Abstract
Integration of a protein into the endoplasmic reticulum (ER) membrane occurs through a series of multi-step reactions that include targeting of ribosome-nascent polypeptide complexes to the ER, attachment of the ribosome to the protein translocation channel, lateral partitioning of α-helical transmembrane spans into the lipid bilayer, and folding of the lumenal, cytosolic and membrane embedded domains of the protein. However, the molecular mechanisms and kinetics of these steps are still not entirely clear. To obtain a better understanding of the mechanism of membrane protein integration, we propose that it will be important to utilize in vivo experiments to examine the kinetics of membrane protein integration and in vitro experiments to characterize interactions between nascent membrane proteins, protein translocation factors and molecular chaperones.
Limitations to in vitro analyses of membrane protein integration
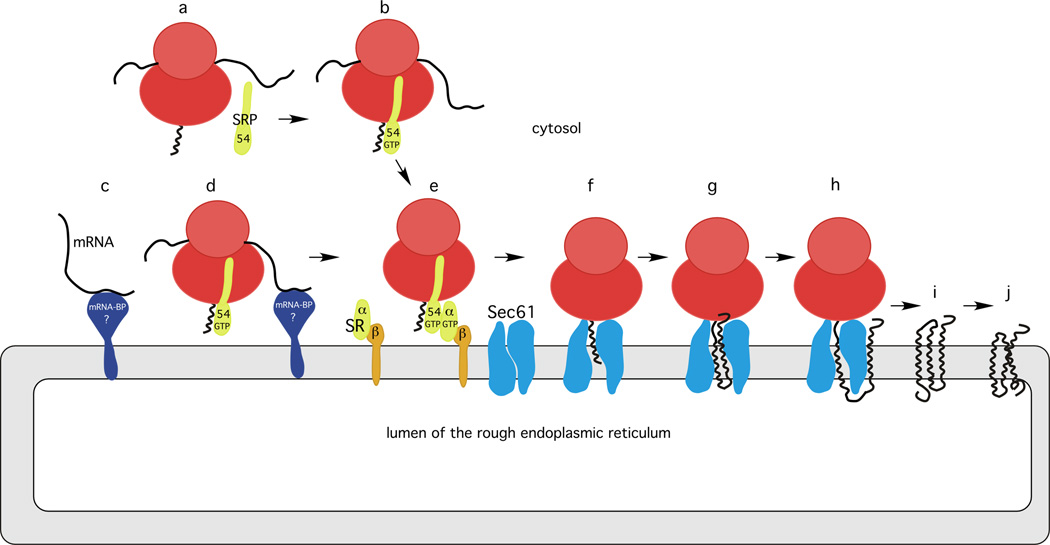
Integral membrane proteins account for roughly 25% of the open reading frames in eubacterial, archaebacterial and eukaryotic organisms [1]. With the exception of proteins destined for the mitochondria and chloroplast, almost all integral membrane proteins in eukaryotic cells are synthesized by membrane-bound ribosomes on the surface of the rough ER (RER). Tail-anchored (TA) membrane proteins, which have a single C-terminal α-helical TM span, are not synthesized by membrane bound ribosomes and so they are not discussed in this article (see [2] for a recent review). Current models describing the mechanism of membrane protein integration are primarily based upon in vitro biochemical and biophysical assays that monitor specific steps in this complex reaction pathway (as reviewed by [2, 3]). Transmembrane (TM) spans, like the cleavable signal sequences on secretory proteins, serve as RER-targeting signals that are recognized by the signal recognition particle (SRP, see Glossary) as the TM span emerges from the large ribosomal subunit (Figure 1a,b). Targeting of the SRP-ribosome-nascent polypeptide complex (RNC) to the SRP receptor (SR) on the ER leads to the formation of a high-affinity GTP stabilized complex between the SRP and SR [4] (Figure 1e). Dissociation of the SRP from the signal sequence then allows RNC to attach to the cytosolic face of the Sec61 complex, which is the eukaryotic cotranslational translocation channel (Figure 1f). The TM spans of integral membrane proteins adopt an α-helical conformation before passing through the lateral gate of Sec61 into the membrane bilayer, thereby minimizing the number of unsatisfied hydrogen bond donors and acceptors. In our opinion, a better understanding of the in vivo kinetics of membrane protein integration will provide novel insight into the mechanism of membrane protein synthesis, and might yield insight into integral membrane protein misfolding diseases.
