Abstract
Dicentric chromosomes are products of genome rearrangement that place two centromeres on the same chromosome. Depending on the organism, dicentric stability varies after formation. In humans, dicentrics occur naturally in a substantial portion of the population and usually segregate successfully in mitosis and meiosis. Their stability has been attributed to inactivation of one of the two centromeres, creating a functionally monocentric chromosome that can segregate normally during cell division. The molecular basis for centromere inactivation is not well under-stood, although studies in model organisms and in humans suggest that genomic and epigenetic mechanisms can be involved. Furthermore, constitutional dicentric chromosomes ascertained in patients presumably represent the most stable chromosomes, so the spectrum of dicentric fates, if it exists, is not entirely clear. Studies of engineered or induced dicentrics in budding yeast and plants have provided significant insight into the fate of dicentric chromosomes. And, more recently, studies have shown that dicentrics in humans can also undergo multiple fates after formation. Here, we discuss current experimental evidence from various organisms that has deepened our understanding of dicentric behavior and the intriguingly complex process of centromere inactivation.
Keywords: CENP-A, heterochromatin, euchromatin, DNA methylation, deletion, fusion
Introduction
The centromere is a complex chromosomal locus where the kinetochore is formed and microtubules attach during cell division. A major component of functional centromeres is CENP-A/CenH3, a histone H3 variant that replaces canonical H3 to create unique centromeric nucleosomes (Palmer et al. 1989; Palmer et al. 1991). CENP-A physically marks centromeres by assembling into largely homotypic nucleosomes (two copies of CENP-A) that have a more rigid conformation than H3-containing nucleosomes (Black and Bassett 2008; Black et al. 2004). The centromeric chromatin of multi-cellular eukaryotes is arranged as multiple subunits of CENP-A nucleosomes periodically interspersed with subunits of H3 nucleosomes that are dimethylated at lysine 4 (H3K4me2) (Blower et al. 2002; Sullivan and Karpen 2004). Together, physically distinct nucleosomes and long-range chromatin organization are thought to create a platform for kinetochore formation and recruitment of additional centromere and kinetochore proteins (Blower and Karpen 2001; Foltz et al. 2006; Hori et al. 2008). In general, each chromosome contains a single region of centromeric DNA where the centromere and kinetochore are assembled. However, genome rearrangements can lead to fusion of two different chromosomes, often resulting in a dicentric chromosome on which two centromeres are physically linked. The prevailing view of dicentric behavior, first described in maize by Barbara McClintock, is that they are inherently unstable, often undergoing successive rounds of anaphase bridge formation and breakage (McClintock 1939; McClintock 1941). Indeed, this model holds true for budding yeast and Drosophila in which dicentrics are largely unstable and lead to broken and rearranged chromosomes (Hill and Bloom 1987; Hill and Bloom 1989; Thrower and Bloom 2001; Thrower et al. 2003). However, it has been known for some time in mammals, and more recently in several model organisms, that dicentric chromosomes can be quite stable in mitosis. In fact, dicentric human chromosomes can be inherited through meiosis. Such stability has been attributed to the phenomenon of centromere inactivation, a poorly understood process by which one of the centromeres becomes nonfunctional or inactive (Earnshaw and Migeon 1985; Sullivan and Schwartz 1995). Consequently, the structurally dicentric chromosome behaves and segregates as a functionally monocentric chromosome during cell division.
Our understanding of centromere inactivation comes largely from observational studies in humans. Cytologically, inactive centromeres lack a constricted appearance on metaphase chromosomes. In addition, a variety of essential centromere and kinetochore proteins and chromosomal and chromatin proteins are absent from inactive centromeres (Earnshaw and Migeon 1985; Sullivan and Schwartz 1995; Page et al. 1995b; Craig et al. 2003). Beyond these cytological observations, however, the molecular basis of centromere inactivation has remained unclear for decades. Testing mechanisms of centromere inactivation has proven difficult for several reasons. First, it was not appreciated until recently that centromere inactivation is a major mechanism of dicentric stability in model organisms. Furthermore, there are few experimental systems to produce de novo dicentric human chromosomes, so it has been difficult to capture the process of inactivation in human cells where it is a common mechanism of dicentric stabilization.
It has been known for two decades that inactive centromeres of dicentric chromosomes lack key centromere and kinetochore proteins, such as CENP-A, CENP-C, and CENP-E (Earnshaw et al. 1989; Sullivan and Schwartz 1995). Additionally, they lack a defined primary constriction on metaphase chromosomes and morphologically resemble chromosome arms. Thus, centromere inactivation is predicted to involve exclusion of centromere proteins and chromatin remodeling so that the two centromeres on the dicentric become functionally distinct. The molecular basis of dicentric stability is further complicated by observations that dicentrics behave in different ways immediately after formation. Most undergo centromere inactivation, but some remain functionally dicentric (Sullivan and Schwartz 1995; Higgins et al. 2005; Page and Shaffer 1998; Sullivan and Willard 1998; Stimpson et al. 2010). The molecular basis for why dicentrics behave in such variable ways remains an area of great interest. In this review, we will discuss dicentric chromosome behavior in various organisms, mechanisms by which these dicentrics are stabilized, and current knowledge regarding features of inactive centromeres.
