Abstract
Background
We sought to validate global microarray results indicating the differential expression of 383 genes in Peripheral Blood Mononuclear Cells (PBMCs) from patients with pancreatic cancer (PC) and to further evaluate their PC diagnostic potential.
Methods and materials
In total, 177 patients were recruited (47 healthy controls (HC), 35 CP patients, and 95 PC patients). PBMC expressions of six genes from our previous study (ANXA3, ARG1, CA5B, F5, SSBP2, and TBC1D8) along with four new genes (MIC1, NGAL, MUC1, and MUC16) were analyzed using multiplex Q-RT PCR.
Results
Differential expressions of 5 of the 6 genes previously identified by PBMC microarray were validated in this study. Multivariate models for PBMC gene expression were attempted to determine if any combination was diagnostically superior to CA19-9 alone. We found that addition of PBMC CA5B, F5, SSBP2, and MIC1 expression levels to CA19-9 significantly improved CA19-9’s diagnostic abilities when comparing resectable PC to CP patients (p = 0.023).
Conclusions
Results of our previous study were validated, indicating reproducibility of PC-associated PBMC expression profiling. We identified a score-based model that can differentiate resectable PC from CP better than CA19-9, potentiating that PBMC differential expression analysis may offer a novel tool for early PC diagnosis.
Keywords: Pancreatic cancer, chronic pancreatitis, PBMC, CA19-9, diagnosis
1. Introduction
Though the incidence of pancreatic cancer (PC) is relatively low compared to other cancers, comprising only 2–3% of cancer diagnoses, it is the fourth leading cause of cancer death. It has a 5-year survival rate of only 5% and a median survival from the time of diagnosis ranging between 3 and 6 months [1]. This poor prognosis is largely a result of the lack of an effective early detection method. Currently tumor resection is the only treatment that offers reasonable hope for a cure though PC is often asymptomatic in the early stages, evading detection until it is advanced beyond resectability. It is estimated that only 8% of PC cases are diagnosed with tumors localized to the pancreas, while only 15–20% are considered resectable [2]. For the remaining 80–85% of PC patients whose tumors are not resectable, clinical treatment is confined to largely ineffective chemo and radiotherapy that do little to extend patient life [3,4]. Consequently, improving clinical ability for the early detection of PC, thus allowing for increased rates of resection, has significant hope to improve the prognosis for this highly lethal disease.
Designing an early diagnostic test for PC however, presents a particular challenge owing to the relative rarity of the disease and its subclinical nature in initial stages. As such, any early diagnostic test will be accomplished via a screening mechanism, necessitating high enough sensitivity to detect most cases of PC while simultaneously having high specificity to reduce the significant risk of false positive results for clinical viability. High specificity alone, however, cannot overcome the unacceptably high ratio of false-positive/true-positive results of such a screening test if used in the general population, thus indicating the need for high-risk populations in which screening can be made practical due to increased PC incidence.
Currently, one of the few populations considered at high risk for PC is patients with chronic pancreatitis (CP). It is well accepted that CP serves as a significant risk factor for the development of PC, with CP patients having up to a 26 fold increase in risk of subsequent PC diagnosis as compared to the general population [5–11]. Given the increased incidence of PC in CP patients and the relatively low incidence of CP, estimated to affect 0.04–5% of the general population [12], this group provides a manageable subpopulation in which to screen for PC. Another important consideration in the differentiation of PC from CP is that often these two pathologies present with similar radiographic and clinical symptoms, such as epigastric pain, weight loss, and jaundice, making PC the primary diagnosis that must be ruled out in a CP differential diagnosis and further adding to the need for a test that can distinguish between the two diagnoses.
CA19-9 is currently the only marker approved by the FDA for use in PC. However, while CA19-9 is useful as a marker of disease burden, its lacks both sensitivity and specificity (approximately 80% and 73% respectively) as a diagnostic marker [13–18]. Nonetheless, it remains the gold standard against which every potential biomarker is compared. Thus, there is great clinical need for novel markers for the early diagnosis of PC.
One promising avenue for establishing such a marker is through the analysis of peripheral blood mononuclear cells (PBMCs). PBMCs comprise circulating mononuclear cells, including monocytes, T-cells, B-cells, and natural killer (NK) cells and have emerged in recent years as surrogate markers of several diseases including inflammatory (e.g. preeclampsia and rheumatoid arthritis) and malignant (chronic lymphocytic leukemia and renal cell carcinoma) diseases [4,19–22]. Our previous work has also suggested the diagnostic utility of PBMCs in the early detection of PC. PBMC expression is particularly promising in early PC detection as the differential expression of these cells may appear as soon as cancer immunogenicity or immune evasion is established, both of which have been shown to occur as early as preneoplastic PC [23,24]. As PBMCs act as the immune system’s first line of defense against cancer, using them as surrogate markers of PC may allow for earlier detection and would not be dependent on the presence of substantial tumor burden, which is a primary limitation of many current biomarkers.
Our previous study showed 383 genes to be differentially expressed in the PBMCs of PC patients as compared to those of healthy controls, with 65 having at least a 1.5 fold change in expression [23]. Furthermore we identified an eight-gene predictor set which could distinguish PC patients from healthy controls with a sensitivity of 83% and a specificity of 75%. As our previous study had a limited patient population, the present study attempted to validate and improve the clinical adaptability of these changes in PBMC gene expression for six genes chosen from the original microarray results (ANXA 3, ARG 1, SSBP2, CA5B, F5, and TBC1D8) as well as four other genes of interest based on previous studies (MIC1 [25–27], NGAL [28, 29] MUC1 [25,30,31], and MUC16 [32,33]) in a larger patient population using a multiplex PCR assay design. In addition, we determined how the expressions of these genes change as the cancer develops by comparing normal controls to both early stage (resectable) and late stage (nonresectable) PC patients. We also evaluated the potential diagnostic utility of this differential gene expression in both healthy individuals and in chronic pancreatitis (CP) patients, as compared to CA19-9, thus further determining the likely diagnostic utility of PBMC expression profiling in PC as well as if different single gene or combination tests will need to be developed for PC diagnosis across various pathological backgrounds.
The results of our study validated the differential expression of 5 of the 6 genes analyzed from our previous work. Additionally, our analysis indicates that the best genes for differentiating PC from HC patients are ARG1, which was found to be upregulated in PC (p = 0.014), and F5, which was found to be downregulated in PC (p = 0.036), while ARG1 (p = 0.043), CA5B (p = 0.0016), F5 (p = 0.0042), MIC1 (p = 0.044), and SSBP2 (p = 0.0053) were best for distinguishing PC from CP. Multivariate models for PBMC gene expression both independent of and in conjunction with plasma CA19-9 levels were attempted to determine if any combination was diagnostically superior to CA19-9 alone. We found that addition of PBMC CA5B, F5, SSBP2, and MIC-1 expression levels to CA19-9 significantly improved the diagnostic abilities of CA19-9 when comparing resectable PC to CP patients (AUC = 0.82 vs. 0.70 respectively, p = 0.023).
2. Materials and methods
2.1. Study population
The study of blood-based biomarkers in PC was approved by the Institutional Review Board (IRB) at the University of Pittsburgh Medical Center (UPMC) (IRB number 491-97-EP) in conjunction with Dr. Randall Brand, M.D. Written informed consent was obtained from all patients and controls before enrollment into the study. Upon collection, samples were shipped by overnight mail to the University of Nebraska Medical Center (UNMC) for processing. After processing samples were coded to blind those conducting the gene expression analysis to diagnosis and stage. All sample analysis was completed at UNMC. For this study, 35 CP patients, 47 healthy controls, 48 early, resectable (stage 1 or 2) PC patients, and 47 late, unresectable (stage 3 or 4) patients were recruited. In order to attain a power of 0.80 with a type-1 error (α) of 0.1, a sample size of 34 patients per group is required for detection of ≤ 1.5-fold differences in gene expression levels.