Figure 1.
Integration of α-helical membrane proteins. (a) Translation of an mRNA encoding an integral membrane protein probably initiates in the vicinity of the rough endoplasmic reticulum (RER). (b) The hydrophobic transmembrane span, like a cleavable signal sequence, is recognized by the 54 kDa subunit of the signal recognition particle (SRP) particle when the signal sequence emerges from the polypeptide exit tunnel on the large ribosomal subunit. (c) mRNA molecules encoding endomembrane resident proteins might be localized to the RER via interactions between an RER-localized mRNA binding protein (mRNA-BP) and sequence motifs in the mRNA. (d) SRP recognition of the signal sequence (transmembrane (TM) span) might occur on these RER-tethered mRNA molecules, thereby simplifying the targeting of ribosome nascent chain complexes (RNC) to the RER. (e) Both targeting pathways converge on the alpha subunit of the SRP receptor (SRα). The GTP-bound conformation of the beta-subunit of the SRP receptor (SRβ) anchors SRα to the RER. The GTP-stabilized SR-SRP-RNC does not hydrolyze GTP until the RNC is delivered to a vacant Sec61 complex. The Sec61 complex is in a closed conformation in the absence of the RNC. (f) Dissociation of SRP54 from the TM span allows the RNC to bind to the Sec61 complex. Current evidence indicates that the TM span is initially inserted in a ‘head-first’ orientation into the lateral gate region of Sec61, with the N terminus of the nascent polypeptide oriented towards the RER lumen. (g) If the N-terminal segment flanking the first TM span has a net positive charge relative to the C-terminal flanking segment, the first TM inverts as the nascent polypeptide elongates. (h) The first TM span passes through the lateral gate of Sec61 to be integrated into the membrane bilayer. (i) Upon termination of protein synthesis, the nascent integral membrane protein is in an incompletely folded, but membrane integrated conformation. (j) Folding of the membrane protein may be facilitated by the cooperative action of lumenal, cytosolic and membrane-embedded chaperones.
An in vitro experimental strategy that has proven to be particularly useful to investigate protein translocation and membrane protein integration is to assemble RNC complexes by in vitro translating a truncated mRNA that lacks a termination codon. Incorporation of a photoreactive amino acid into the nascent polypeptide has led to the identification and characterization of the mammalian Sec61 complex as the protein translocation channel [5], consistent with evidence that Sec61p is the core subunit of the yeast protein translocation channel [6]. Site-specific incorporation of photoreactive amino acids [7] or fluorescent probes[8] has allowed a detailed examination of the integration pathway for multi-spanning membrane proteins [9–11]. Intramolecular fluorescence energy transfer (FRET) between fluorophores that flank a TM span indicate that the TM spans of membrane proteins adopt a more compact, perhaps α-helical, conformation before they emerge from the large ribosomal subunit, and upon doing so alter the junction between the RNC and the translocation channel [12, 13].
One limitation of in vitro analysis of membrane protein integration is that eukaryotic translation systems synthesize proteins at roughly 5–10% of the in vivo translation elongation rate (5–7 residues/second, [14]), hence in vitro experiments provide little information about the kinetics of membrane protein integration in cells. This caveat is compounded when one considers the time required to purify and analyze membrane protein integration intermediates assembled by translation of a truncated mRNA. Consequently, in vitro assembled integration intermediates might correspond to equilibrium conformations of the RNC-translocon complex rather than kinetically relevant reaction pathway intermediates.
In vivo experiments are also particularly informative for proteins that are either targeted to multiple organelles or are targeted to an unexpected cellular location. A recent noteworthy example concerns the biosynthesis of peroxisomal membrane proteins, which were previously thought to be synthesized by free polysomes in the cytosol. Recent in vivo evidence indicates that all peroxisomal membrane proteins are initially integrated into the RER in yeast cells [15]. In this article, we review recent conceptual advances in the analysis of membrane protein integration, many of which have been made possible by in vivo experimental approaches. We also highlight some of the major open questions in this research field, which we believe can best be addressed by in vivo studies.