Dicentrics in the budding yeast Saccharomyces cerevisiae
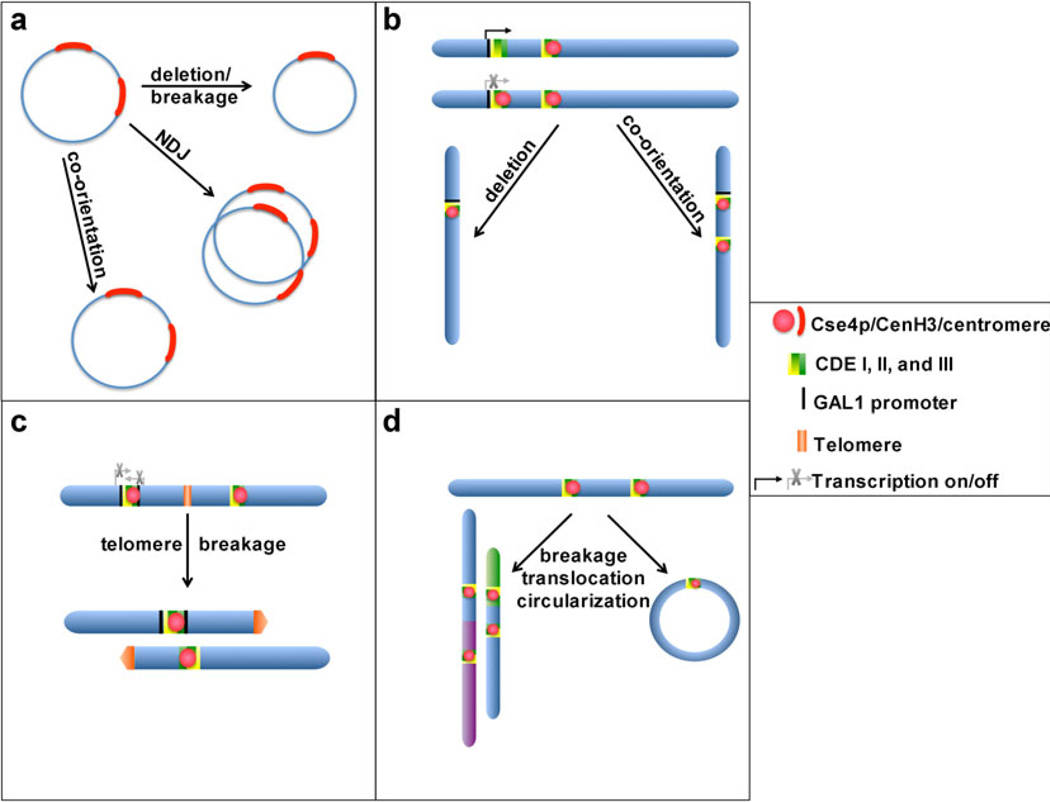
Studies in the budding yeast S. cerevisiae were among the first to demonstrate both the mitotic instability and fate of dicentric chromosomes in a model organism other than maize. Yeast centromeres contain distinct structural elements CDEI, CDEII, and CDEIII that are absolutely required for proper centromere assembly and function. Dicentrics in yeast have been engineered using (1) nonessential minichromosomes; (2) endogenous chromosomes containing an extra-conditional centromere encoded on an integrated GAL1-CEN cassette; (3) endogenous, conditional centromeres (those flanked by GAL1 promoter sequences); (4) spontaneous or induced loss of specific markers integrated into the genome; or (5) specific kinetochore, DNA replication, or DNA damage mutants (Fig. 1). The minichromosome studies revealed that mutations in CDEII or CDEIII could effectively stabilize the dicentrics so that they functioned as monocentric structures (Koshland et al. 1987). Additionally, if inter-centromeric distances were decreased, both centromeres could remain functional if they co-oriented to the same spindle poles and/or behaved cooperatively (Fig. 1b).
Fig. 1.
Dicentric chromosome fate in the budding yeast S. cerevisiae. a Circular supernumerary minichromosomes containing two centromere regions (red) are stabilized by deletion of one centromere or cooperation of the two centromeres if closely spaced. The majority of dicentric minichromosomes undergo non-disjunction (NDJ) events, leading to chromosome loss. b Dicentrics can be formed by insertion of GAL1-CEN cassette (black) onto endogenous chromosomes. The second centromere is kept inactive by GAL1 transcription. When transcription is turned off, both centromeres are active and the dicentric can be stabilized if one centromere is deleted or both co-orient to the same spindle pole. c Dicentrics can also be formed by telomere fusion after an endogenous centromere of one chromosome is flanked by GAL1 promoter sequences (black). Like in (b), the second flanked centromere is kept inactive by GAL1 transcription, but after telomere fusion to create the dicentric, transcription is turned off. The dicentrics can be stabilized if breakage occurs at the telomeric sequences (orange), liberating the two chromosomes. d Dicentric chromosome breakage is common in budding yeast, often leading to secondary rearrangements such as non-reciprocal translocations with other chromosomes. Alternatively, after breakage, the ends of the derivative chromosome may fuse, creating a circular monocentric chromosome
Dicentrics created using endogenous chromosomes have revealed more complex mitotic behaviors. In assays using the GAL1-CEN construct, the second CEN was kept inactive by driving GAL1 transcription through the cassette. However, when GAL1 transcription was turned off, leading to activation of the second centromere, the dicentric chromosome became unstable during cell division, resulting in chromosome lag and DNA breakage via breakage–fusion–bridge (BFB) cycles. However, the dicentrics could be stabilized if one of the centromeres underwent breakage and recombination that physically deleted one centromere (Hill and Bloom 1989; Jager and Philippsen 1989) (Fig. 1b). More recently, this assay was modified so that the GAL1-CEN strain was combined with a conditional Rap1 mutant allele. Rap1 is important for telomere stability and prevents NHEJ of chromosome ends. The Rap1 conditional mutation produced chromosome end-fusions and those between a normal chromosome and a GAL1-CEN marked chromosome were selected for analysis (Pobiega and Marcand 2010). Upon induction of end-fusions, the GAL1-CEN was activated and dicentric behavior and fate evaluated. A little over half of the fusions were unstable and resulted in cell death since the cells were unable to complete cytokinesis. However, in cells that did not die (40%), the dicentrics had broken at the site of telomere fusion, resulting in liberation of each parental chromosome involved in the rearrangement (Fig. 1c).