The diagnoses of PC and CP were made as per standard clinical practice. All PC samples were obtained pre-treatment. PC staging was either surgical based on operative pathology or biopsy of metastatic disease or clinical based on results of radiographic imaging studies. All patient demographic information can be found in Table 1.
Table 1.
Patient demographic information
| Healthy controls | Chronic pancreatitis | Early stage PC | Late stage PC | p-valuea | |
|---|---|---|---|---|---|
| N | 47 | 35 | 48 | 47 | |
| Mean age (SE) | 56.9 (2.05) | 55.3 (2.55) | 68.8 (1.34) | 66.7 (1.55) | < 0.001 |
| Sex (N (%) | 0.01 | ||||
| Mal | 13 (27.7) | 19 (54.3) | 21 (43.8) | 26 (55.3) | 0.019 |
| Femal | 34 (72.3) | 16 (45.7) | 27 (56.2) | 19 (40.4) | |
| Missing | 0 (0) | 0 (0) | 0 (0) | 2 (4.3) | |
| Race (N (%)) | 0.044 | ||||
| Caucasian | 44 (93.6) | 32 (91.4) | 48 (100) | 45 (95.7) | |
| African America | 2 (4.3) | 3 (8.6) | 0 (0) | 0 (0) | |
| Asian | 1 (2.1) | 0 (0) | 0 (0) | 0 (0) | |
| Missing | 0 (0) | 0 (0) | 0 (0) | 2 (4.3) | |
| PC Stage (N (%)) | |||||
| Stage 1 | 5 (10.4) | ||||
| Stage 2a | 13 (27.1) | ||||
| Stage 2b | 30 (62.5) | ||||
| Stage 3 | 17 (36.2) | ||||
| Stage 4 | 30 (63.8) |
P-values for age was calculated using an ANOVA model. Race and gender distributions were compared between the groups using Fisher’s exact test and chi-square test, respectively. Missing values were not included in the significance testing.
2.2. Isolation of total RNA from PBMCs
Upon sample arrival at UNMC, PBMCs were isolated from the whole blood using the PharmLyse RBC lysis solution (BS, San Jose, CA) according to the manufacturer’s instructions. Total RNA was extracted using the Qiagen RNAeasy RNA isolation kit (QIAGEN, Valencia, CA, USA) and then converted to cDNA using the SuperScript II cDNA synthesis kit (Invitrogen, Carlsbad, CA) according to a previously published protocol [34].
2.3. Q-RT PCR analysis of select gene expression
Samples were analyzed using quantitative real time PCR (Q-RT PCR) for gene expression using multiplexed Taqman based chemistry and normalized to β-actin. Six of the genes (ANXA3, ARG1, CA5B, F5, SSBP2, and TBC1D8) from our previous study’s 8 gene predictor set as well as MUC1, MUC16, NGAL, and MIC1 were analyzed. Multiplexing was performed for 2 genes at a time, consisting of β-actin (using the Cy5 fluorophore) and a gene of interest (using the FAM fluorophore). Quenching was accomplished for all probes via BHQ. The primers and probes (Integrated DNA Technologies) used in this study can be found in Supplementary Table 1. Fold-change in gene expression was determined using the 2−ΔΔCt method using human reference RNA (Agilent Stratagene Products, Cedar Creek, TX) as a standard.
2.4. CA19-9 radioimmunoassay assay
CA 19-9 antigen concentration was determined by a solid phase radioimmunoassay (Centocor, Malvern, PA, USA), using the manufacturers recommendation. All samples were analyzed in duplicate and the quantities of CA 19-9 were expressed in arbitrary units (U/ml) where one unit activity corresponds to approximately 0.8 ng of purified antigenic protein for CA 19-9 in a solid phase radioimmunoassay [35].
2.5. Statistical analysis
Interplate and intraplate variation were calculated using the coefficient of variance (CV = where = standard deviation and μ = mean). Due to the skewed nature inherent to the results of biomarker studies, all data was log-transformed prior to analysis. For ease of interpretation, all data presented is reverse-log-transformed with all values reported in Relative Expression Units (REU), defined as PBMC expression levels normalized to expression levels found in the employed universal human reference, unless otherwise stated. Samples were analyzed for statistically significant differences (p ≤ 0.05) between groups using ANOVA models, with Tukey’s adjustment for pairwise comparisons. As detection of early-stage PC is of greater consequence than late-stage disease, the ability of genes to distinguish between early PC and the two control groups (CP and healthy controls) was determined through cutoffs, derived through analysis of the Area Under the Curve (AUC) using Receiver Operating Characteristic (ROC) curve analysis, using a fixed specificity of 80% due to the fact that specificity is of greater importance than sensitivity for PC diagnostic biomarkers. Multivariate models were fit comparing resectable PC to both CP and healthy controls, with differentiating abilities compared to CA19-9 alone based on ROC curve analyses. For demographic information, age was compared between the 4 groups using an ANOVA model while race and gender distributions were compared between the groups using chi-square tests, with Fisher’s exact tests utilized for small sample size situations. SAS software Version 9.2 (SAS Institute Inc., Cary, NC) was used for all data analysis.
3. Results
3.1. Differential expression of genes in PBMCs of pancreatic cancer patients
A total of 177 samples were analyzed comprised of 95 (53%) pancreatic cancer (PC), 35 (20%) chronic pancreatitis (CP), and 47 (27%) healthy control (HC) patients. Of the analyzed PC patients, 48 (27%) were diagnosed with resectable disease (early PC, EPC) and 47 (26%) were diagnosed as unresectable (late PC, LPC). All analyzed demographic variables, consisting of age, gender, and race, were found to differ significantly between the groups (Table 1). Patients in the EPC and LPC groups were significantly older than those in the HC and CP groups (p < 0.001). Neither EPC and LPC ages nor HC and CP differed significantly. Race distribution was marginally different between the groups, with EPC and LPC groups having all Caucasian members. The HC group was also found to have a significantly larger proportion of females than the CP and LPC groups (both p < 0.05), while no other pairwise comparisons showed differences in gender.
For our multiplex Q-RT PCR assay, a total of 10 genes were selected for analysis. Six (60%)were genes found in our earlier study to be significantly altered in expression in PBMCs of PC patients compared to healthy controls. Four (MUC1, MUC16, MIC1, and NGAL) were selected based on previous reports of their differential upregulation in PC tissues compared to normal pancreata. After normalization, we found four genes to be qualitatively downregulated (CA5B, F5, MIC1, and SSBP2) and six to be qualitatively upregulated (ANXA3, ARG1, MUC1, MUC16, NGAL, and TBC1D8) in PC patients as compared to healthy controls (Table 2). Upon subdividing the PC population into early (Stage 1–2) and late (Stage 3–4) disease, we found that PBMC gene expression levels generally decreased from healthy controls to early PC patients (ANXA3, CA5B, F5, SSBP2, TDC1D8, and MIC1). Further, gene expression levels were generally found to increase with PC progression; with ANXA3, ARG1, F5, TBC1D8, MIC1, MUC1, and MUC16 all being expressed to a greater extent in LPC than EPC. Despite these observed trends, alteration of gene expression varied significantly with PC disease progression for F5 only (p = 0.028); whereas the trend remained non-significant for the other genes examined (p > 0.05).
Table 2.