In vivo targeting of mRNAs and RNC complexes to the RER
The SRP and the SR mediate targeting of RNCs that are synthesizing membrane proteins to the Sec61 complex. However, a growing body of evidence suggests that trafficking of mRNAs to the vicinity of the RER can also occur by a translation-independent process (Figure 1c). In mammalian cells, many mRNAs encoding resident proteins of the exocytic and endocytic membrane systems are bound to the ER by a ribosome-independent mechanism, suggesting that mRNA targeting to the vicinity of the RER might precede SRP-SR mediated RNC targeting to the Sec61 complex [16] (Figure 1d). Additionally, mRNAs encoding a subset of nucleocytoplasmic proteins were recovered in the membrane bound polysome fraction [17]. Additional research might lead to the identification of cis-acting mRNA motifs that interact with novel RER-associated trans-acting factors to mediate mRNA targeting or binding to the ER.
A panel of secretory protein reporter constructs consisting of a cleavable signal sequence and a transcription factor reporter domain were expressed in cultured cells to evaluate the in vivo efficiency of protein translocation [18]. Interestingly, translocation of the reporter constructs varied considerably (60 – 95% translocation) depending upon the signal sequence and showed additional variability between different mammalian cell lines, implying that trans-acting factors could impact translocation efficiency.
In vivo experiments conducted using the budding yeast Saccharomyces cerevisiae have provided insight into the physiological role of the SRP-SR targeting pathway. Despite the well-characterized role for SRP and the SR in RNC targeting to the Sec61 complex, budding yeast can tolerate disruption of the genes that encode the SR or SRP subunits, or the RNA component of the SRP [19, 20]. Adaption of budding yeast to the loss of the SRP-SR targeting pathway is accompanied by a 4-fold decrease in growth rate, a loss of the ability to grow on a non-fermentable carbon source and a global change in gene expression profile [21]. A reduction in ribosome biogenesis in SRP-deficient cells reduces the protein synthesis rate several fold [21]. Integration of membrane proteins in SRP or SR-deficient yeast cells occurs by partitioning of nascent integral membrane proteins between a posttranslational translocation pathway and a SRP-SR independent cotranslational pathway [22]. Direct binding of RNCs to the Sec61 complex, via the an interaction between the cytosolic loops of Sec61p and the large ribosomal subunit [23], is the most likely mechanism to explain SRP-independent targeting of RNCs to the Sec61 complex. Unlike budding yeast, the fission yeast Schizosaccharomyces pombe cannot tolerate disruption of the genes that encode the RNA or protein subunits of the SRP [24, 25] highlighting the importance of conducting in vivo experiments using several model systems.
In vivo kinetics of precursor targeting to the ER
Differential accessibility of nascent polypeptides to protein modification enzymes that are either localized in the cytosol or within the ER lumen has allowed investigators to devise assays that monitor the in vivo kinetics of targeting of precursor proteins to the ER. For example, coexpression of the BirA biotin ligase in the cytosol, and a precursor polypeptide appended with a 13-residue biotinylation tag, can be used to identify cytosolically exposed domains of membrane proteins [26]. Recently, this method was used to analyze posttranslational translocation of a very small secretory protein in mammalian cells [27]. Asparagine linked (e.g. N-linked) glycosylation of proteins by the oligosaccharyltransferase in the RER lumen has been widely used to detect lumenally exposed domains of integral membrane proteins [28, 29]. N-linked glycosylation is primarily cotranslational in mammalian cells [30, 31], therefore acquisition of an N-linked oligosaccharide can provide kinetic information about membrane protein integration. The in vivo kinetics of SRP-SR mediated targeting of RNCs to the ER has also been monitored using a protein modification technique. In this case, a protein kinase A phosphorylation site (LRRASLG) was inserted into the N-terminal lumenal domain of a model type 1 membrane protein [32]. Phosphorylation of the serine residue, as detected using 32P orthophosphate, can only occur when the protein kinase A site is in the cytosol. Transient exposure of the lumenal domain in the cytosol allowed the calculation of the time required for SRP-mediated RNC targeting (~3–4 sec) and translocation of the N-terminal 85 residue lumenal domain (~10 sec).