Other studies of dicentric budding yeast chromosomes have revealed even more complex means of reducing the unstable dicentric to a monocentric state. Repeated BFB cycles lead to multiple secondary rearrangements (Pennaneach and Kolodner 2009) (Fig. 1d). The dicentrics can break at repetitive elements and acquire other chromosomal sequences, yielding a healed chromosome with partial duplications or amplifications. If repetitive elements are present between the two centromeres, the dicentric can also break and non-reciprocally translocate to another chromosome. Finally, broken dicentrics can form circular chromosomes if the site of interstitial breakage fuses to the end of the same chromosome (Fig. 1d). Taken together, existing data from multiple groups emphasize that dicentric chromosomes in budding yeast are generally very unstable and are primarily associated with secondary rearrangement and cell death.
Dicentrics in fission yeast Schizosaccharomyces pombe
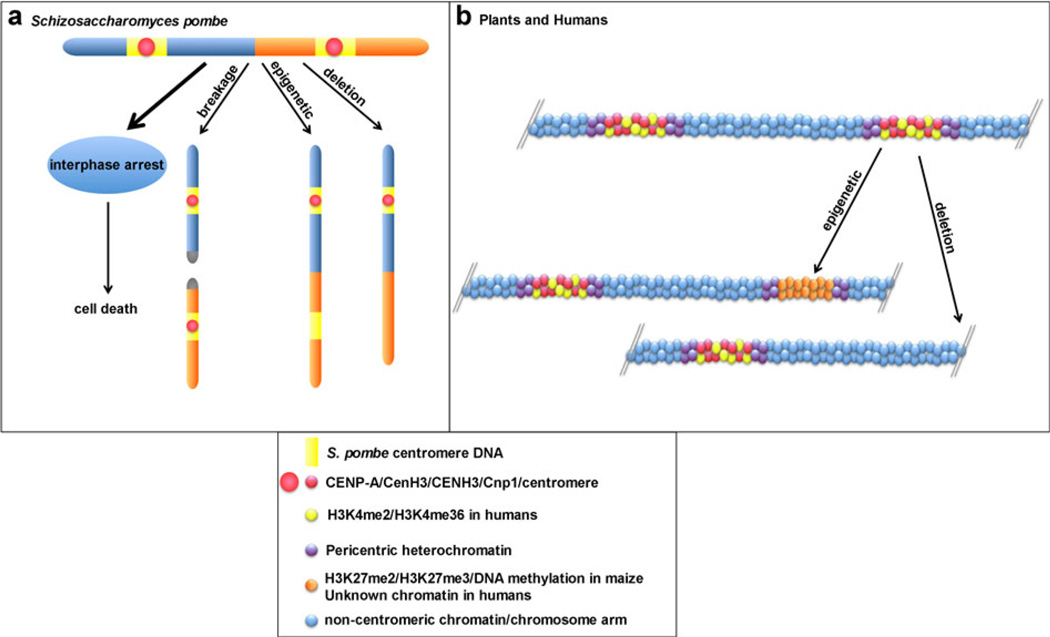
The fission yeast S. pombe contains three chromosomes (I, II, and III), and each centromere is organized as a unique central domain containing CenH3 chromatin flanked by shared repetitive sequences that are packaged into heterochromatin. Dicentrics have not been reported to occur naturally in this organism, but because of the similarities in centromere organization between S. pombe and larger eukaryotes, a recent study has engineered dicentric chromosomes between chromosomes II and III using site-specific or meiotic recombination (Sato et al. 2012). The formation of dicentrics were tracked using high molecular weight pulsed field gel electrophoresis (PFGE), since the S. pombe chromosomes can be resolved as three distinct electrophoretic entities. The PFGE karyotypes of strains containing dicentrics showed two bands, one representing the normal chromosome I and a larger band that was the product of fusion between chromosomes II and III. Like dicentrics in budding yeast, the majority (99%) of cells containing dicentrics died. Surprisingly, though, the defects were not due to chromosome missegregation or breakage, because the cells arrested indefinitely in interphase (Fig. 2a). This result suggested that dicentrics typically trigger DNA damage and replication cell cycle checkpoints and not checkpoints associated with improper spindle dynamics or faulty chromosome segregation during cell division.
Fig. 2.