Differential expression of selected genes between pancreatic cancer (PC) patients and control
| Marker | Previous studya | Present study | |||||
|---|---|---|---|---|---|---|---|
| (PC/Normal) | (PC/Normal) | (Early PC/Normal) | (Late PC Normal) | (PC/CP) | (Early PC/CP) | (Late PC/CP) | |
| Anxa3b | 1.79 | 1.13 | 0.87 | 1.40 | 0.89 | 0.68 | 1.09 |
| Arg1b | 2.11 | 1.49 | 1.14 | 1.83 | 1.33 | 1.02 | 1.64 |
| Ca5bb | 0.62 | 0.57 | 0.56 | 0.58 | 0.33 | 0.32 | 0.34 |
| F5b | 1.59 | 0.99 | 0.70 | 1.31 | 0.65 | 0.46 | 0.86 |
| SSBP2b | 0.69 | 0.79 | 0.80 | 0.78 | 0.62 | 0.62 | 0.61 |
| TBC1D8b | 1.44 | 1.54 | 0.92 | 2.19 | 2.10 | 1.26 | 2.99 |
| MUC1 | 1.00 | 0.96 | 1.05 | 1.03 | 0.98 | 1.07 | |
| MUC1 | 1.17 | 1.15 | 1.19 | 0.94 | 0.93 | 0.96 | |
| NGAL | 1.02 | 1.03 | 1.01 | 0.87 | 0.88 | 0.86 | |
| MIC1 | 0.84 | 0.76 | 0.93 | 0.59 | 0.53 | 0.66 | |
The previous study did not divide samples into early PC and late PC but rather grouped all PC together. Furthermore, it contained only 6 out of 35 samples that were considered early PC.
Genes chosen for validation in the expanded patient set.
Additionally, we also found that eight genes were upregulated in CP as compared to healthy controls (ANXA3, ARG1, CA5B, F5, SSBP2, NGAL, MIC1, and MUC16). Likewise seven genes (ANXA3, CA5B, F5, MIC1, MUC16, NGAL, and SSBP2) were expressed at higher levels in CP as compared to early PC while CP patients were found to express levels of 5 genes (F5, SSBP2, NGAL,MIC1, and MUC16) greater than those found in late PC (Table 3). Importantly, the intraplate and interplate coefficients of variation were found to be 7.58 × 10−4 and 1.07 × 10−2 respectively, indicating the reliability of our results.
Table 3.
Relative expression levels of tested genes in PBMCs of PC, CP, and HC patients
| Label | Group | Mean | Lower 95% CI | Upper 95% CI | Minimum | Maximum | p-valuea |
|---|---|---|---|---|---|---|---|
| ANXA3 | HC | 0.457 | 0.319 | 0.656 | 0.038 | 5.333 | 0.120 |
| CP | 0.584 | 0.362 | 0.944 | 0.038 | 11.353 | 0.120 | |
| EPC | 0.398 | 0.296 | 0.536 | 0.044 | 3.149 | ||
| LPC | 0.639 | 0.470 | 0.868 | 0.059 | 6.727 | ||
| ARG1 | HC | 4.444 | 3.321 | 5.945 | 0.812 | 44.942 | 0.014 |
| CP | 4.955 | 3.528 | 6.962 | 0.809 | 30.274 | 0.043 | |
| EPC | 5.049 | 3.725 | 6.844 | 0.465 | 38.319 | ||
| LPC | 8.140 | 6.034 | 10.984 | 0.856 | 47.505 | ||
| CA5B | HC | 3.465 | 2.143 | 5.603 | 0.150 | 32.786 | 0.140 |
| CP | 5.967 | 3.673 | 9.692 | 0.289 | 34.896 | 0.002 | |
| EPC | 1.924 | 1.254 | 2.951 | 0.285 | 21.481 | ||
| LPC | 2.003 | 1.268 | 3.163 | 0.164 | 22.943 | ||
| F5 | HC | 7.830 | 5.488 | 11.168 | 0.264 | 41.788 | 0.036 |
| CP | 11.901 | 8.060 | 17.578 | 0.287 | 51.625 | 0.004 | |
| EPC | 5.445 | 3.668 | 8.082 | 0.162 | 67.182 | ||
| LPC | 10.239 | 8.168 | 12.842 | 0.438 | 36.378 | ||
| SSBP2 | HC | 0.169 | 0.145 | 0.196 | 0.023 | 0.371 | 0.170 |
| CP | 0.216 | 0.177 | 0.263 | 0.053 | 0.616 | 0.005 | |
| EPC | 0.134 | 0.108 | 0.168 | 0.008 | 0.390 | ||
| LPC | 0.131 | 0.105 | 0.163 | 0.005 | 0.354 | ||
| TBC1D8 | HC | 0.314 | 0.164 | 0.601 | 0.000 | 1.641 | 0.087 |
| CP | 0.230 | 0.083 | 0.632 | 0.000 | 2.969 | 0.073 | |
| EPC | 0.288 | 0.150 | 0.554 | 0.000 | 2.071 | ||
| LPC | 0.686 | 0.444 | 1.061 | 0.000 | 4.028 | ||
| MUC1 | HC | 0.057 | 0.040 | 0.082 | 0.001 | 0.563 | 0.940 |
| CP | 0.056 | 0.039 | 0.079 | 0.010 | 0.688 | 0.930 | |
| EPC | 0.055 | 0.042 | 0.072 | 0.010 | 0.280 | ||
| LPC | 0.060 | 0.040 | 0.089 | 0.002 | 1.404 | ||
| MUC16 | HC | 0.402 | 0.298 | 0.542 | 0.009 | 2.462 | 0.700 |
| CP | 0.500 | 0.328 | 0.760 | 0.011 | 9.781 | 0.950 | |
| EPC | 0.463 | 0.353 | 0.607 | 0.076 | 5.696 | ||
| LPC | 0.478 | 0.343 | 0.666 | 0.091 | 6.566 | ||
| NGAL | HC | 11.337 | 9.017 | 14.259 | 0.796 | 45.728 | 0.980 |
| CP | 13.297 | 9.674 | 18.286 | 0.333 | 76.639 | 0.690 | |
| EPC | 11.705 | 9.238 | 14.826 | 0.530 | 61.181 | ||
| LPC | 11.448 | 9.349 | 14.021 | 1.366 | 75.322 | ||
| MIC1 | HC | 30.910 | 22.928 | 41.671 | 7.235 | 570.072 | 0.400 |
| CP | 43.956 | 28.383 | 68.116 | 4.823 | 643.591 | 0.044 | |
| EPC | 23.344 | 17.447 | 31.216 | 5.796 | 414.434 | ||
| LPC | 28.860 | 21.033 | 39.577 | 5.598 | 184.183 |
CI, Confidence Interval; HC, Healthy Controls; CP, Chronic Pancreatitis; EPC, Early (resectable) Pancreatic Cancer; LPC, Late (unresectable) Pancreatic Cancer;
P-values based on ANOVA analyses comprised of EPC, LPC, and the corresponding control group (CP or healthy controls).
Based on ANOVA analyses, ARG1 and F5 showed the best abilities to differentiate early PC, late PC, and healthy control groups, having p-values of 0.014 and 0.036, respectively. Better differentiation abilities were observed for PBMCs in early PC, late PC, and CP groups, with ARG1 (p = 0.043), CA5B (p = 0.002), F5 (p = 0.004), SSBP2 (p = 0.005), and MIC1 (0.044) all showing significant differential expressions. Additionally, differential expression of marginal significance was observed for TBC1D8 between these groups (p = 0.073) (Table 3).
3.2. Diagnostic potential of individual genes comparing EPC patients to healthy and CP controls
As differentiation of early PC from control groups is of the greatest consequence for diagnostic purposes, diagnostic abilities were analyzed in this group as compared to the healthy controls. Since specificity is of greater importance than sensitivity in PC diagnostic testing due to the relative rarity of the disease and significant psychological, financial, and physical consequence of false-positive diagnoses, individual cutoffs for each gene were designed based on a specificity ≥ 80%.
