ± 0.5 | D0S6 | 16 ± 1 | ||||
| D2S6 | 4.9 ± 0.5 | D2S6 | 3.7 ± 0.9 | D2S6 | 25 ± 1 | D2S6 | 22 ± 1 |
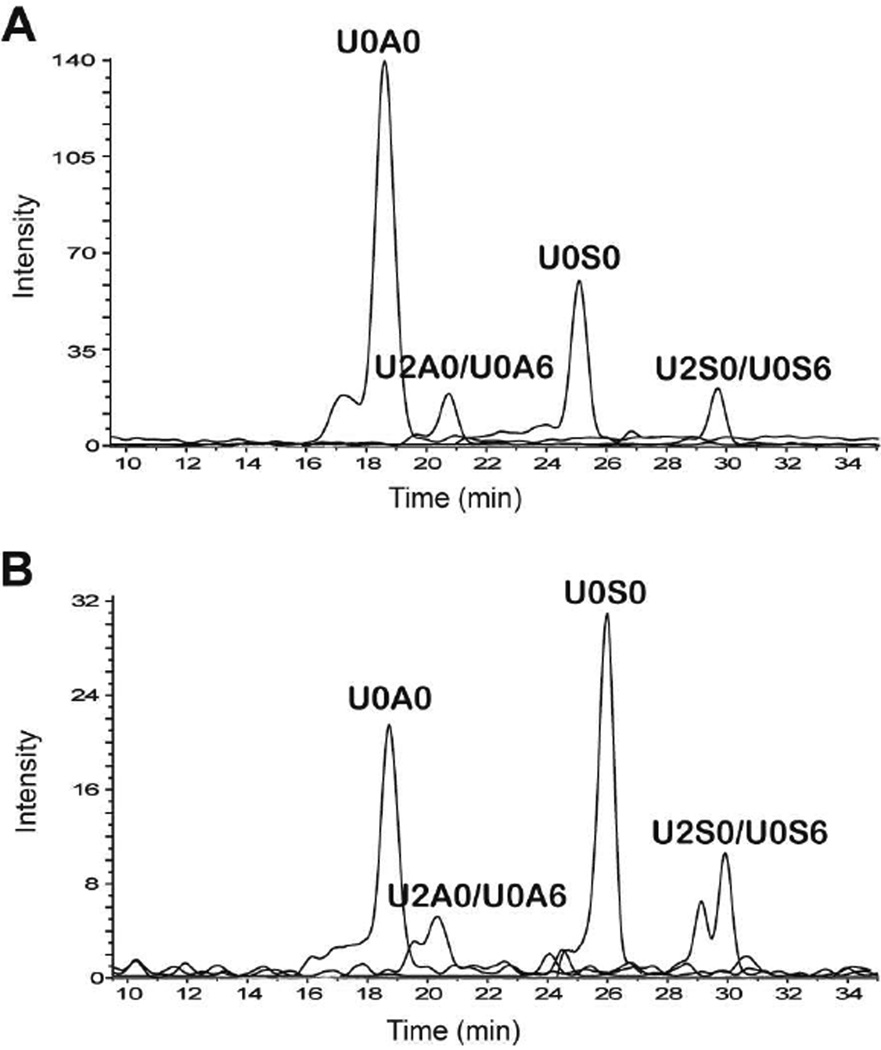
Figure 3.
Extracted ion chromatograms of saturated heparin lyase-derived HS disaccharides analyzed by HILC-MS from (A) HSBK and (B) HSPIM
The same enzyme-treated HSBK and HSPIM samples were also analyzed by SEC-MS. This method has been used extensively by our group for disaccharide analysis.26,43–47 A comparison of the relative quantities of the 4,5-Δ-unsaturated disaccharides from the HS samples obtained using HILIC-MS and SEC-MS is shown in Table 2. The table also illustrates the capability of HILIC-MS to determine the relative amounts of 2-O- from 6-O-sulfated disaccharides without any additional steps (i.e. tandem MS to determine relative amounts of O-sulfation when using SEC-MS). Saturated disaccharide digestion products from the nonreducing end were observed in both SEC-MS and HILIC-MS. The ability of the HILIC-MS and SEC-MS methods to determine average chain length is compared in (Supporting Information Table S-1). The two methods produce the same average chain lengths to within standard deviation of replicate analyses.