Mechanisms of dicentric stability in fission yeast and multicellular eukaryotes. a In the fission yeast, S. pombe, engineered dicentrics primarily result in cell cycle arrest in interphase (heavy black arrow), followed by cell death. A small proportion of cells can maintain structurally dicentric chromosomes because one centromere has undergone inactivation. In ~10 % of dicentrics, one centromere is physically deleted. In another ~10 % of dicentrics, a breakage event occurs that splits the dicentric into the two monocentric parental chromosomes. However, two thirds of retained dicentrics undergo epigenetic centromere inactivation in which centromeric DNA (yellow) is retained but Cnp1/CENP-A/CENH3 (red) is absent. The chromatin of the inactive centromere also acquires epigenetic features of heterochromatin. b In plants and humans, engineered and naturally occurring dicentric chromosomes appear to be stabilized by two mechanisms: partial deletion of one centromere or epigenetic inactivation of one centromere. In plants, the chromatin of the inactive centromere loses CENP-A/CENH3 (red spheres), CENP-C, and PH3S10 and acquires heterochromatic features (orange spheres) such as H3K27me2/3 and DNA methylation. In humans, epigenetically inactivated centromeres lose CENP-A/CENH3 and H3K4me2 (yellow and red spheres)
Although the majority of cells with dicentrics were arrested or died, it was noted that a small proportion (~1%) of dicentrics were stably maintained. Within this population, about 10 % of the dicentrics were stabilized by a breakage event that split the dicentric, returning the karyotype to three chromosomes, as denoted by the reappearance of three normal-sized chromosomal bands on agarose gels (Fig. 2a). Another ~10 % of the dicentrics remained fused, and the cells divided normally. These dicentrics were stabilized because one of the two centromeres had been physically deleted. Notably, these two categories of stable fission yeast dicentrics indicate that ~30 % of the dicentrics are stabilized by mechanisms that have also been observed primarily in budding yeast. Intriguingly, however, two thirds of dicentrics were stabilized by epigenetic mechanisms (Sato et al. 2012). The central core regions of the inactive centromeres no longer contained Cnp1/CENP-A/CenH3 and instead became enriched for H3K9me2, a heterochromatin-associated histone modification, and depleted for the euchromatic acetylated histones H3K9ac and H3K14ac. None of the epigenetically inactivated centromeres showed evidence of DNA changes (deletions or mutations). These studies are exciting because they provide evidence in a unicellular model organism for epigenetic centromere inactivation that was previously thought to only occur in larger multi-cellular organisms. The results also raise the question of how the cell identifies a chromosome with two centromeres and suspends the cell cycle before mitosis even occurs. Finally, these studies leave open the possibility that other organisms may detect dicentrics prior to metaphase and could trigger similar early cell cycle checkpoints that have been previously underappreciated.
Dicentrics in plants
Given that dicentric chromosomes were first described in plants, it seems fitting that much of what we know about centromere inactivation has come from studies in maize and other crop plants in the past 10 years. Although McClintock first reported on the instability of dicentric chromosomes in maize, more recently stable dicentrics have been described in both maize and wheat (Gao et al. 2011; Han et al. 2006; Zhang et al. 2010). The stability of these chromosomes occurs by inactivation of one centromere, resulting in a functionally monocentric chromosome. Naturally occurring and engineered dicentrics in plants have revealed several features of inactive centromeres and emphasize key similarities and several differences in how centromere inactivation is achieved in plants compared with other organisms.
Plant centromeres generally consist of a mix of both tandem arrays of simple repeats and centromeric retrotransposons, forming a complex chromosome locus (Ma et al. 2007). The 20 endogenous chromosomes in maize are typically referred to as A chromosomes. But maize also contains small, extranumerary B chromosomes. B chromosomes have aided in the study of active and inactive centromeres due to their unique qualities, including being entirely dispensable and containing the B-chromosome-specific ZmB repeats. Naturally occurring B-A translocation chromosomes have undergone BFB cycles to yield a collection of minichromosomes. Fluorescence in situ hybridization (FISH) using a ZmB probe on 23 of these minichromosomes revealed that five minichromosomes had two B centromeres. However, only one centromere was active, as determined by immunostaining for CENH3, indicating that centromere inactivation had occurred (Han et al. 2006). Additional studies of a dicentric chromosome with large and small versions of the B centromere showed that the smaller of the two was preferentially inactivated based on immunostaining for CENP-C, CENH3, and H3 phosphorylated at Ser-10 (PH3S10) that distinguish active centromeres (Han et al. 2009). These dicentric minichromosomes were stable over two generations, with small centromeres remaining inactivated. However, when separated by intrachromosomal recombination, the smaller centromeres could be reactivated, regaining the molecular attributes of an active centromere, such as CENH3, CENP-C, and PH3S10. Thus, these studies emphasize the epigenetic nature of centromere inactivation, since both inactivation and reactivation could occur without changing the underlying DNA sequences of the small centromere.