The kinetics of translocation channel gating can be monitored through the processing of an ubiquitin precursor protein by a cytosol-localized ubiquitin specific protease (UbP) [33]. The ubiquitin translocation assay (UTA) reporter consists of an N-terminal ER signal sequence, a spacer segment preceding a ubiquitin domain, followed by a UbP processing site and a C-terminal reporter domain appended with an epitope tag for immunoprecipitation of the pulse-labeled protein. If the C terminus of the ubiquitin domain emerges from the ribosome before the N terminus enters the translocation channel, rapid folding of ubiquitin in the cytosol leads to UbP cleavage of the UTA reporter. The UTA reporters were initially used to provide in vivo evidence documenting the yeast posttranslational translocation pathway [33]. More recently, UTA reporters with variable length spacer segments have been used to analyze the kinetics of membrane protein integration [34]. The translocon gating assay revealed that translocation of the lumenal domain initiates within 15 sec after the signal sequence emerges from the large ribosomal subunit in yeast cells [34]. During integration of more complex model integral membrane proteins, lumenal domains and transmembrane spans are transiently exposed on the cytosolic surface of the membrane [34]. We speculate that cytosolic chaperones or accessory translocation components might prevent aggregation or premature folding of these segments within the cytosol. The UTA assay reporters have also been used to analyze partitioning of precursors between cotranslational and posttranslational pathways in yeast strains bearing mutations in the Sec61 complex, the SRP or the SR [22, 34, 35]. The in vivo approaches described above have confirmed a central role for the SRP-SR targeting pathway for membrane protein integration, and have provided insight into the kinetics and mechanism of membrane protein biosynthesis. After targeting of SRP-RNCs to the membrane has been completed, the next event in the biosynthesis of a membrane protein is the recognition and subsequent integration of TM spans by the Sec61 complex.
Recognition and orientation of transmembrane spans
The orientation of membrane proteins is primarily determined by the distribution of charged residues that flank the first TM span in accord with the ‘positive-inside’ rule [36, 37]. A net positive charge in the N-terminal flanking sequence favors the type 2 (Ncyt-Cexo) orientation, whereas a net positive charge in the C-terminal flanking sequence favors the type 1 (Nexo- Ccyt) orientation. Using model membrane proteins that were designed to yield mixed topologies in vivo, Goder and Spiess obtained evidence that type 2 (Ncyt-Cexo) membrane proteins are initially inserted into the Sec61 complex in a type 1 orientation (Figure 1f), but invert to obtain a type 2 topology within the first 50 sec of biosynthesis [38] (Figure 1g). TM span inversion occurred more rapidly when positively charged residues precede the TM span and less rapidly as the hydrophobic core of the TM span was increased from 13 residues to 25 residues, consistent with previous evidence that longer TM spans favor a type 1 orientation in model proteins that have similarly charged N-terminal and C-terminal flanking regions [39]. Based upon this analysis, a typical type 2 membrane protein is expected to adopt the correct orientation rapidly (< 5 sec) after insertion into the Sec61 complex. Consistent with the hypothesis that TM span orientation occurs after contact between the nascent membrane protein and Sec61, mutations in yeast Sec61 influence the orientation of model proteins with ambiguous topologies [40]. A recent in vitro analysis of a model type 2 membrane protein provided support for the hypothesis that the TM span of a type 2 protein is initially inserted into the Sec61 complex in a type 1 orientation [41].