The one disadvantage of the HILIC-MS method for HS disaccharide analysis is its inability to determine the amount of D0H6 and D2H6 dp2s. Standards of these dp2s show that they co-elute with the D0S0 and D2S0 disaccharides, respectively. Apart from this, Tables 2 and S-1 demonstrate that the results from the two methods are comparable, thus, confirming the validity of the HILIC-MS method for the analysis heparin lyase-derived HS disaccharides.
Commercially available CS/DS standards (CS-4S, DS, and CS-6S) and CS from shark cartilage (CS-SC) were digested to reaction completion using excess chondroitinase ABC, which specifically degrades all linkages found within both chondroitin and dermatan sulfate without acting on other polysaccharides. The resulting CS disaccharides were quantitatively analyzed by HILIC-MS. Different gradient and isocratic conditions were tested to optimize separation of the CS/DS disaccharide standards at 60 °C. However, D0a4 and D0a6 co-eluted under all gradient conditions. Decreasing the temperature is expected to increase the affinity of analytes to the stationary phase and lowering the column temperature to 30 °C did allow sufficient separation of the isomeric CS/DS disaccharides. The best results were obtained using 80% B for 4 min followed by an isocratic elution of 71% B for 25 min (Supporting Information Figure S-5). Because an isocratic mobile phase condition was used to elute the disaccharides, ionization efficiency correction factors were not required. The MS profiles indicated a composition dominated by mono-sulfated disaccharides, except for CS-SC, which has a significant amount of di-sulfated dp2s. Also, the CS-4S and DS standards were contaminated with hyaluronan, an unsulfated GAG consisting of repeating 4GlcAβ1–3GlcNAcβ1- disaccharide units that is also cleaved by chondroitinase enzymes. The observed non-sulfated (m/z 339) peak at ~15.8 min has the same retention time as the D0A0 HS disaccharide. Comparison of the disaccharide composition determined by HILIC-MS to that from SEC-MS reveals that the results are in good agreement (Supporting Information Table S-2). Hence, HILIC-MS is an effective method for the analysis of enzyme-derived HS and CS/DS disaccharides.
Separation of HONO-Derived Disaccharides
GAG samples were subjected to N-deacetylation by hydrazinolysis and deaminative cleavage by nitrous acid. These conditions led to cleavage at GalN, GlcN as well as GlcNS units followed by reduction of the products with NaBH4. This results in depolymerization to HexA- 2,5-anhydromannose (HexA-aManR) and HexA-2,5-anhydrotalose (HexA-aTalR) disaccharide units for HS and CS chains, respectively (Supporting Information Figure S-6).
