Abstract
To determine which species of Culicoides biting midges carry Schmallenberg virus (SBV), we assayed midges collected in the Netherlands during autumn 2011. SBV RNA was found in C. scoticus, C. obsoletus sensu stricto, and C. chiopterus. The high proportion of infected midges might explain the rapid spread of SBV throughout Europe.
Keywords: Orthobunyaviridae, Schmallenberg virus, Culicoides obsoletus sensu stricto, Culicoides scoticus, Culicoides chiopterus, RT-PCR, viruses, biting midges, RNA, the Netherlands, SBV, Culicodes spp
During early summer 2011, Schmallenberg virus (SBV), a novel orthobunyavirus of the Simbu serogroup, spread across much of northern Europe, infecting ruminant livestock. The Simbu serogroup (family Bunyaviridae, genus Bunyavirus) includes Shamonda virus, Akabane virus, Sathuperi virus, and Aino virus. These viruses cause teratologic effects in ruminants and are arthropod-borne, and most have been isolated in the Old World from mosquitoes and Culicoides spp. biting midges (1). Recent preliminary studies indicate that ≥1 species of Culicoides midges act as field vectors for SBV in Europe (2). To determine which Culicoides midge species harbor SBV, we analyzed midges collected from 3 livestock holdings in eastern and northeastern parts of the Netherlands.
The Study
Throughout September and early October 2011, Culicoides spp. biting midges were trapped almost daily at a dairy in the municipality of Ermelo (eastern Netherlands) by various methods, including the standard Onderstepoort-type blacklight trap. In addition, during several days in August and September 2011, Culicoides spp. biting midges were trapped near sheep flocks in the municipalities of Bilthoven (central Netherlands) and Midden-Drenthe (northeastern Netherlands) by using the Onderstepoort-type trap and a drop-tent cage. Captured midges were stored in 70% ethanol.
Female midges were categorized as nulliparous, parous, gravid, or freshly blood fed (engorged) (3); only midges belonging to the first 3 categories were assayed. The 6,100 selected midges were divided into 610 species-specific pools, 10 midges per pool. Under a dissecting microscope, the heads were separated from abdomens by use of a scalpel; 10 heads were then pooled and assayed for SBV, whereas the corresponding abdomens (also pooled) were stored in 70% ethanol.
All midges were identified morphologically, but because female C. obsoletus sensu stricto midges cannot be separated with confidence from C. scoticus midges, they were pooled and are referred to jointly as the C. obsoletus complex. The number of pools assayed for each species was as follows: C. obsoletus complex (230), C. chiopterus (144), C. dewulfi (130), C. punctatus (105), and C. pulicaris (1). After assays were conducted, the species identity of each SBV-positive midge pool was established by using molecular techniques.
Only when a pool of 10 heads was found SBV positive was the corresponding pool of dissected abdomens retrieved and assayed. In this instance, the 10 abdomens were assayed singly, so that the individual abdomen that was SBV-positive could be identified molecularly, to establish exactly which of the 2 species of the C. obsoletus complex was involved and to confirm or refute the morphologic identifications that had been made for the remaining Culicoides species.