The TM spans of multi-spanning membrane proteins are often less hydrophobic than the TM spans of type 1 or type 2 membrane proteins. Nonetheless, the protein translocation channel must be able to recognize bona fide TM spans and partition these segments into the membrane bilayer, while allowing complete translocation of marginally hydrophobic segments from exoplasmic domains of membrane proteins. To obtain a better understanding of TM span recognition, the von Heijne lab devised a multi-TM span reporter protein to systematically test the integration efficiency of a large series of synthetic transmembrane spans that are flanked by diagnostic N-linked glycosylation sites [42]. The synthetic TM spans were derived from a 19-residue polyalanine test segment, wherein variable numbers of alanine residues were replaced by leucine residues to increase the hydrophobicity of the synthetic TM span, or by combinations of leucine and more hydrophilic amino acids to decrease the hydrophobicity of the synthetic TM span. The results of in vitro experiments using mammalian microsomes suggest that lateral exit of TM spans from the Sec61 complex is primarily governed by partitioning of the TM span between the relatively polar interior of the Sec61 translocation pore and the more hydrophobic environment of the membrane bilayer [42]. Importantly, expression of these multi-TM span constructs in yeast cells yielded similar results to those obtained using the mammalian in vitro system [43].The in vitro analysis of the synthetic TM spans allowed von Heijne and colleagues to derive an in vivo hydrophobicity scale for amino acids, wherein the experimentally determined ΔGapp values correspond to the free energy of membrane insertion. Subsequent studies from von Heijne and colleagues have shown that the orientation of the tested TM span (Ncyt vs. Ccyt) does not significantly alter the experimentally determined ΔGapp values for each of the amino acids [43].
von Heijne and colleagues have now applied this strategy to investigate integration of proteins into the mitochondrial inner membrane by the Tim23 complex. In this case, the in vivo assay exploits processing of the GTPase Mgm1p by the rhomboid protease Pcp1p [44]. Although the biological hydrophobicity scales for Sec61- and Tim23- mediated integration of TM spans are in general similar, certain aromatic residues (tryptophan and tyrosine) that favor TM span integration into the ER are less favorable for integration into the inner mitochondrial membrane. It is not clear whether these differences in hydrophobicity scale are explained by differences in membrane phospholipid composition or by structural differences in the import channel (Sec61 versus Tim23). Although the recognition and correct orientation of TM spans can be viewed as an initial step in the folding of an α-helical membrane protein, acquisition of the native folded structure will depend upon interactions between the individual TM spans within the membrane bilayer and upon folding of lumenal or cytoplasmic domains.
Folding and misfolding of integral membrane proteins
Misfolding of integral membrane proteins, and their subsequent degradation via the ER associated degradation (ERAD) pathway, is an area of great research interest because certain disease-causing mutations result in the misfolding of membrane proteins, such as the cystic fibrosis transmembrane conductance regulator (CFTR; see [45] for a recent review of protein misfolding diseases).
Comparing the topology diagram of a membrane protein, derived from a hydropathy plot, to the three-dimensional structure, determined by crystallography, strikingly demonstrates that the final fold of a multi-spanning membrane protein is not readily predictable based upon the location of hydrophobic segments within the protein sequence. Transmembrane spans are often tilted with respect to the phospholipid bilayer, and have kinks. Membrane proteins that serve as channels can have hydrophilic reentrant loops that are within the plane of the membrane, but do not contact the lipid bilayer. A recent analysis of the Pyrococcus hirokoshi glutamate transporter GltPh by the von Heijne lab [46] revealed that transmembrane span boundaries calculated using the biological hydrophobicity scale [42], or detected in the context of a reporter protein, do not match the final folded structure of GltPh. These results indicate that TM spans in multi-pass membrane proteins can be repositioned with respect to the membrane bilayer as the protein folds [46].
Concluding remarks
The combination of in vitro and in vivo assays to monitor specific reaction steps in membrane protein integration has led to a better understanding of how transmembrane spans are recognized by the translocation channel and partitioned between the relatively hydrophilic environment of the Sec61 pore and the hydrophobic environment of the membrane bilayer. In some cases, in vitro and in vivo approaches have yielded conflicting rather than complementary results, so it clear that there are many open questions (Box 1) that remain to be addressed. In our opinion, further progress in this research field will be aided by the continued use of a multi-faceted experimental approach that incorporates both in vitro and in vivo methods.
Box 1. Outstanding questions.