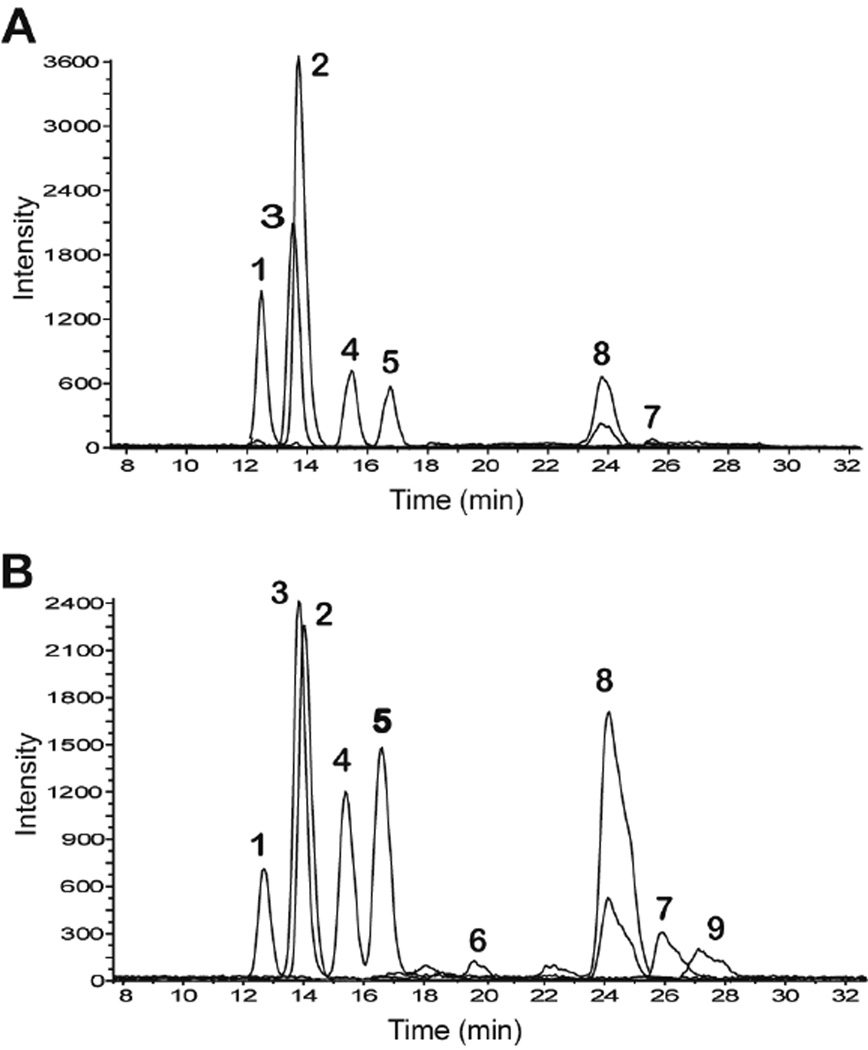
In a previous study, we had ascertained the identities of the HONO-derived disaccharides using PGC-MS.23 Since no HexA-aMan standards are available commercially, this PGC-MS method was used to purify each deaminative cleavage-derived disaccharide. These purified disaccharides were analyzed by HILIC to establish their retention times (Supporting Figure S-7). The best separation was obtained using an isocratic elution of 80% B for 29 min. Much like the enzyme-derived CS disaccharides, some of the HONO-derived HS dp2s co-eluted at 60 °C, thus, 30 °C was used for the HILIC separation. Extracted ion chromatograms (EIC) for the nonsulfated (m/z 339), mono-sulfated (m/z 419), and di-sulfated (m/z 499) HexA-aManR disaccharides derived from HONO treatment of HSBK and HSPIM separated by HILIC are shown in Figure 4. The disaccharides were baseline resolved except for G0M0 and I0M6. Ionization efficiency correction factors were not required because an isocratic gradient was used to elute the disaccharides. The relative quantities of the nine chromatographically separated HexA-aManR disaccharides extracted from the HILIC-MS data are summarized in Table 3. This HILIC-MS method produced profies of disaccharides from deaminative cleavage of HS extracted from bovine tissue samples very similar to those obtained using PGC-MS (Supporting Information Table S-3).
Figure 4.
Extracted ion chromatograms of the HONO-derived disaccharides analyzed by HILIC-MS. From (A) HSBK and (B) HSPIM. The disaccharide peaks are: 1, I0M0; 2, G0M0; 3, I0M6; 4, G0M6; 5, I2M0; 6, G0M3/6; 7, G0M3; 8, I2M6; 9, G0M9.
Table 3.