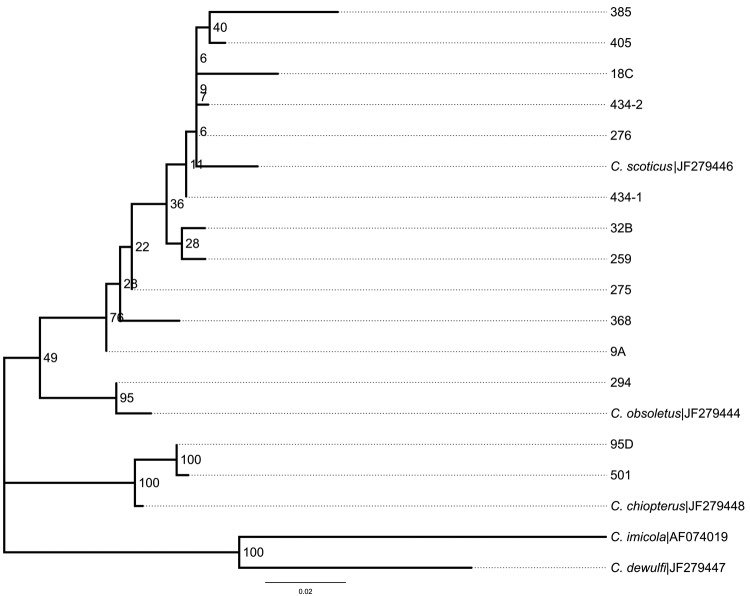
RNA extraction was performed according to a protocol developed by CODA-CERVA (Centrum voor Onderzoek in Diergeneeskunde en Agrochemie, Centre d’Étude et de Recherches Vétérinaires et Agrochimiques), Brussels, Belgium; whereas, reverse transcription PCR (RT-PCR) was performed according to a method recently developed to detect the small segment of SBV (4). The RT-PCR cutoff value for the pooled heads was set at a cycle threshold (Ct) value of 35. Pools with Ct >35 were retested and considered positive when confirmed. Reported Ct values for blood samples from infected cattle in Germany, tested by using the same RT-PCR, were 24–35 (5) and were used as the guide for our choice of cutoff value. If a specific pool of midge heads tested positive, individual abdomens from the corresponding stored pool were tested separately by RT-PCR. For molecular identification of the SBV-positive midges, the 18S internal transcribed spacer 1 (ITS1) 5.8S region was amplified by using the PanCulF and PanCulR primer set, adapted from Cêtre-Sossah et al. (6). The ITS1 sequences obtained from the SBV-positive abdomens were used to develop a Culicoides spp. phylogeny (Figure), which includes GenBank sequences representing all 5 species of the subgenus Avaritia (including C. imicola) known to be involved in the transmission of arboviruses in western Europe.
Figure.
Phylogenetic tree comparing Schmallenberg virus–positive Culicoides spp. biting midge abdomens isolated in different regions in the Netherlands, 2011, with reference sequences from Deblauwe et al. (7). C. imicola was used as an outgroup. Bootstrap values are indicated at the significant nodes. Scale bar indicates nucleotide substitutions per site.
Of the 610 Culicoides midge head pools, 14 (2.3%) were SBV positive according to RT-PCR (Table 1): 11 C. scoticus, 1 C. obsoletus s.s., and 2 C. chiopterus. Of the 14 pools, 13 comprised midges from the dairy in Ermelo; midges in the remaining C. chiopterus pool came from Midden-Drenthe. Ct values for 12 of the 14 pools ranged from 19.6 to 30.44; Ct values for the remaining 2 pools were 34.98 and 36.78 (Table 2). Ct values for 13 of the individual midge abdomens linked to each pool of SBV-positive heads were lower (meaning a higher viral load) than those obtained for their corresponding heads. In 1 pool of C. obsoletus complex midges, 2 of 10 abdomens were positive for SBV, 1 strongly and 1 weakly. RT-PCRs for SBV were negative for all 130 pools of C. dewulfi, 105 pools of C. punctatus, and the 1 pool of C. pulicaris midges.
Table 1. Schmallenberg virus RNA in Culicoides spp. biting midges collected August–September 2011, the Netherlands.
| Municipality (Province) | Pools, no. positive/no. tested* |
|||||
|---|---|---|---|---|---|---|
| C. obsoletus complex | C. dewulfi | C. chiopterus | C. punctatus | C. pulicaris | Total | |
| Bilthoven (Utrecht) | 0/10 | 0 | 0 | 0 | 0 | 0/10 |
| Midden-Drenthe (Drenthe) | 0/5 | 0 | 1/39 | 0 | 0 | 1/44 |
| Ermelo (Gelderland) | 12/215 | 0/130 | 1/105 | 0/105 | 0/1 | 13/556 |
| Total | 12/230 | 0/130 | 2/144 | 0/105 | 0/1 | 14/610 |
*Tested by reverse transcription PCR.