Using these criteria, none of the examined genes appeared individually promising for the differentiation of EPC from healthy controls, with AUCs consistently less than 0.60 (range = 0.503–0.584) (Table 4). Improved differentiation abilities of PBMC genetic expressions was observed, however, when comparing EPC to CP patients (AUCs = 0.517–0.700) (Table 5). Interestingly, CA5B was the most effective individual gene for differentiating EPC from both healthy and CP patients, giving AUC(SE)s of 0.584 (0.063) and 0.700 (0.066) with a sensitivities of 20.8% and 47.9% at its optimal cutoffs. F5 was found to provide the best sensitivity in comparing EPC to CP patients (61.7%), with an AUC(SE) similar to that of CA5B (0.678 (0.067)).
Table 4.
Diagnostic abilities of individual PBMC genetic expression analyses for differentiating early PC from healthy controls
| AUC 95% CI | Estimated | Estimated | ||||
|---|---|---|---|---|---|---|
| Gene | AUC | SE | Lower | Upper | Threshold* | Sensitivity* |
| ANXA3 | 0.526 | 0.063 | 0.402 | 0.649 | < 0.152 | 0.125 |
| ARG1 | 0.567 | 0.063 | 0.444 | 0.689 | ≥ 9.32 | 0.295 |
| CA5B | 0.584 | 0.063 | 0.461 | 0.706 | < 0.39 | 0.208 |
| F5 | 0.561 | 0.063 | 0.438 | 0.685 | < 2.43 | 0.319 |
| SSBP2 | 0.535 | 0.063 | 0.412 | 0.659 | < 0.13 | 0.292 |
| TBC1D8 | 0.501 | 0.064 | 0.376 | 0.626 | < 0.35 | 0.292 |
| MUC1 | 0.539 | 0.063 | 0.415 | 0.663 | < 0.019 | 0.146 |
| MUC16 | 0.503 | 0.063 | 0.379 | 0.627 | < 0.23 | 0.196 |
| NGAL | 0.504 | 0.063 | 0.38 | 0.628 | < 8.57 | 0.271 |
| MIC1 | 0.574 | 0.063 | 0.452 | 0.697 | < 12.17 | 0.261 |
AUC, Area Under the Curve; SE, Standard Error; CI, Confidence Interval;
All thresholds and sensitivities were estimated using a fixed specificity of 0.80.
Table 5.
Diagnostic abilities of individual PBMC genetic expression analyses for differentiating early PC from CP patients
| AUC 95% CI | Estimated | Estimated | ||||
|---|---|---|---|---|---|---|
| Gene | AUC | SE | Lower | Upper | Threshold* | Sensitivity* |
| ANXA3 | 0.592 | 0.071 | 0.452 | 0.731 | < 0.20 | 0.271 |
| ARG1 | 0.516 | 0.071 | 0.376 | 0.656 | > 14.67 | 0.205 |
| CA5B | 0.700 | 0.066 | 0.572 | 0.829 | ≤ 1.43 | 0.479 |
| F5 | 0.678 | 0.067 | 0.547 | 0.808 | ≤ 9.99 | 0.617 |
| SSBP2 | 0.685 | 0.067 | 0.554 | 0.817 | ≤ 0.17 | 0.479 |
| TBC1D8 | 0.537 | 0.074 | 0.391 | 0.682 | < 0.23 | 0.167 |
| MUC1 | 0.536 | 0.072 | 0.394 | 0.677 | < 0.024 | 0.208 |
| MUC16 | 0.537 | 0.072 | 0.397 | 0.678 | < 0.28 | 0.261 |
| NGAL | 0.557 | 0.071 | 0.419 | 0.696 | < 9.25 | 0.333 |
| MIC1 | 0.640 | 0.069 | 0.504 | 0.776 | ≤ 12.82 | 0.37 |
AUC, Area Under the Curve; SE, Standard Error; CI, Confidence Interval;
All thresholds and sensitivities were estimated using a fixed specificity of 0.80.
3.3. Combination testing is superior to CA19-9 alone in the differentiation of early PC from CP
While PBMC expression profiling is highly promising for PC diagnostics in theory, the complexities of the immune system and the immune-neoplastic interaction make it unlikely that the monitoring of any single gene will provide any diagnostic benefit. This observation, further highlighted by the above results, necessitates the consideration of PBMC genetic expression levels for PC diagnosis as a combination test consisting of multiple genes either with or without other PC surrogate biomarkers. Towards these ends, multivariate analyses were performed to determine the odds ratios (OR) of a PC diagnosis using the given univariate-derived cutoffs for the best 4 (by AUC) in comparing early PC to the CP and healthy controls (Tables 4 and 5). Additionally, plasma levels of CA19-9 were analyzed for each sample and AUCs and ORs were calculated for comparing the patient groups. ORs for the genes and CA19-9 were then rounded to the nearest whole number and combined into a score-based formula for early PC diagnosis and compared to the gold-standard CA19-9 alone based on AUC.
For differentiating early PC from healthy controls, the 4 genes with the highest AUCs based on univariate analysis were MIC1 (0.574), ARG1 (0.567), F5 (0.561), and CA5B (0.584). Upon multivariate analysis, the ORs for these genes were found to be 1.253, 1.638, 1.949, and 0.914 respectively. In comparison, CA19-9 was found to have an AUC of 0.719 with an OR of 8.565 at a cutoff (to provide 80% specificity) of 61.7 U/mL. From this, the following formula was constructed:
Score (early PC vs. healthy Controls) = 1*(MIC1 < 12.17 REU) + 2*(ARG1 ≥ 9.32 REU) + 2*(F5 < 2.43 REU) + 1*(CA5B < 0.39 REU) + 9*(CA19-9 ≥ 61.7 U/mL) with a score calculated such that if MIC1 < 12.2 then assign 1 point, otherwise 0; if ARG1 ≥ 9.32 assign 2 points, otherwise 0; if F5 < 2.43 assign 2 points, otherwise 0; etc. Then add up the total points assigned for each biomarker to determine the total score for a patient. If the total score is found to be ≤ 4, a diagnosis of healthy control is predicted, but if the total score > 4 then we predict the patient has early PC. For ease of reporting, we have termed this combination test “Panel A”.
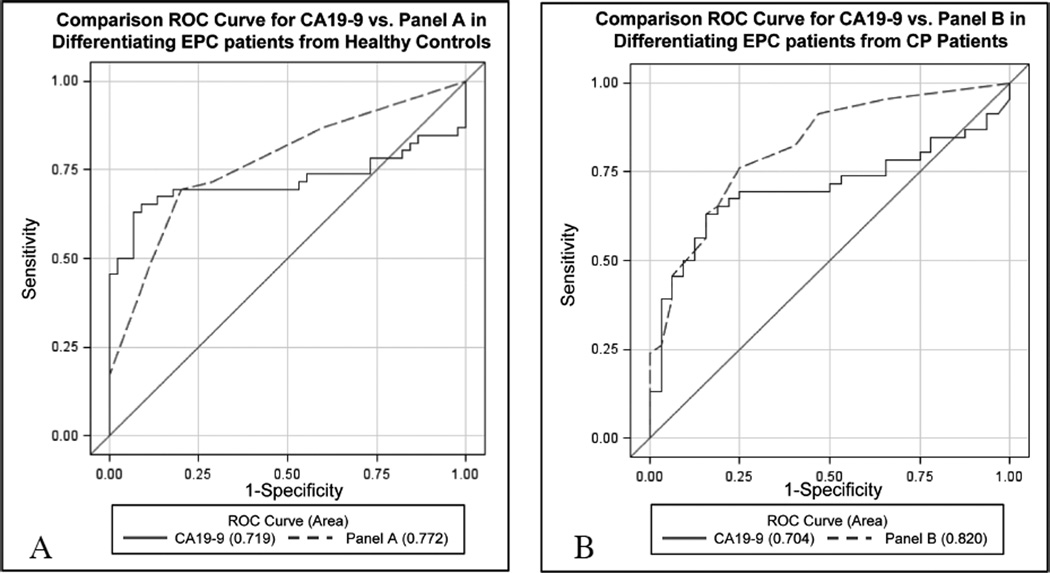
Considering both the total score and CA19-9 as continuous variables, the AUC for the total score model was found to be 0.772 (95% CI: 0.68–0.87),which was not found to be significantly greater than the AUC for CA19-9 alone (AUC diff = 0.053, 95% CI: −0.02 to 0.13, p = 0.17) (Fig. 1, Table 6).