Studies of dicentrics in S. pombe have shown that heterochromatin formation is important for, or indicative of, centromere inactivation. This also appears to be true for centromere inactivation in plants. Recent work in maize has shown distinct differences in DNA methylation patterns between active and inactive centromeres on B chromosomes (Koo et al. 2011). Using a DNA fiber-based technique, cytosine methylation was mapped within highly repetitive DNA sequences, revealing that active centromeres are hypomethylated, while inactive centromeres are hypermethylated. Active centromeres were enriched for 5-methylcytosine (5mC) signals at the ZmB satellite arrays but hypomethylated at the regions containing CentC-CRM retrotransposons that were interspersed between the satellite sequences. Conversely, inactive centromeres had nearly uniform 5mC signals throughout both the ZmB arrays and flanking regions, creating an overall contiguously hypermethylated state. The differential methylation of ZmB and CentC-CRM blocks at active centromeres may favor loading and propagation of CENH3 given that CENH3 nucleosomes are associated with the CentC-CRM blocks and ZmB repeats associate with canonical H3 nucleosomes (Jin et al. 2005). Propagation of CENH3 nucleosomes may prevent methylation of ZmB sequences at active B centromeres, maintaining their hypomethylation. Alternatively, CENH3 could recruit demethylases to ZmB sites during the cell cycle to establish an undermethylated state. Consequently, during B-centromere inactivation, loss of CENH3 may leave underlying CentC-CRM sequences unprotected so that they can now become heavily methylated.
Inactive plant centromeres, based on studies in maize, lack CENH3, CENP-C, PH3S10, and are hypermethylated (Fig. 2b). These modifications suggest that, like inactive centromeres in fission yeast, inactive centromeres in plants adopt a heterochromatic structure. Studies in wheat have supported this conclusion. Wheat has been utilized for centromere investigation since 1946 when Sears first reported a transmissible dicentric wheat chromosome that was found to contain one primary centromere and a weaker secondary centromere (Sears and Camara 1952). In contemporary studies that evaluated CENH3 immunofluorescence and microtubule attachment, this “dicentric chromosome” was actually identified as tricentric, with one large and two small centromeres (Zhang et al. 2010). This chromosome was often functionally tricentric, but stable, presumably due to the dominant pulling capacity of the large centromere during meiotic anaphase. However, in those versions that did not remain functionally tricentric, both of the small centromeres were inactivated. While there were no sequence changes at the inactive centromeres, they were epigenetically distinct from the active in that they appeared cytologically to have higher levels of the heterochromatic histone modifications H3K27me2 and H3K27me3. Collectively, these studies in plants have revealed that inactive centromeres exhibit many features of heterochromatin, both at the level of DNA and chromatin.
Dicentrics in humans
The occurrence of dicentric chromosomes in humans has been appreciated since the 1960s (De la Chapelle et al. 1966; Ockey et al. 1966). Many of these dicentrics were associated with birth defects such as Turner Syndrome and Down Syndrome and with reproductive abnormalities. Although dicentrics can occur between any two chromosomes, some types are more prevalent than others in the human population. These include Robertsonian translocations (ROBs) and isochromosome X [i(X)] (De la Chapelle and Stenstrand 1974;Warburton et al. 1973). ROBs involve any two of the ten acrocentric chromosomes (13, 14, 15, 21, and 22), although rob(13; 14) and rob(14; 21), account for approximately 85 % of dicentric ROBs ascertained from patients (Therman et al. 1989). Early studies of patient-derived ROBs revealed that centromere inactivation occurred in the majority, particularly in those that were inherited. The inactive centromeres were shown to lack CENP-A, CENP-C, and CENP-E, and these features are shared by inactive centromeres of other non-ROB dicentric human chromosomes (Warburton et al. 1997; Sullivan and Schwartz 1995; Earnshaw and Migeon 1985; Fisher et al. 1997; Page et al. 1995a). In a series of patient-derived ROBs, it was also observed that chromosome 14 remained active most often, irrespective of the other acrocentric involved in the ROB while the centromere of chromosome 15 was more likely to be inactivated (Sullivan et al. 1994). These results imply that some centromeres are “stronger” or less amenable to centromere inactivation. However, studies of both de novo ROBs and i(X)s have also revealed that that some dicentric chromosomes remain functionally dicentric (Lange et al. 2009; Higgins et al. 1999; Sullivan and Willard 1998). It was suggested that shorter inter-centromeric distances promoted the retention of two functional centromeres, similar to what had been observed in budding yeast. It is presumed that steric constraints on dicentrics with small inter-centromeric distances may reduce the possibility for improper microtubule attachments or orientation of centromeres to opposite spindle poles.
A limitation of studying patient-derived dicentrics has been that most are evaluated months to decades after the dicentric has formed. By this time, centromere fate has been determined, established, and is simply being maintained. Furthermore, the dicentrics obtained from patient samples presumably represent the most stable versions of dicentric chromosomes, so that the spectrum of dicentric fates in humans may not be fully appreciated. However, two studies of engineered i(X)s and ROBs have been important in revealing dicentric behavior after formation. The first assay involved creating dicentric isochromosomes of the human X short arm [i(Xp)] in a rodent background (Higgins et al. 1999). The dicentric i(Xp)s that were produced exhibited a range of inter-centromeric distances, from a few megabases to over 20 Mb. These studies revealed several classes/fates of dicentric chromosomes, including (1) functionally monocentric chromosomes, in which one of the two genetically identical centromeres was inactivated; (2) functionally dicentric chromosomes, in which both centromeres remained active; and (3) dicentric chromosomes that exhibited heterogeneous centromere activity (Higgins et al. 2005). Serial single cell clones from this latter class revealed that centromere activity was usually clonal, but centromere state (i.e., functionally monocentric or dicentric) could switch within a population of proliferating cells. Molecular analysis showed that the size of the alpha satellite DNA at the centromeres of these chromosomes did not change, suggesting that inactivation occurred primarily due to epigenetic changes. The i(Xp)s exhibited both short and large inter-centromeric distances, but those with more distantly located centromeres could undergo centromere inactivation or maintain two functional centromeres suggesting that there was not an absolute correlation with inter-centromeric distances and centromere fate.