Table 2. Ct values of Schmallenberg virus–positive Culicoides spp. biting midges collected August–September 2011, the Netherlands*.
| Pool no. | Pooled heads |
Individual abdomens |
|||||
|---|---|---|---|---|---|---|---|
| Species identification by morphologic examination | Ct value |
Species identification by DNA sequencing | Ct value |
||||
| First test | Second test | First test | Second test | ||||
| 95-D | C. chiopterus | 27.88 | NA | C. chiopterus | 24.59 | NA | |
| 501 | C. chiopterus | 35.36 | 34.98 | C. chiopterus | 36.45 | 35.07 | |
| 9-A | C. obsoletus complex | 30.44 | NA | C. scoticus | 24.75 | NA | |
| 18-C | C. obsoletus complex | 28.24 | NA | C. scoticus | 24.95 | NA | |
| 32-B | C. obsoletus complex | 21.84 | NA | C. scoticus | 18.32 | NA | |
| 259 | C. obsoletus complex | 19.60 | NA | C. scoticus | 18.16 | NA | |
| 275 | C. obsoletus complex | 20.72 | NA | C. scoticus | 20.39 | NA | |
| 276 | C. obsoletus complex | 36.02 | 36.78 | C. scoticus | 36.68 | NA | |
| 293 | C. obsoletus complex | 20.43 | NA | No reliable sequence | 19.95 | NA | |
| 294 | C. obsoletus complex | 24.60 | NA | C. obsoletus sensu stricto | 20.06 | NA | |
| 368 | C. obsoletus complex | 25.21 | NA | C. scoticus | 21.80 | NA | |
| 385 | C. obsoletus complex | 20.67 | NA | C. scoticus | 20.25 | NA | |
| 405 | C. obsoletus complex | 23.38 | NA | C. scoticus | 21.64 | NA | |
| 434† | C. obsoletus complex | 23.68 | NA | C. scoticus | 23.10 | NA | |
*Ct values determined by reverse transcription PCR. All midges were collected from cattle in the Ermelo municipality except no. 95-D, which was collected from sheep in the Midden-Drente municipality. Ct, cycle threshold; NA, not applicable. †Two abdomens from this pool were positive; Ct values for the second abdomen were 35.75 and 35.37.
The species of all but 1 midge abdomen could be molecularly identified on the basis of ITS1 (Table 2). Not only did the molecular results confirm most of the morphologic identifications, but they also showed that C. scoticus seems to have played a more prominent role than C. obsoletus s.s. in transmission of SBV. The ITS1 sequences obtained from samples 95-D and 501 were almost identical to those published for C. chiopterus; the same applies to sample 294, which represented C. obsoletus s.s. (Table 2) (7). Although sequence polymorphism in C. scoticus was diverse, we were able to unambiguously assign each of the 11 SBV-positive abdomens to this species (Figure).
Prevalence of SBV among the Culicoides spp. midges was 0.25% (15/6,100 midges tested). More specifically, the prevalence of SBV in the 2 species that comprised the C. obsoletus complex was 0.56% (13/2,300 tested). This prevalence is similar to that obtained for Akabane virus in C. brevitarsis midges from Australia (8–11) but about 10× higher than that reported for bluetongue virus (12). For C. chiopterus midges, prevalence of SBV was 0.14% (2/1,440 tested), ≈5× higher than prevalence of bluetongue virus (13).
Conclusions
Our results demonstrate that SBV was harbored in 3 species of field-collected Culicoides biting midges: C. scoticus, C. chiopterus, and C. obsoletus s.s. These species were among the more abundant of the 15 species found at the livestock holdings sampled. The holdings were situated in the center of the epidemic area, and of the ≈100 animals at the dairy in Ermelo, >96% had seroconverted to SBV. The low Ct values indicate that concentrations of the virus in most SBV-positive Culicoides midges were high. The fact that the Ct values for the heads of midges matched closely with those from the associated abdomens renders it certain that SBV had replicated to transmissible levels in these midges and supports the contention that 2 species of the C. obsoletus complex, along with C. chiopterus, act as natural vectors for SBV. Despite the relatively large numbers of SBV-negative pools, our findings should not be interpreted to exclude the involvement of other species, such as C. dewulfi or C. punctatus, in field transmission of SBV. We conclude that the high proportion of SBV-positive Culicoides spp. midges and the multiple vector species could help explain the rapid spread of SBV throughout much of Europe during 2011.
Acknowledgments
We thank Maykel van Gent, Connie Nijenhuis (Hulshorst), Petra de Jong, Rob Andriessen, Jesse Erens, Roos Stoop, and Marianne Wessels for helping trap Culicoides spp. midges; Eefje and Eric Meihuizen for excellent and friendly cooperation and generously providing access to their pastures and animals; and Els de Boer-Luijtze, Marga van Zetten, and Ruth Bossers-de Vries for decapitating the midges.