Fig. 1.
Comparison receiver operating characteristic (ROC) curves comparing combination testing to CA19-9 for the differentiation of early pancreatic cancer patients from controls. Comparison ROC curves were constructed analyzing combination tests consisting of multiplex Q-RT PCR-based PBMC expression profiles combined with plasma CA19-9 levels versus plasma CA19-9 alone for the differentiation of Early (resectable) Pancreatic Cancer (EPC) from A) Healthy Controls and B) Chronic Pancreatitis (CP) patients. Panel A consists of PBMC-based expression of CA5B, F5, MIC1, and ARG1 along with plasma CA19-9 levels and was not found to be significantly superior to CA19-9 alone for differentiating EPC from healthy control patients (p = 0.17) Panel B consists of CA5B, F5, MIC1, and SSBP2 expression analysis along with plasma CA19-9 levels. Panel B was found to distinguish EPC from CP patients significantly better than CA19-9 alone (P = 0.023). (Colours are visible in the online version of the article; http://dx.doi.org/10.3233/CBM-2012-0260)
Table 6.
Diagnostic improvement in differentiating resectable PC (EPC) from controls observed when combining PBMC expression analysis with the gold standard CA19-9
| EPC vs HC | EPC vs CP | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test | Composition | AUC | Sensitivity | Specificity | P-value | Composition | AUC | Sensitivity | Specificity | P-value |
| Gold standard | CA19-9 clinical cutoff > 37U/mL | 0.719 | 74% | 27% | 0.17 | CA19-9 clinical cutoff > 37U/mL | 0.704 | 74% | 34% | 0.023 |
| CA19-9 optimal cutoff ≥; 61.7U/mL | 70% | 80% | CA19-9 optimal cutoff > 74.0 U/mL | 65% | 80% | |||||
| Combination test | Panel Aa | 0.772 | 67% | 81% | Panel Bb | 0.820 | 67% | 83% | ||
EPC, Early Pancreatic Cancer; HC, Healthy Controls; CP, Chronic Pancreatitis; AUC, Area Under the ROC Curve
Panel A represents a combination test consisting of PBMC expression analysis of CA5B, F5, MIC1, and ARG1 along with plasma CA19-9 levels;
Panel B represents a combination test consisting of PBMC expression analysis of CA5B, F5, MIC1, and SSBP2 along with plasma CA19-9 levels.
Multi-gene testing for differentiating early PC from CP yielded more promising results. Multivariate analysis of the best 4 genes (AUC) based on univariate analysis, CA5B (0.700), F5 (0.678), SSBP2 (0.685), and MIC1 (0.640), were found to give ORs of 2.19, 3.38, 2.56, and 1.78, respectively. Comparatively, the AUC for CA19-9 was found to be 0.704 with an OR of 5.66 at a cutoff of > 74.0 U/mL. Using these ORs, the formula:
Score (early PC vs. CP) = 2*(CA5B ≤ 1.43 REU) + 3*(F5 ≤ 9.99 REU) + 3*(SSBP2 ≤ 0.17 REU) + 2*(MIC1 ≤ 12.82 REU) + 6*(CA19-9 > 74.0 U/mL) was made, with a total score of ≤ 6 resulting in a predicted diagnosis of CP, while a total score > 6 leads to a prediction of a early PC diagnosis. We have termed this combination test “Panel B”. The AUC for this formula was found to be 0.820 (95% CI: 0.73–0.91), a significant improvement over CA19-9 alone (AUC diff = 0.116, 95% CI: 0.016 to 0.220, p = 0.023) (Fig. 1). As further illustration of the superiority of this PBMC expression/CA19-9 combination test over CA19-9 alone, sensitivity and specificity were calculated for each using the designed test cutoff values. The combination test was able to improve all aspects of early PC diagnosis in the context of CP, providing a 2% increase in sensitivity (67% vs. 65%) and a 3% increase in specificity (83% vs. 80%) over CA19-9 alone (Table 6).
3.4. Validation of previous results
A unique feature of this study is the fact that we used a set of patient PBMC samples unique from those previously studied to validate the results of our previous report [23]. Six of the genes from our previously established 8-gene predictor set were chosen for validation by Q-RT PCR in this expanded population. The mean fold-change in expression for five of the genes (ANXA3, ARG1, CA5B, SSBP2, and TBC1D8) was in the same direction as the previous study when comparing the entire population of PC patients to normal controls. When the PC population was restricted to nonresectable patients, as was the case for the majority (29/35) of samples analyzed in our original study, the median fold expression change for all six of the genes was in the same direction as found in the previous study, strengthening the validity of our earlier results (Table 2).
4. Discussion
Several recent studies have demonstrated that gene expression in PBMCs is altered in the context of malignancy [4,19–23,36,37]. A differential gene expression profile in the PBMCs of PC patients was first reported by Huang et al. in their study using microarray and Q-RT PCR validation to identify 48 differentially expressed genes as potential biomarkers for differentiating newly diagnosed diabetic patients with PC from diabetic patients without PC. These results, showing PBMC gene expression alterations, were further bolstered by research conducted in our lab demonstrating that 383 genes are differentially expressed between PC patients and healthy controls. Further, we developed an eight gene predictor set comprised of SSBP2, Ube2b-rs1, CA5B, F5, TBC1D8, ANXA3, ARG1, and ADAMTS20 that could distinguish between PC and HCs with a sensitivity and specificity of 83% and 75% respectively. However, this initial study was conducted using the SYBR green method employing a standard human reference RNA. This method however is difficult to employ clinically for several reasons. The most important is the difficulty in maintaining a stable, large stock of the same human reference RNA for multiple centers for repeated use.
To overcome this problem, we investigated in this study the use of absolute quantification methods employing TaqMan chemistry to develop a practically applicable set of genes that can be employed as an adjunct in the diagnosis of PC patients. We found that CA5B, one of the eight genes in the original predictor set could distinguish early PC patients from both CP and HC patients with the highest accuracy (based on AUC). Further, a mathematical model developed from the gene combination was found to be better at differentiating resectable PC from CP than CA19-9. Additionally, the present study validated five genes (ANXA3, ARG1, CA5B, SSBP2, and TBC1D8) found to be differentially expressed between PC patients and healthy controls from our previous work. Importantly however, our previous study did not distinguish between early and late PC, containing only 6 of 35 patients with an early PC diagnosis. When the differential expression compared to healthy controls was limited to only include late PC patients, the five previously listed genes as well as F5 were found to be differentially expressed in a similar manner to that found in the previous study, providing a 100% validation of results. This consistency in differential expression across an expanded patient population and multiple testing platforms adds significant credence to PBMC transcriptional profiling as a valid diagnostic option in PC.