In a more recent study, dicentric chromosomes were produced in a human cell background by transiently expressing a mutant version of telomere protein TRF2, TRF2ΔBΔM or dnTRF2 (Stimpson et al. 2010). The mutant protein behaves as a dominant-negative, binding to endogenous TRF2 and displacing it from chromosome ends, resulting in chromosome end-fusions (van Steensel et al. 1998). Expression of dnTRF2 can be controlled by doxycyline in the cell culture media, so that after its expression is shut off, cells containing dicentrics continue to proliferate (Stimpson et al. 2010). Hundreds of dicentrics were produced in this in vitro assay, and unexpectedly, the most were fusions between the human acrocentric chromosomes (i.e., induced ROBs). The dicentrics varied in structure so that inter-centromeric distances could be studied in conjunction with the fates of the de novo dicentric fusions. These studies revealed that ~50 % of dicentrics maintain two active centromeres, even after 150 cell divisions. Even dicentrics with large (>20 Mbp) inter-centromeric distances were stable through at least 20 cell divisions. The remaining dicentric fusions underwent centromere inactivation between 4 days and 20 weeks after formation. In about half of these induced dicentrics, centromere inactivation was accompanied by the temporary appearance of small, marker-sized chromosome fragments that were shown by immunostaining and FISH to contain both CENPs and alpha satellite DNA homologous to the inactive centromere of the dicentric. Using semi-quantitative FISH, it was observed that the alpha satellite array of the inactive centromere became reduced in size after centromere inactivation. These results suggested that one mechanism of dicentric stabilization and centromere inactivation in humans involves partial deletion of the alpha satellite array (Fig. 2b). Since CENP-A chromatin assembles on only a portion of the multi-megabase alpha satellite arrays at endogenous human centromeres (Sullivan et al. 2011), centromere inactivation could be explained by removal of the CENP-A chromatin portion of the array. Indeed, decreases in alpha satellite array size at inactivated centromeres have been suggested from studies of patient-derived dicentrics (Fisher et al. 1997; Maraschio et al. 1990). In the remainder of induced dicentric fusions with inactive centromeres, there was no evidence of centromeric deletion, suggesting that similar to the engineered i(Xp)s, epigenetic mechanisms are also involved in centromere inactivation (Fig. 2b).
Concluding remarks and future challenges
It is clear from studies spanning the early days of McClintock’s work in maize to contemporary experiments on engineered dicentrics in yeast and mammals that dicentric behavior is complex and is associated with multiple outcomes. In general, dicentrics are unstable in most organisms, with the exception of humans. But, irrespective of the organisms, those that are retained experience similar fates. Dicentrics in budding and fission yeasts, plants, and mammals can be stabilized by total or partial centromeric deletion. The mechanisms could involve recombination, break-induced replication, or non-homologous end joining, since all of these processes have been associated with centromeres (Pennaneach and Kolodner 2009; Pobiega and Marcand 2010; Thrower et al. 2003). A notable difference between centromeric deletion in yeast and human dicentrics is that alpha satellite DNA, the genomic marker of the human centromere, is not completely removed during inactivation. Instead, only the portion associated with CENP-A/CENH3, which presumably identifies the site of kinetochore assembly, appears to be eliminated. Spatial and temporal incorporation of the centromeric H3 variant CENP-A/CENH3 maintains the location and function of the centromere. Newly synthesized CENP-A/CENH3 is loaded into chromatin in late telophase/early G1 by the escort protein HJURP (Dunleavy et al. 2009; Foltz et al. 2009). Thus, removal of CENP-A/CENH3, and other centromere/kinetochore proteins, from a centromere destined for inactivation must occur, in addition to blocking the loading of new CENP-A/CENH3. It is not known if existing CENP-A/CENH3 nucleosomes, recruitment of additional factors, or H3-containing nucleosomes within centromeric chromatin guide incorporation of new CENP-A/CENH3 or if such factors are recognized by HJURP or intermediates. It would be consistent with any of these models if centromere inactivation occurred by simultaneously deleting existing CENP-A/CENH3 and nearby accessory chromatin that targets new CENPA/CENH3 deposition.
Studies of dicentrics in S. pombe, plants, and humans also provide strong support for epigenetic mechanisms of centromere inactivation that include dramatic changes in the chromatin organization of the inactive centromere. Inactive centromeres exhibit markers of heterochromatin and lose euchromatic modifications such as histone acetylation and H3K4me2. The loss of euchromatic histone modifications such as H3K4me2 or H3K36me2 during centromere inactivation makes sense in the context of how centromeric chromatin is normally organized. H3K4me2 and H3K36me2 nucleosomes are present within the CENP-A-containing region of the alpha satellite array (Sullivan and Karpen 2004; Bergmann et al. 2011). Removal of H3K4me2 and H3K36me2 may signify other changes at inactive centromeres, such as a decrease in alpha satellite transcription. Future experiments will be needed to determine if changes in centromeric transcription are required for or accompany inactivation.