Comparison of the relative abundances of disaccharides from HSBK and HSPIM after deaminative cleavage by nitrous acid analyzed using PGC-MS and HILIC-MS
| Peak | Disaccharide | HSBK | HSPIM | ||
|---|---|---|---|---|---|
| PGCMS | HILICMS | PGCMS | HILICMS | ||
| 1 | I0M0 | 5.2 ± 0.7 | 5.5 ± 0.8 | 2.7 ± 0.7 | 2.4 ± 0.3 |
| 2 | G0M0 | 39 ± 1 | 40 ± 1 | 17 ± 1 | 18 ± 1 |
| 3 | I0M6 | 13 ± 1 | 14 ± 1 | 10 ± 1 | 13 ± 1 |
| 4 | G0M6 | 14 ± 1 | 11 ± 1 | 11 ± 1 | 9.5 ± 0.4 |
| 5 | I2M0 | 12 ± 1 | 13 ± 1 | 9.5 ± 0.4 | 11 ± 1 |
| 6 | G0M3/6 | 0.2 ± 0.1 | 0.3 ± 0.1 | 1.6 ± 0.1 | 1.0 ± 0.1 |
| 7 | G0M3 | 0.6 ± 0.2 | 0.7 ± 0.1 | 6.3 ± 0.6 | 5.4 ± 0.9 |
| 8 | I2M6 | 16 ± 1 | 15 ± 1 | 38 ± 2 | 36 ± 1 |
| 9 | G0M9 | 0 | 0.3 ± 0.3 | 4.2 ± 0.4 | 3.4 ± 0.2 |
As discussed in our previous study23, peaks 6 and 9 in Figure 4 were not detected in HSBK and the other bovine tissue samples as these peaks are likely specific to heparinoids from porcine intestinal mucosa. Tables 3 and Table S-3 demonstrate that the relative disaccharide abundance determined using PGC and HILIC are comparable. There is a slight difference in the percentage of G0M6. Compared to the PGC method, this disaccharide has lower relative abundance when analyzed using HILIC. This is because this disaccharide co-elutes with the salts present in the sample solution, resulting in a degree of ion suppression. Therefore, it is important to ensure that all samples are desalted prior to HILIC analysis.
A set of CS/DS preparations were also subjected to hydrazinolysis and nitrous acid treatment. The resulting HexA-aTalR disaccharides were quantitatively analyzed by both PGCMS and HILIC-MS. The HONO-derived CS disaccharides were separated by HILIC using an isocratic elution of 80%B for 29 minutes (same gradient as HS HONO-derived disaccharides) at 30 °C. The disaccharides generated are mostly mono-sulfated (m/z 419). Since no HexA-aTal standards are available commercially, PGC was used to purify each deaminative cleavagederived CS mono-sulfated disaccharide as all the disaccharides are baseline-resolved using PGC23. These purified disaccharides were analyzed by HILIC to establish their retention times (Supporting Figure S-8). The data show that only G0T4 and G0T6 are well-resolved using HILIC. Also, consistent with the results obtained using chondroitinase ABC digestion, CS-4S and DS were found to contain small amounts of glucosamine containing GAGs as evidenced by the presence of non-sulfated (m/z 339) peaks at 12.6 and 13.7 min (data not shown), the same retention time as the I0M0 and G0M0 HS deaminative cleavage disaccharides, respectively. Comparison of the disaccharide composition determined by HILIC-MS to that from SEC-MS reveals that the results are in good agreement (Supporting Information Table S-2).
Conclusions
We have demonstrated the effectiveness of 1.9 micron amide-HILIC MS for analysis of GAG disaccharides produced by either polysaccharide lyase enzymes or by deaminative cleavage. The HILIC-MS conditions are MS compatible and therefore appropriate for routine use. Because the disaccharides need not be derivatized, there are no sample losses from cleanup procedures. The fact that the same chromatography system can be used for HS and CS/DS GAGs will help maximize sample throughput. The 1.9 micron stationary phase produces chromatographic resolution adequate for separation of most GAG positional isomers. Thus, it is now possible to produce complete GAG disaccharide analysis, i.e. from both lyase enzymes and deaminative cleavage using a single LC-MS platform. HILIC using 3–5 micron stationary phases is readily scaled down to capillary and nano-flow formats. It is therefore expected that the described 1.9 micron HILIC chromatography will also be scalable. If so, this will greatly increase the sensitivity of disaccharide analysis.