The early diagnostic potential of PBMC differential gene expression is theoretically quite high when it is considered that the two mechanisms most likely responsible for the observed differential expression are the immune system’s recognition of the cancer and the evasion of the immune system by the cancer. As immune system evasion has been shown to occur as early as pre-malignant disease in PC, differential PBMC expression may also occur in this early, resectable stage posing a significant advantage over current biomarkers, such as CA19-9, whose concentrations are dependent on tumor burden [24]. Further, this likelihood of detectable changes in early disease provides theoretical superiority to currently available PC imaging techniques which often fail to detect small or premalignant lesions and though which it is difficult to differentiate PC from CP-induced pancreatic masses. Substantiation of this premise is bolstered by our observation that PBMC differential expression is detectable in early stage PC patients.
As it is not clinically practical to screen the entire population for early detection of PC given the small percentage of the population that it affects, it would be more efficient to identify a test to screen solely those individuals who have a high risk of developing PC, such as patients with chronic pancreatitis, patients with type II diabetes mellitus, chronic smokers, individuals with a working history in the rubber industry, patients with a history of tropical or idiopathic pancreatitis or cystic fibrosis, and patients with a family history of PC [9].
To date, no current biomarker has been conclusively shown to aid in the differentiation of PC from CP. Current studies using CA19-9 to stratify PC and CP patients are contradictory with some showing its ability to successfully differentiate between the two patient groups while others show CA19-9 to be high not only in PC patients (90%) but in many CP patients (66%) as well [38,39]. The utility of other biomarkers in differentiating PC and CP has also been explored. A Western blot array (Powerblot, BD Biosciences) analysis of pooled protein samples from normal, CP and PC derived pancreatic tissues identified more than 50 proteins that were upregulated in PC compared to CP, while an almost equal number were downregulated [40,41]. Proteomics based approaches have identified Maspin [42], MUC4-p53 combination [43] Annexin-2, Insulin-like growth factor binding protein 2 (IGFBP-2) [44], and Zinc-alpha-2-glycoprotein (AZGP1) [45] as possible biomarkers to distinguish PC from CP. Another study identified a set of seven genes that could discriminate between CP and PC with an accuracy of 92% in a randomly assigned training set. However, the applicability of the results of many of these studies are limited due to small samples sizes, no distinction between resectable and unresectable PC, a lack of validation in a test set, and no direct comparison with plasma CA19-9 levels. Additionally, most of these studies used pancreatic tissue as a source for biomarkers, making such tests invasive and thereby limiting the patient population in which such a test could be used.
By contrast, from the ten genes analyzed in PBMCs through this study, a combination test consisting of PBMC expression levels of CA5B, F5, SSBP2, and MIC1 combined with plasma levels of CA19-9 was identified that can distinguish resectable PC from CP patients with a sensitivity and specificity of 67% and 83%, respectively. This represents a significant (p = 0.023) improvement over plasma CA19-9 alone, even when used at its optimal cutoff of 74.0 U/mL which is substantially greater than the cutoff currently used clinically (37 U/mL).
Importantly, as PC cannot be attributed solely to one high-risk group but rather to many, with each contributing a small proportion of PC patients to the general PC population, knowledge as to the universality of early PC diagnostic testing across patients of different backgrounds is imperative. Based on our results, it appears that diagnostic testing based upon PBMC expression profiling may require different diagnostic formulas, employing differing gene sets and/or expression cutoffs as directions and magnitudes of differential expression vary based on the control group that PC is compared against. However, of the respective four genes employed in the two diagnostic formulas designed in this study, three (CA5B, F5, and MIC1) were shared; though the cutoffs used in the early PC vs. healthy control and the early PC vs. CP tests were dissimilar (0.39 vs. 1.43, 2.43 vs. 9.99, and 12.2 vs. 12.8 REU). These results highlight the necessity for further testing in other high-risk groups in addition to CP, such as patients with a significant family history of PC, to establish both the diagnostic potential and respective diagnostic formulas for PBMC expression analysis within each as well as to discover latent PBMC expression similarities across high-risk groups that may be exploitable for designing a test with universal applicability.
Adding to the diagnostic interpretability of the results of this work, the present study utilizes a technique that is clinically viable and could easily be added to the repertoire of tests already provided in a standard clinical lab. PBMC gene expression analysis is no more invasive of a test than CA19-9, both being amendable to a simple venopuncture; thus providing practical diagnostic superiority over techniques currently utilized in PC diagnosis such as endoscopic ultrasound-guided fine needle aspiration and endoscopic retrograde colangiopancreatography, through which early PC may be detectable but the invasiveness of which limit utility solely to confirmatory testing. In addition, adapting PBMC testing to a multiplex Q-RT PCR format is particularly promising as it provides a quantitative means by which complex assays can be streamlined to produce multiple results with minimal effort and still maintain a high level of specificity. Additionally, such assays do not require a large quantity of starting template, which is essential as PBMCs are not highly concentrated in blood. Multiplex Q-RT PCR is an ideal diagnostic assay because it is rapid, does not require multiple rounds of confirmatory testing, has a relatively low setup and operation cost, and can analyze multiple genes in a single reaction [46–48]. Further, multiplex Q-RT PCR assays have already proven viable in a clinical context, as they have been developed for the detection of a broad spectrum of bacteria and fungi in human blood (LightCycler SeptiFast test) [49] and for the detection of metallo-β-lactamase genes in gram negative bacteria (hyplex MBL ID Multiplex PCR-ELISA) [50]. Multiplex Q-RT PCR has also been shown to be successful in the diagnosis of severe combined immunodeficiency disorder (SCID) [51], as well as the detection of circulating tumor cells in breast cancer [52].
Beyond the diagnostic potential of this PBMC differential gene expression profile, the normal functions and direction of differential expression of each of the genes hints at a potential pathophysiological mechanism. It appears that PC is characterized by a significant decrease in the ability of the immune system to respond to non-self-antigens, including tumor associated antigens, which may in part be a result of the upregualtion of ARG1 which was further validated in this study [23]. ARG1 expression has been closely associated with an increase in the presence of myeloid derived suppressor cells (MDSCs) [53] which are classically known to decrease CTL response, mostly through destabilization of T-cell receptors and decreased expression of certain CD3 subtypes, ultimately leading to CTL apoptosis.
Another gene of interest that may play a role in decreasing the body’s immunological function is F5, which was shown to be down regulated in early PC. F5 associates with the VH3 domain of IgE to increase the release of IL4, which is associated with the proliferation, differentiation, chemotaxis, and development of B cells and T cells. Therefore, a decrease in F5 may result in a decreased release of IL4, ultimately resulting in the down regulation of the body’s humoral and cellular immune response machinery. While our study did not directly investigate the expression of IL4, our results do suggest the need for further research to explore this possible mechanism.
A shortcoming of the study is the knowledge of whether a correlation exists between expression of a gene in the PBMCs and the same gene in tissues from PC patients. The problem of such an analysis would be obtaining the pancreas of matched HCs and CP. A solution would be to investigate in a mouse model of spontaneous PC wherein the expression of homologous genes is compared in wild type and transgenic mice, thus mimicking the comparison of PC to HCs. Another possibility is to conduct a retrospective analysis using samples of HCs and CP patients banked in tissue banks. This might give us the opportunity to use blood and tissue from the same patient. However, the issue of RNA stability introduces an additional variability and thus would make the results difficult to interpret. Other possibilities that can be explored in future studies include developing gene sets to distinguish smokers who develop PC from those who do not develop PC and a set of genes that reflect the risk of development of PC in long standing CP.
Another minor pitfall of the study is that the genes used to differentiate PC from CP were all down regulated, potentiating that the effects of CP on PBMC gene expression could overshadow the effects of PC. However, the use of multiple genes and the quantitative nature of the technique should help overcome this problem, further strengthening the argument in favor of a multiplexed assay.