Studies of dicentrics in plants and human ROBs have suggested that the choice of which centromere is inactivated may be nonrandom (Han et al. 2009; Sullivan et al. 1994). This implies that there are differences in the “strength” of functional centromeres or in the ease of inactivating certain centromeres. Mechanistically, this phenomenon could be linked to genomic or epigenetic features of individual centromeres, such as satellite array size or chromatin composition. In plants, smaller centromeres are preferentially inactivated (Han et al. 2009). The effect of alpha satellite array size on centromere inactivation remains to be tested in humans. It is well known that alpha satellite array size can vary by 10– to 20-fold among chromosomes, so it is possible that chromosomes with consistently smaller arrays are more likely to be inactivated. Still, other factors, such as sequence characteristics, repeat monomer length, CENP-B box density, nucleosome positioning, or DNA looping, may equally influence the “choice” of which centromere is inactivated on a dicentric human chromosome.
An aspect of dicentric behavior that appears to be unique to humans is the observation that dicentrics are equally likely to exist as functionally dicentric, versus functionally monocentric, chromosomes (Higgins et al. 1999; Sullivan and Willard 1998; Lange et al. 2010). In patient-derived dicentrics (i.e., dicentric Xs and many de novo ROBs) short inter-centromeric distances have been proposed to influence centromere function, so that dicentrics with closely spaced centromeres are more likely to remain functionally dicentric. However, over 80 % of patient-derived ROBs undergo centromere inactivation and even dicentric isochromosome Xs with closely spaced centromeres experience centromere inactivation (Page and Shaffer 1998; Sullivan and Schwartz 1995; Sullivan et al. 1994). Data on engineered dicentric human chromosomes argue that centromeric distance may not be the strongest predictor of the functional state of a centromere on a dicentric chromosome (Higgins et al. 2005; Higgins et al. 1999; Stimpson et al. 2010). Centromere inactivation might rely instead on chromosome-specific features or may even occur differently in each cell. Dissecting the mechanism(s) of centromere inactivation is an important, yet difficult, aspect of chromosome biology. Other experimental systems to produce dicentric chromosomes will be important for defining the mechanisms of genomic versus epigenetic centromere inactivation and determining the underlying basis of centromere function.
Acknowledgments
KMS was supported by NIH Ruth L. Kirschstein National Research Service Award F31 AG034749 (NIA). BAS and research in the Sullivan lab are supported by the National Institutes of Health grant R01 GM098500 (NIGMS) and the March of Dimes Foundation (06-FY10-294). We apologize to our colleagues whose work on centromeres and dicentric chromosomes could not be included due to space constraints.
Abbreviations
- 5mC
5-methylcytosine
- BFB
Breakage–fusion–bridge
- CDE
Centromere DNA element
- CEN
Centromere
- CenH3/CENH3
Centromere-specific histone H3 variant (also known as CENP-A)
- CENP-A
Centromere protein A
- CENP-C
Centromere protein C
- CENP-E
Centromere protein E
- CentC
Maize A-chromosome centromere specific satellite repeat
- Cnp1
S. pombe CENP-A/CenH3
- CRM
Maize centromeric retrotransposon element
- Cse4p
S. cerevisiae CENP-A/CenH3
- GAL1
Galactose 1 (gene or promoter)
- H3K4me2
Histone H3 dimethylated at lysine 4
- H3K9ac
Histone H3 acetylated at lysine 9
- H3K9me2
Histone H3 dimethylated at lysine 9
- H3K14ac
Histone H3 acetylated at lysine 13
- H3K27me2/me3
Histone H3 di- or tri-methylated at lysine 27
- HJURP
Holliday junction protein
- i(Xp)
Isochromosome of the X short arm
- i(Xq)
Isochromosome of the X long arm
- isoX/i(X)
Isochromosome X
- FISH
Fluorescence in situ hybridization
- NDJ
Non-disjunction
- NHEJ
Non-homologous end joining
- PFGE
Pulsed field gel electrophoresis
- PH3S10
Histone H3 phosphorylated at serine 10
- ROB/rob
Robertsonian translocation
- TRF2
Telomere repeat-binding factor 2
- ZmB
Maize B-chromosome-specific satellite repeat
Contributor Information
Kaitlin M. Stimpson, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27708, USA
Justyne E. Matheny, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27708, USA
Beth A. Sullivan, Email: beth.sullivan@duke.edu, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27708, USA; Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, NC 27710, USA.