Supplementary Material
Acknowledgements
The authors acknowledge NIH grants P41GM104603 and R01HL098950.
Footnotes
Supporting Information
Additional information as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Morgan MR, Humphries MJ, Bass MD. Nat. Rev. Mol. Cell. Biol. 2007;8:957–969. doi: 10.1038/nrm2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esko JD, Lindahl U. J. Clin. Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volpo N. Adv. Pharmacol. 2006;53:1–568. [Google Scholar]
- 4.Bulow HE, Hobert O. Annu. Rev. Cell Dev. Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 5.Silbert JE, Sugumaran G. IUBMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 6.Conrad HE. Heparin Binding Proteins. New York: Academic Press; 1988. [Google Scholar]
- 7.Capila I, Linhardt RJ. Angew Chem. Int. Ed. Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Fuster MM, Esko JD. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 9.Sasisekharan R, Shriver Z, Venkataraman G. Nat. Rev. Cancer. 2002;2:521–528. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 10.Catlow KR, Deakin JA, Wei Z, Delehedde M, Fernig DG, Gheraradi E, Gallagher JT, Pavao MSG, Lyon M. J. Biol.Chem. 2008;283:5235–5248. doi: 10.1074/jbc.M706589200. [DOI] [PubMed] [Google Scholar]
- 11.Kreuger J, Spillmann D, Li J, Lindahl U. J. Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, Nakato H. J. Cell Biol. 2006;174:773–778. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst R, Langer R, Cooney CL, Sasisekharan R. Crit. Rev. Biochem. Mol. Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 14.Shaklee PN, Conrad HE. Biochem. J. 1984;217:187–197. doi: 10.1042/bj2170187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Conrad HE. Anal. Biochem. 1989;176:96–104. doi: 10.1016/0003-2697(89)90278-9. [DOI] [PubMed] [Google Scholar]
- 16.Karousou EG, Viola M, Genasetti A, Vigetti D, Luca GD, Karamanos NK, Passi A. Biomed. Chromtogr. 2005;19:761–765. doi: 10.1002/bmc.511. [DOI] [PubMed] [Google Scholar]
- 17.Calabro A, Midura R, Wang A, West L, Plaas A, Hascall VC. Osteoathr. Cartil. 2001;9:S16–S22. doi: 10.1053/joca.2001.0439. [DOI] [PubMed] [Google Scholar]
- 18.Hitchcock AM, Bowman MJ, Staples GO, Zaia J. Electrophoresis. 2008;29:4538–4548. doi: 10.1002/elps.200800335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Militsopoulou M, Lamari FN, Hjerpe A, Karamanos NK. Electrophoresis. 2002;23:1104–1109. doi: 10.1002/1522-2683(200204)23:7/8<1104::AID-ELPS1104>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Toyoda H, Kinoshita-Toyoda A, Selleck SB. J. Biol. Chem. 2000;275:21856–21861. doi: 10.1074/jbc.M003540200. [DOI] [PubMed] [Google Scholar]
- 21.Babu P, Victor XV, Nelsen E, Thao KNN, Raman K, Kuberan B. Anal. Bioanal. Chem. 2011;401:237–244. doi: 10.1007/s00216-011-5087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. J. Biol. Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 23.Gill VL, Wang Q, Shi X, Zaia J. Anal. Chem. 2012 doi: 10.1021/ac3016054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babu P, Victor XV, Nelsen E, Thao KNN, Raman K, Kuberan B. Anal. Bioanal. Chem. 2011;401:237–244. doi: 10.1007/s00216-011-5087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CJ, Membreno N, Larive CK. Chromatogr. A. 2010;1217:479–488. doi: 10.1016/j.chroma.2009.11.064. [DOI] [PubMed] [Google Scholar]