In conclusion, we have investigated the expression of six genes previously identified as being differentially expressed in PBMCs of PC patients. Our results, performed on a unique set of patient samples distinct from the earlier study reveal that the six genes were affected in the same direction as previously reported. We observed that a combination of genes was able to significantly improve the diagnostic abilities of CA19-9 for resectable PC in the context of CP. The study is significant in that it demonstrates that use of PBMCs is a reproducible, quantitative method which can be applied clinically. Future studies will aim to validate the model using samples from other patient groups commonly encountered in the differential diagnosis of PC including other solid tumor malignancies, benign tumors and inflammatory conditions. Further, we will also examine the functions of these genes in animal models of PC to elucidate their role in the pathogenesis of the malignancy.
Supplementary Material
Acknowledgments
Financial support
This work was in part supported by grants from the National Institutes of Health (RO1 CA131944, EDRN UO1 CA111294, and SPORE P50 CA127297).
Role of the funding source
Study sponsors played no role in the design, sample collection, analysis, data interpretation or decision to submit this work.
Footnotes
Conflict of interest
The authors declare that no conflicts of interest exists for this work
References
- 1.Cancer Facts and Figures 2011. Atlanta: American Cancer Society; 2011. [Google Scholar]
- 2.National Cancer Institute. Surveillance Epidemiology and End Results Cancer Statistics Review 1975–2006. National Cancer institute. 2011 Ref Type: Electronic Citation. [Google Scholar]
- 3.Ghaneh P, Costello E, Neoptolemos JP. Biology and management of pancreatic cancer. Postgrad Med J. 2008;84:478–497. doi: 10.1136/gut.2006.103333. [DOI] [PubMed] [Google Scholar]
- 4.Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhanot UK, Moller P. Mechanisms of parenchymal injury and signaling pathways in ectatic ducts of chronic pancreatitis: implications for pancreatic carcinogenesis. Lab Invest. 2009;89:489–497. doi: 10.1038/labinvest.2009.19. [DOI] [PubMed] [Google Scholar]
- 6.Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, Yeo C, Fishman EK, Brune K, Axilbund J, Griffin C, Ali S, Richman J, Jagannath S, Kantsevoy SV, Kalloo AN. Screening for early pancreatic neoplasia in high-risk individuals: a prospective controlled study. Clin Gastroenterol Hepatol. 2006;4:766–781. doi: 10.1016/j.cgh.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Howes N, Greenhalf W, Stocken DD, Neoptolemos JP. Cationic trypsinogenmutations and pancreatitis. Clin Lab Med. 2005;25:39–59. doi: 10.1016/j.cll.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, DiMagno EP, Andren-Sandberg A, Domellof L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group N Engl J Med. 1993;328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 10.Pho-Iam T, Thongnoppakhun W, Yenchitsomanus PT, Limwongse C. A Thai family with hereditary pancreatitis and increased cancer risk due to a mutation in PRSS1 gene. World J Gastroenterol. 2005;11:1634–1638. doi: 10.3748/wjg.v11.i11.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uomo G, Rabitti PG. Chronic pancreatitis: relation to acute pancreatitis and pancreatic cancer. Ann Ital Chir. 2000;71:17–21. [PubMed] [Google Scholar]
- 12.Tsirambidis JV, Conwell DL, Zuccaro G. Chronic pancreatitis. MedGenMed. 2003;5:17. [PubMed] [Google Scholar]
- 13.Tumour markers in gastrointestinal cancers – EGTM recommendations. European Group on Tumour Markers. Anticancer Res. 1999;19:2811–2815. [PubMed] [Google Scholar]
- 14.DiMagno EP, Reber HA, Tempero MA. AGA technical review on the epidemiology, diagnosis, and treatment of pancreatic ductal adenocarcinoma. American Gastroenterological Association. Gastroenterology. 1999;117:1464–1484. doi: 10.1016/s0016-5085(99)70298-2. [DOI] [PubMed] [Google Scholar]
- 15.Goggins M. Molecular markers of early pancreatic cancer. J Clin Oncol. 2005;23:4524–4531. doi: 10.1200/JCO.2005.19.711. [DOI] [PubMed] [Google Scholar]
- 16.Ni XG, Bai XF, Mao YL, Shao YF, Wu JX, Shan Y, Wang CF, Wang J, Tian YT, Liu Q, Xu DK, Zhao P. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur J Surg Oncol. 2005;31:164–169. doi: 10.1016/j.ejso.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Pleskow DK, Berger HJ, Gyves J, Allen E, McLean A, Podolsky DK. Evaluation of a serologic marker, CA19-9, in the diagnosis of pancreatic cancer. Ann Intern Med. 1989;110:704–709. doi: 10.7326/0003-4819-110-9-704. [DOI] [PubMed] [Google Scholar]
- 18.Steinberg W. The clinical utility of the CA 19-9 tumor-associated antigen. Am J Gastroenterol. 1990;85:350–355. [PubMed] [Google Scholar]
- 19.Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, Larsen G, Foxwell BM, Brennan FM, Feldmann M, Pittman DD. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol Med. 2007;13:40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun CJ, Zhang L, Zhang WY. Gene expression profiling of maternal blood in early onset severe preeclampsia: identification of novel biomarkers. J Perinat Med. 2009;37:609–616. doi: 10.1515/JPM.2009.103. [DOI] [PubMed] [Google Scholar]
- 21.Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W, Logan T, Dutcher J, Hudes G, Dorner AJ, Slonim DK, Trepicchio WL, Burczynski ME. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res. 2003;63:6069–6075. [PubMed] [Google Scholar]
- 22.Bluth M, Lin YY, Zhang H, Viterbo D, Zenilman M. Use of gene expression profiles in cells of peripheral blood to identify new molecular markers of acute pancreatitis. Arch Surg. 2008;143:227–233. doi: 10.1001/archsurg.2007.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baine MJ, Chakraborty S, Smith LM, Mallya K, Sasson AR, Brand RE, Batra SK. Transcriptional profiling of peripheral blood mononuclear cells in pancreatic cancer patients identifies novel genes with potential diagnostic utility. PLoS One. 2011;6:e17014. doi: 10.1371/journal.pone.0017014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao F, Obermann S, von WR, Haile L, Manns MP, Korangy F, Greten TF. Increase in frequency of myeloid-derived suppressor cells in mice with spontaneous pancreatic carcinoma. Immunology. 2009;128:141–149. doi: 10.1111/j.1365-2567.2009.03105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grote T, Logsdon CD. Progress on molecular markers of pancreatic cancer. Curr Opin Gastroenterol. 2007;23:508–514. doi: 10.1097/MOG.0b013e3282ba5724. [DOI] [PubMed] [Google Scholar]
- 26.Bunger S, Laubert T, Roblick UJ, Habermann JK. Serum biomarkers for improved diagnostic of pancreatic cancer: a current overview. J Cancer Res Clin Oncol. 2011;137:375–389. doi: 10.1007/s00432-010-0965-x. [DOI] [PubMed] [Google Scholar]