Are mRNAs targeted to the RER by a translation-independent mechanism? Are there mRNA binding proteins on the RER that tether mRNAs to the organelle to facilitate translation in the vicinity of the RER?
Does the 80S ribosome, or just the 40S small subunit, detach from the Sec61 complex following termination of protein synthesis? The high affinity between the ribosome and the Sec61 complex (~5 nM) raises the possibility that the large ribosomal subunit needs to be actively detached from Sec61.
Are there membrane-embedded chaperones that assist protein folding of membrane proteins? A potential candidate for a general membrane protein chaperone in mammalian cells is the translocon-associated membrane (TRAM) protein [47], which contacts integral membrane proteins as they exit the translocation channel [48].
Do membrane protein sequences and topologies influence the kinetics of TM-span exit from the Sec61 complex? Are marginally hydrophobic TM spans retained in the Sec61complex for a longer time?
Acknowledgements
Research on protein translocation in the Gilmore laboratory is supported by National Institutes of Health Grant GM35687.
Glossary Box
- Endomembrane
The membranes in a eukaryotic cell that are biosynthetically derived from the ER. These membranes include the nuclear envelope, the peroxisome, the lysosome, the Golgi, the plasma membrane, endosomes and vesicles that traffic between these destinations.
- FRET
Fluorescence resonance energy transfer between donor and acceptor fluorophores. FRET efficiency is dependent upon the distance between the two fluorescent probes and the overlap between the emission and excitation spectra of the donor and acceptor fluorophores. FRET measurements can be used to estimate distances between two sites on a protein.
- ΔGapp
The ΔGapp can be viewed as the equilibrium for partitioning of an amino acid between an aqueous environment and the hydrophobic core of the membrane bilayer. The calculated ΔGapp values of the in vivo hydrophobicity scale include TM-span location specific differences in the partitioning of amino acid side chains into the membrane bilayer.
- Positive-inside rule
The cytosolically exposed loops of membrane proteins have a net positive charge relative to the exoplasmic loops of membrane proteins. As a predictive method, one determines the net charge on the 10 amino acid residues on both sides that flank the first TM span in a protein. The flanking segment that has the greater net positive charge will face the cytosol.
- RNC
Ribosome-nascent chain complex. This term is used to refer to ribosomes synthesizing full length polypeptides and RNCs assembled in vitro by translation of a truncated mRNA that lacks a termination codon.
- Sec61 complex
Eukaryotic protein translocation channel in the rough RER. The Sec61 complex, and associated proteins, is often referred to as the translocon.
- SR
SRP receptor. A heterodimeric RER protein composed of two GTPases. The GTP-bound form of SRβ anchors SRα to the membrane. SRα and SRP54 hydrolyze GTP in a cooperative manner to allow dissociation of the SRP-SR complex.
- SRP
Signal recognition particle. The 54kD subunit of the signal recognition particle (SRP). SRP54 is a GTPase that has a C-terminal methionine-rich domain (M-domain) that is the site for signal sequence binding.
- Tim23 complex
The Tim23 complex is a protein translocase located in the inner membrane of the mitochondria that is primarily involved in transporting proteins with mitochondrial presequences into the matrix. A subset of mitochondrial inner membrane proteins, including Mgm1p, are integrated by the Tim23 complex.
- UbP
Ubiquitin specific protease that is responsible for processing the polyubiquitin precursor into mature ubiquitin.
- UTA
ubiquitin translocation assay. The ubiquitin translocation assay can be used to analyze the in vivo kinetics of protein translocation channel gating. Quantification of the cleaved and uncleaved forms of the UTA reporter indicates the fraction of RNCs that gate the Sec61 complex before the complete ubiquitin domain emerges from the large ribosomal subunit. Increasing the length of the spacer segment that precedes the Ub domain provides additional time (~0.125 sec per added residue) for translocon gating.