This study was commissioned and funded by the Dutch Ministry of Economic Affairs, Agriculture, and Innovation (WOT program 01) and the Directorate General for Health and Consumers, European Commission, Brussels, Belgium.
Biography
Dr Elbers is a veterinary epidemiologist and senior scientist in the Department of Epidemiology, Crisis Organisation and Diagnostics, Central Veterinary Institute, part of Wageningen University and Research Centre, Lelystad. His research interests are notifiable animal diseases and surveillance and early detection systems.
Footnotes
Suggested citation for this article: Elbers ARW, Meiswinkel R, van Weezep E, Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis [Internet]. 2013 Jan [date cited]. http://dx.doi.org/10.3201/eid1901.121054
References
- 1.Saeed MF, Li L, Wang H, Weaver SC, Barrett ADT. Phylogeny of the Simbu serotype of the genus Bunyavirus. J Gen Virol. 2001;82:2173–81. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, Bødker R, et al. Culicoids as vectors of Schmallenberg virus [letter]. Emerg Infect Dis. 2012;18:1204–6. 10.3201/eid1807.120385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goffredo M, Meiswinkel R. Entomological surveillance of bluetongue in Italy: methods of capture, catch analysis and identification of Culicoides biting midges. In: MacLachlan NJ, Pearson JE, editors. Bluetongue, part 1, proceedings of the Third International Symposium, Taormina, 2003 Oct 26–29. Veterinaria Italiana. 2004;40:260–65. [PubMed] [Google Scholar]
- 4.Bilk S, Schulze C, Fischer M, Beer M, Hlinak A, Hoffmann B. Organ distribution of Schmallenberg virus RNA in malformed newborns. Vet Microbiol. 2012;159:236–8. 10.1016/j.vetmic.2012.03.035 [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–72. 10.3201/eid1803.111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cêtre-Sossah C, Baldet T, Delecolle JC, Mathieu B, Perrin A, Grillet C, et al. Molecular detection of Culicoides spp. and Culicoides imicola, the principal vector of bluetongue (BT) and African horse sickness (AHS) in Africa and Europe. Vet Res. 2004;35:325–37. 10.1051/vetres:2004015 [DOI] [PubMed] [Google Scholar]
- 7.Deblauwe I, De Witte JC, De Deken G, De Deken R, Madder M, Van Erk S, et al. A new tool for the molecular identification of Culicoides species of the Obsoletus group: the glass slide microarray approach. Med Vet Entomol. 2012;26:83–91. 10.1111/j.1365-2915.2011.00979.x [DOI] [PubMed] [Google Scholar]
- 8.St George TD, Standfast HA, Cybinski DH. Isolation of Akabane virus from sentinel cattle and Culicoides brevitarsis. Aust Vet J. 1978;54:558–61. 10.1111/j.1751-0813.1978.tb02412.x [DOI] [PubMed] [Google Scholar]
- 9.St George TD, Standfast HA. Simbu group viruses with teratogenic potential. In: Monath TP, editor. The aboviruses: epidemiology and ecology. Vol. 4. Boca Raton (FL): CRC Press; 1989. [Google Scholar]
- 10.Standfast HA, Dyce AL. Arthropods biting cattle during an epizootic of ephemeral fever in 1968. Aust Vet J. 1972;48:77–80. 10.1111/j.1751-0813.1972.tb02219.x [DOI] [PubMed] [Google Scholar]
- 11.Doherty RL, Carley JG, Standfast HA, Dyce AL, Snowdon WA. Virus strains isolated from arthropods during an epizootic of bovine ephemeral fever in Queensland. Aust Vet J. 1972;48:81–6. 10.1111/j.1751-0813.1972.tb02220.x [DOI] [PubMed] [Google Scholar]
- 12.Savini G, Goffredo M, Monaco F, Di Gennaro A, Cafiero MA, Baldo L, et al. Bluetongue virus isolation from midges belonging to the Obsoletus complex (Culicoides, Diptera: Ceratopogonidae) in Italy. Vet Rec. 2005;157:133–9. [DOI] [PubMed] [Google Scholar]
- 13.Dijkstra E, van der Ven IJK, Hölzel DR, van Rijn PA, Meiswinkel R. Culicoides chiopterus as a potential vector of bluetongue virus in Europe. Vet Rec. 2008;162:422. 10.1136/vr.162.13.422-a [DOI] [PubMed] [Google Scholar]