- 27.Brown DA, Lindmark F, Stattin P, Balter K, Adami HO, Zheng SL, Xu J, Isaacs WB, Gronberg H, Breit SN, Wiklund FE. Macrophage inhibitory cytokine 1: a new prognostic marker in prostate cancer. Clin Cancer Res. 2009;15:6658–6664. doi: 10.1158/1078-0432.CCR-08-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Zhao ZS, Tao HQ. Expressions of neutrophil gelatinase-associated lipocalin in gastric cancer: a potential biomarker for prognosis and an ancillary diagnostic test. Anat Rec (Hoboken) 2010;293:1855–1863. doi: 10.1002/ar.21230. [DOI] [PubMed] [Google Scholar]
- 29.Porta C, Paglino C, De AM, Quaglini S, Sacchi L, Imarisio I, Canipari C. Predictive value of baseline serum vascular endothelial growth factor and neutrophil gelatinase-associated lipocalin in advanced kidney cancer patients receiving sunitinib. Kidney Int. 2010;77:809–815. doi: 10.1038/ki.2009.552. [DOI] [PubMed] [Google Scholar]
- 30.Uen YH, Lin SR, Wu CH, Hsieh JS, Lu CY, Yu FJ, Huang TJ, Wang JY. Clinical significance of MUC1 and c-Met RT-PCR detection of circulating tumor cells in patients with gastric carcinoma. Clin Chim Acta. 2006;367:55–61. doi: 10.1016/j.cca.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Cheng JP, Yan Y, Wang XY, Lu YL, Yuan YH, Jia J, Ren J. MUC1-positive circulating tumor cells and MUC1 protein predict chemotherapeutic efficacy in the treatment of metastatic breast cancer. Chin J Cancer. 2011;30:54–61. doi: 10.5732/cjc.010.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiss A, Timms JF, Smith C, Devetyarov D, Gentry-Maharaj A, Camuzeaux S, Burford B, Nouretdinov I, Ford J, Luo Z, Jacobs I, Menon U, Gammerman A, Cramer R. Highly accurate detection of ovarian cancer using CA125 but limited improvement with serum matrix-assisted laser desorption/ionization time-of-flight mass spectrometry profiling. Int J Gynecol Cancer. 2010;20:1518–1524. [PubMed] [Google Scholar]
- 33.Chang ST, Zahn JM, Horecka J, Kunz PL, Ford JM, Fisher GA, Le QT, Chang DT, Ji H, Koong AC. Identification of a biomarker panel using a multiplex proximity ligation assay improves accuracy of pancreatic cancer diagnosis. J Transl Med. 2009;7:105. doi: 10.1186/1479-5876-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 35.Del Villano BC, Brennan S, Brock P, Bucher C, Liu V, McClure M, Rake B, Space S, Westrick B, Schoemaker H, Zurawski VR., Jr Radioimmunometric assay for a monoclonal antibody-defined tumor marker, CA 19-9. Clin Chem. 1983;29:549–552. [PubMed] [Google Scholar]
- 36.Burczynski ME, Twine NC, Dukart G, Marshall B, Hidalgo M, Stadler WM, Logan T, Dutcher J, Hudes G, Trepicchio WL, Strahs A, Immermann F, Slonim DK, Dorner AJ. Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res. 2005;11:1181–1189. [PubMed] [Google Scholar]
- 37.Huang H, Dong X, Kang MX, Xu B, Chen Y, Zhang B, Chen J, Xie QP, Wu YL. Novel blood biomarkers of pancreatic cancer-associated diabetes mellitus identified by peripheral blood-based gene expression profiles. Am J Gastroenterol. 2010;105:1661–1669. doi: 10.1038/ajg.2010.32. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka N, Okada S, Ueno H, Okusaka T, Ikeda M. The usefulness of serial changes in serum CA19-9 levels in the diagnosis of pancreatic cancer. Pancreas. 2000;20:378–381. doi: 10.1097/00006676-200005000-00007. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe H, Kawakami H, Yamakawa O, Satomura Y, Ohta H, Motoo Y, Okai T, Sawabu N. Clinical usefulness of tumor markers associated with pancreatic cancer. Rinsho Byori. 1994;42:127–138. [PubMed] [Google Scholar]
- 40.Chang MC, Chang YT, Su TC, Yang WS, Chen CL, Tien YW, Liang PC, Wei SC, Wong JM. Adiponectin as a potential differential marker to distinguish pancreatic cancer and chronic pancreatitis. Pancreas. 2007;35:16–21. doi: 10.1097/MPA.0b013e3180547709. [DOI] [PubMed] [Google Scholar]
- 41.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, Costello E, Neoptolemos J, Lemoine NR. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Zheng B, Robbins DH, Lewin DN, Mikhitarian K, Graham A, Rumpp L, Glenn T, Gillanders WE, Cole DJ, Lu X, Hoffman BJ, Mitas M. Accurate discrimination of pancreatic ductal adenocarcinoma and chronic pancreatitis using multimarker expression data and samples obtained by minimally invasive fine needle aspiration. Int J Cancer. 2007;120:1511–1517. doi: 10.1002/ijc.22487. [DOI] [PubMed] [Google Scholar]
- 43.Nash JW, Bhardwaj A, Wen P, Frankel WL. Maspin is useful in the distinction of pancreatic adenocarcinoma from chronic pancreatitis: A tissue microarray based study. Appl Immunohistochem Mol Morphol. 2007;15:59–63. doi: 10.1097/01.pai.0000203037.25791.21. [DOI] [PubMed] [Google Scholar]
- 44.Bhardwaj A, Marsh WL, Jr, Nash JW, Barbacioru CC, Jones S, Frankel WL. Double immunohistochemical staining with MUC4/p53 is useful in the distinction of pancreatic adenocarcinoma from chronic pancreatitis: a tissue microarray-based study. Arch Pathol Lab Med. 2007;131:556–562. doi: 10.5858/2007-131-556-DISWPI. [DOI] [PubMed] [Google Scholar]
- 45.Pan S, Chen R, Crispin DA, May D, Stevens T, McIntosh MW, Bronner MP, Ziogas A, Anton-Culver H, Brentnall TA. Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J Proteome Res. 2011;10:2359–2376. doi: 10.1021/pr101148r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond) 2005;109:365–379. doi: 10.1042/CS20050086. [DOI] [PubMed] [Google Scholar]
- 47.Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med. 2003;9:189–195. doi: 10.1016/s1471-4914(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 48.Mocellin S, Rossi CR, Marincola FM. Quantitative real-time PCR in cancer research. Arch Immunol Ther Exp (Warsz) 2003;51:301–313. [PubMed] [Google Scholar]
- 49.Lamoth F, Jaton K, Prod’hom G, Senn L, Bille J, Calandra T, Marchetti O. Multiplex blood PCR in combination with blood cultures for improvement of microbiological documentation of infection in febrile neutropenia. J Clin Microbiol. 2010;48:3510–3516. doi: 10.1128/JCM.00147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avlami A, Bekris S, Ganteris G, Kraniotaki E, Malamou-Lada E, Orfanidou M, Paniara O, Pantazatou A, Papagiannitsis CC, Platsouka E, Stefanou I, Tzelepi E, Vagiakou H, Miriagou V. Detection of metallo-beta-lactamase genes in clinical specimens by a commercial multiplex PCR system. J Microbiol Methods. 2010;83:185–187. doi: 10.1016/j.mimet.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Gerstel-Thompson JL, Wilkey JF, Baptiste JC, Navas JS, Pai SY, Pass KA, Eaton RB, Comeau AM. High-throughput multiplexed T-cell-receptor excision circle quantitative PCR assay with internal controls for detection of severe combined immunodeficiency in population-based newborn screening. Clin Chem. 2010;56:1466–1474. doi: 10.1373/clinchem.2010.144915. [DOI] [PubMed] [Google Scholar]
- 52.Lianidou ES, Markou A. Circulating Tumor Cells in Breast Cancer: Detection Systems, Molecular Characterization, and Future Challenges. Clin Chem. 2011 doi: 10.1373/clinchem.2011.165068. [DOI] [PubMed] [Google Scholar]
- 53.Talmadge JE. Pathways mediating the expansion and immunosuppressive activity of myeloid-derived suppressor cells and their relevance to cancer therapy. Clin Cancer Res. 2007;13:5243–5248. doi: 10.1158/1078-0432.CCR-07-0182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.