- TRAM
The translocon-associated membrane protein is an accessory component of the mammalian translocation channel that is in contact with nascent TM spans that exit the Sec61 complex. The exact role of TRAM in membrane protein integration is not well understood. Budding yeast do not have an obvious homologue of TRAM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shao S, Hegde RS. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 2011;27:25–56. doi: 10.1146/annurev-cellbio-092910-154125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park E, Rapoport TA. Mechanisms of Sec61/SecY-Mediated Protein Translocation Across Membranes. Annu. Rev. Biophys. 2012;41 doi: 10.1146/annurev-biophys-050511-102312. in press. [DOI] [PubMed] [Google Scholar]
- 4.Song W, et al. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 5.Görlich D, et al. A mammalian homologue of Sec61p and SecYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- 6.Deshaies RJ, et al. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- 7.Mothes W, et al. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. Embo J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigh NG, Johnson AE. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinrich SU, et al. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 10.Heinrich SU, Rapoport TA. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. Embo J. 2003;22:3654–3663. doi: 10.1093/emboj/cdg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadlish H, et al. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 12.Woolhead CA, et al. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 13.Lin PJ, et al. Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J. Cell Biol. 2011;195:55–70. doi: 10.1083/jcb.201103118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey JWB. Translational control in mammalian cells. Annu. Rev. Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 15.van der Zand A, et al. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 2010;21:2057–2065. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q, et al. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol. Biol. Cell. 2011;22:2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerner RS, et al. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine CG, et al. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 20.Ogg SC, et al. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, et al. An interaction between the SRP receptor and the translocon is critical during cotranslational protein translocation. J. Cell. Biol. 2008;180:1149–1161. doi: 10.1083/jcb.200707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Z, et al. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 2005;168:67–77. doi: 10.1083/jcb.200408188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennwald P, et al. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol. Cell. Biol. 1988;8:1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Althoff SM, et al. The SRP54 GTPase is essential for protein export in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14:7839–7854. doi: 10.1128/mcb.14.12.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emerman AB, et al. Compartment-restricted biotinylation reveals novel features of prion protein metabolism in vivo. Mol. Biol. Cell. 2010;21:4325–4337. doi: 10.1091/mbc.E10-09-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao S, Hegde RS. A calmodulin-dependent translocation pathway for small secretory proteins. Cell. 2012;147:1576–1588. doi: 10.1016/j.cell.2011.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hresko RC, et al. Topology of the Glut 1 glucose transporter deduced from glycosylation scanning mutagenesis. J. Biol. Chem. 1994;269:20482–20488. [PubMed] [Google Scholar]
- 29.Popov M, et al. Mapping the ends of transmembrane segments in a polytopic membrane protein. Scanning N-glycosylation mutagenesis of extracytosolic loops in the anion exchanger, band 3. J. Biol. Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 30.Chen W, et al. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc. Natl. Acad. Sci. USA. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Canada C, et al. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goder V, et al. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. Embo J. 2000;19:6704–6712. doi: 10.1093/emboj/19.24.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. Embo J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Z, Gilmore R. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat. Struct. Mol. Biol. 2006;13:930–936. doi: 10.1038/nsmb1146. [DOI] [PubMed] [Google Scholar]
- 35.Trueman SF, et al. Translocation channel gating kinetics balances protein translocation efficiency with signal sequence recognition fidelity. Mol. Biol. Cell. 2011;22:2983–2993. doi: 10.1091/mbc.E11-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. Embo J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartmann E, et al. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. Embo J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahlberg JM, Spiess M. Multiple determinants direct the orientation of signal–anchor proteins: the topogenic role of the hydrophobic signal domain. J. Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goder V, et al. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devaraneni PK, et al. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell. 2011;146:134–147. doi: 10.1016/j.cell.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 43.Lundin C, et al. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) trans-membrane helices. Proc. Natl. Acad. Sci. USA. 2008;105:15702–15707. doi: 10.1073/pnas.0804842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Botelho SC, et al. TIM23-mediated insertion of trans-membrane alpha-helices into the mitochondrial inner membrane. EMBO J. 2011;30:1003–1011. doi: 10.1038/emboj.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Powers ET, et al. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 46.Kauko A, et al. Repositioning of trans-membrane alpha-helices during membrane protein folding. J. Mol. Biol. 2010;397:190–201. doi: 10.1016/j.jmb.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 47.Görlich D, et al. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- 48.Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]