References
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Stenstrand K. Dicentric human X chromosomes. Hereditas. 1974;76:259–268. doi: 10.1111/j.1601-5223.1974.tb01344.x. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Wennstrom J, Hortling H, Ockey CH. Isochromosome-X in man. I. Hereditas. 1966;54:260–276. doi: 10.1111/j.1601-5223.1966.tb02021.x. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie HD, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Fisher AM, Al-Gazali L, Pramathan T, Quaife R, Cockwell AE, Barber JC, Earnshaw WC, Axelman J, Migeon BR, Tyler-Smith C. Centromeric inactivation in a dicentric human Y;21 translocation chromosome. Chromosoma. 1997;106:199–206. doi: 10.1007/s004120050240. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Fu S, Dong Q, Han F, Birchler JA. Inactivation of a centromere during the formation of a translocation in maize. Chromosome Res. 2011;19:755–761. doi: 10.1007/s10577-011-9240-5. [DOI] [PubMed] [Google Scholar]
- Han F, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci U S A. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21:1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Schueler MG, Willard HF. Chromosome engineering: generation of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosoma. 1999;108:256–265. doi: 10.1007/s004120050376. [DOI] [PubMed] [Google Scholar]
- Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13:745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Jager D, Philippsen P. Stabilization of dicentric chromosomes in Saccharomyces cerevisiae by telomere addition to broken ends or by centromere deletion. EMBO J. 1989;8:247–254. doi: 10.1002/j.1460-2075.1989.tb03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Lamb JC, Vega JM, Dawe RK, Birchler JA, Jiang J. Molecular and functional dissection of the maize B chromosome centromere. Plant Cell. 2005;17:1412–1423. doi: 10.1105/tpc.104.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Han F, Birchler JA, Jiang J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011;21:908–914. doi: 10.1101/gr.116202.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, Oates RD, Silber S, Repping S, Page DC. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Gadzicki D, Schlegelberger B, Gohring G. Recurrent involvement of heterochromatic regions in multiple myeloma-a multicolor FISH study. Leuk Res. 2010;34:1002–1006. doi: 10.1016/j.leukres.2009.10.027. [DOI] [PubMed] [Google Scholar]
- Ma J, Wing RA, Bennetzen JL, Jackson SA. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 2007;23:134–139. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Maraschio P, Zuffardi O, Caiulo A, Dainotti E, Piantanida M, Rivera H, Tupler R. Deletion of specific sequences or modification of centromeric chromatin are responsible for Y chromosome centromere inactivation. Hum Genet. 1990;85:491–494. doi: 10.1007/BF00194222. [DOI] [PubMed] [Google Scholar]
- McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci U S A. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockey CH, Wennstrom J, de la Chapelle A. Isochromosome-X in man. II. Hereditas. 1966;54:277–292. doi: 10.1111/j.1601-5223.1966.tb02022.x. [DOI] [PubMed] [Google Scholar]
- Page SL, Shaffer LG. Chromosome stability is maintained by short intercentromeric distance in functionally dicentric human Robertsonian translocations. Chromosome Res. 1998;6:115–122. doi: 10.1023/a:1009286929145. [DOI] [PubMed] [Google Scholar]
- Page SL, Earnshaw WC, Choo KH, Shaffer LG. Further evidence that CENP-C is a necessary component of active centromeres: studies of a dic(X; 15) with simultaneous immunofluorescence and FISH. Human Mol Genet. 1995a;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- Page SL, Earnshaw WC, Choo KH, Shaffer LG. Further evidence that CENP-C is a necessary component of active centromeres: studies of a dic(X; 15) with simultaneous immunofluorescence and FISH. Hum Mol Genet. 1995b;4:289–294. doi: 10.1093/hmg/4.2.289. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Margolis RL. Biochemical analysis of CENP-A, a centromeric protein with histone-like properties. Prog Clin Biol Res. 1989;318:61–72. [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One. 2009;4:e6389. doi: 10.1371/journal.pone.0006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobiega S, Marcand S. Dicentric breakage at telomere fusions. Genes Dev. 2010;24:720–733. doi: 10.1101/gad.571510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S. Epigenetic Inactivation and subsequent heterochromatinization of a centromere stabilize dicentric chromosomes. Curr Biol. 2012;22:658–667. doi: 10.1016/j.cub.2012.02.062. [DOI] [PubMed] [Google Scholar]
- Sears ER, Camara A. A transmissible dicentric chromosome. Genetics. 1952;37:125–135. doi: 10.1093/genetics/37.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001061. e1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Wolff DJ, Schwartz S. Analysis of centromeric activity in Robertsonian translocations: implications for a functional acrocentric hierarchy. Chromosoma. 1994;103:459–467. doi: 10.1007/BF00337384. [DOI] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therman E, Susman B, Denniston C. The nonrandom participation of human acrocentric chromosomes in Robert-sonian translocations. Ann Hum Genet. 1989;53:49–65. doi: 10.1111/j.1469-1809.1989.tb01121.x. [DOI] [PubMed] [Google Scholar]
- Thrower DA, Bloom K. Dicentric chromosome stretching during anaphase reveals roles of Sir2/Ku in chromatin compaction in budding yeast. Mol Biol Cell. 2001;12:2800–2812. doi: 10.1091/mbc.12.9.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower DA, Stemple J, Yeh E, Bloom K. Nuclear oscillations and nuclear filament formation accompany single-strand annealing repair of a dicentric chromosome in Saccharomyces cerevisiae. J Cell Sci. 2003;116:561–569. doi: 10.1242/jcs.00251. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Warburton D, Henderson AS, Shapiro LR, Hsu LY. A stable human dicentric chromosome, t dic(12;14)(p13;p13) including an intercalary satellite region between centromeres. Am J Hum Genet. 1973;25:439–445. [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Zhang W, Friebe B, Gill BS, Jiang J. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma. 2010;119:553–563. doi: 10.1007/s00412-010-0278-5. [DOI] [PubMed] [Google Scholar]