- 26.Shi X, Zaia J. J. Biol. Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solakyildirim K, Zhang Z, Linhardt RJ. Anal. Biochem. 2010;397:24–28. doi: 10.1016/j.ab.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson NG, Schulz BL, Packer NH, Whitelock JM. J. Chromtatogr. B. 2005;824:139–147. doi: 10.1016/j.jchromb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Oguma T, Tomatsu S, Montano AM, Okazaki O. Anal. Biochem. 2007;368:79–86. doi: 10.1016/j.ab.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Hitchcock AM, Costello CE, Zaia J. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behr JR, Matsumoto Y, White FM, Sasisekharan R. Rapid Commun. Mass Spectrom. 2005;19:2553–2562. doi: 10.1002/rcm.2079. [DOI] [PubMed] [Google Scholar]
- 32.Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Glycobiology. 2005;15:818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- 33.Staples GO, Shi X, Zaia J. J. Biol. Chem. 2010;285:18336–18343. doi: 10.1074/jbc.M110.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staples GO, Shi X, Zaia J. PLoS ONE. 2011;6:e16689. doi: 10.1371/journal.pone.0016689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thelin M, Svensson KJ, Shi X, Bagher M, Axelssom J, Isinger-Ekstrand A, van Kuppevelt TH, Johansson J, Nilbert M, Zaia J, Belting M, Maccarana M, Malmstrom A. Cancer Res. 2012;72:1943–1952. doi: 10.1158/0008-5472.CAN-11-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher VA, Schlotzer-Schrehardt U, Karumanchi SA, Shi X, Zaia J, Jeruschke S, Zhang D, Pavenstaedt H, Drenckhan A, Amann K, Ng C, Hartwig S, Ng KH, Ho J, Kreidberg JA, Taglienti M, Royer-Pokora B, Ai X. J. Am. Soc. Nephrol. 2011;22:1286–1296. doi: 10.1681/ASN.2010080860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langsdorf V, Schumacher V, Shi X, Tran T, Zaia J, Jain S, Taglienti M, Kreidberg JA, Fine A, Ai X. Glycobiology. 2011;21:152–161. doi: 10.1093/glycob/cwq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alpert AJ. J Chromatogr. 1990;499:177–182. doi: 10.1016/s0021-9673(00)96972-3. [DOI] [PubMed] [Google Scholar]
- 39.Mauko L, Nordborg A, Hutchinson JP, Lacher NA, Hilder EF. Anal. Biochem. 2011;408:235–241. doi: 10.1016/j.ab.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 40.Ahn J, Bones J, Yu YQ, Rudd PM, Gilar MJ. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2010;878:403–408. doi: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Pompach P, Chandler KB, Lan R, Edwards N, Glodman R. J. Proteome Res. 2012;11:1728–1740. doi: 10.1021/pr201183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y, Shi X, Yu X, Leymarie N, Staples GO, Yin H, Killeen K, Zaia J. Anal. Chem. 2011;83:8222–8229. doi: 10.1021/ac201964n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staples GO, Naimy H, Yin H, Kileen K, Kraiczek K, Costello CE, Zaia J. Anal. Chem. 2010;82:516–522. doi: 10.1021/ac901706f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagaki K, Munakata H, Kakizaki I, Iwafune M, Itabashi T, Endo M. J. Biol. Chem. 2002;277:8882–8889. doi: 10.1074/jbc.M106479200. [DOI] [PubMed] [Google Scholar]
- 45.Takegawa Y, Araki K, Fujitani N, Furukawa J, Sugiyama H, Sakai H, Shinohara Y. Anal. Chem. 2011;83:9443–9449. doi: 10.1021/ac2021079. [DOI] [PubMed] [Google Scholar]
- 46.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. J. Biol. Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 47.Laremore TN, Ly M, Zhang Z, Solakyildirim K, McCallum SA, Owens RT, Linhardt RJ. Biochem. J. 2010;431:199–205. doi: 10.1042/BJ20100788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naimy H, Leymarie N, Zaia J. Biochemistry. 2010;49:3743–3752. doi: 10.1021/bi100135d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Churms SC. Carbohydrates. In: Heftmann E, editor. Chromatography: Applications. Amsterdam, The Netherlands: Elsevier B.V.; 2004. pp. 839–893. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.