Abstract
Single-cell measurements allow a unique glimpse into cell-to-cell heterogeneity; even small changes in selected cells can have a profound impact on an organism’s physiology. Here an integrated approach to single-cell chemical sampling and assay are described. Capillary electrophoresis (CE) with laser-induced native fluorescence (LINF) has the sensitivity to characterize natively-fluorescent indoles and catechols within individual cells. While the separation and detection approaches are well established, the sampling and injection of individually selected cells requires new approaches. We describe an optimized system that interfaces a single-beam optical trap with CE and multichannel LINF detection. A cell is localized within the trap and then the capillary inlet is positioned near the cell using a computer-controlled micromanipulator. Hydrodynamic injection allows cell lysis to occur within the capillary inlet, followed by the CE separation and LINF detection. The use of multiple emission wavelengths allows improved analyte identification based on differences in analyte fluorescence emission profiles and migration time. The system enables injections of individual rat pinealocytes and quantification of their endogenous indoles, including serotonin, N-acetyl-serotonin, 5-hydroxyindole-3-acetic acid, tryptophol and others. The amounts detected in individual cells incubated in 5-hydroxytryptophan ranged from 10−14 mol to 10−16 mol, an order of magnitude higher than observed in untreated pinealocytes.
Introduction
Single-cell analysis is becoming more important as even so-called “homogenous” cell populations have shown significant cell-to-cell heterogeneity.1,2 However, measuring the contents of individual cells presents several challenges. Cells contain a broad range of compounds that span many orders of magnitude. When working with a tissue homogenate, much of the information on individual cell composition is lost. Addressing these concerns requires technologies that enable single-cell sampling and analysis. Capillary electrophoresis (CE) with laser-induced fluorescence detection (LIF) is particularly appropriate for single-cell chemical analysis due to its ability to probe both mass- and volume-limited samples, produce high separation efficiencies, and detect and quantitate low-abundance analytes.3–5 While CE-LIF has the sensitivity to characterize trace analytes within individual cells and subcellular components, and is a well-established approach, robust single-cell sample preparation methodologies are still evolving. As highlighted in this issue6 options include manual7–9 and automated mechanical10,11 cell handling, microfluidics,12,13 laser-assisted cell lysis,14,15 and, the focus of this article, optical trapping.16–18
Optical trapping was first demonstrated by Ashkin in 1970;19 in 1987 he extended the method to examining single bacteria, viruses, and cells.20,21 Subsequently, its use for studying biological structures has grown dramatically and trapping has become an invaluable technique for studying micrometer- and nanometer-sized objects not only in biology, but in physics and chemistry as well.22,23 Optical trapping is considered a non-contact method as it minimizes outside mechanical interferences that may affect the analytical measurements. There are also a variety of optical trap (OT) types and possible modifications that can be used to optimize the technique for a particular application and sample type.24–26 OT designs allow their hyphenation with other technologies, such as microfluidics,27–29 CE,30,31 Raman32,33 and fluorescence spectroscopy.34,35 Overall, OTs are a versatile tool that can be used to study, isolate, and manipulate single cells and subcellular organelles on the micron scale with a minimum of mechanical interference.
The successful development and optimization of a bioanalytical approach depends on choosing the appropriate biological model. The size and shape of the cells of interest, their biochemical content, and the surrounding extracellular media can add complexity to optical trapping experiments. Located in the pineal gland, pinealocytes are an attractive model for developing an approach that utilizes hyphenated optical trapping and CE laser induced native fluorescence (LINF). The cells of the mammalian pineal gland contain several natively fluorescent indolamines, including serotonin and melatonin, in high concentrations compared to other parts of the central nervous system.36,37 A critical feature of the pineal gland is the large fluctuations in chemical content depending on the time of day. During the day, serotonin is present at a relatively constant basal level whereas N-acetylserotonin (NAS) (a metabolite of serotonin and the precursor to melatonin) and melatonin are present at low levels, if at all. Once the retina signals that darkness has fallen, serotonin synthesis increases by several fold; within an hour, NAS and melatonin levels increase dramatically as they are synthesized, and melatonin is secreted into the blood. Within 1–3 h of daylight, NAS and melatonin production ceases. A number of other indolamines have been reported in the pineal gland, including 5-methoxytryptophan and 5-methoxytryptamine.38,39
In this work we describe an OT multichannel (MC) CE-LINF system optimized for the native fluorescence detection of catecholamines and indolamines.40–42 Analytes are identified by the combination of their unique spectral characteristics and migration times. The OT-MC-CE-LINF instrument enables analysis of a single pinealocyte with a minimum of sample handling and disruption, provides a separation step to reduce chemical complexity, and separates similar analytes and potentially concentrates them. Once the cell is localized within the OT, the capillary inlet is moved adjacent to the trap using a computer-controlled micromanipulator and microscope combination. The cell is released from the OT and quickly injected into the capillary, where it is chemically lysed and its chemical components are separated and detected. The ability of the OT-MC-CE-LINF system to inject individual cells and separate and characterize their indolamines is demonstrated.
Materials and methods
Chemicals
Chemicals, unless otherwise noted, were purchased from Sigma Aldrich (St. Louis, MO) at reagent grade or higher. Citric acid sheath buffer (25 mM, pH 2.25) was made by dissolving 5.25 g of C6H8O7•H2O in 1 L of ultrapure deionized (DI) water (Elga Purelab Ultra, Siemens Water Technologies, Warrendale, PA). Electrophoresis buffers were made by diluting a stock solution of 50 mM borate buffer, pH 8.8, prepared by dissolving 9.2 g of Na2B4O7•10 H2O and 3.0 g of B(OH)3 in 1 L of ultrapure DI water. For surfactant-containing electrophoresis buffers, 0.54 g of sodium dodecyl sulfate (SDS) was added to 50 mL of diluted borate buffer, pH 8.8, sonicated for 2 min to dissolve, and filtered with a 0.22 μm syringe filter (Nalgene, Rochester, NY). Serotonin (5-HT) (Alfa Aesar, Ward Hill, MA) and tyrosine (Tyr) were dissolved in 2.5 mM citric acid, pH 2.5, and sonicated on ice for 30 min. Tryptophan (Trp), NAS, 5-hydroxyindole acetic acid (HIAA), melatonin (MT), 5-hydroxytryptophan (HTP), 5-methoxytrytophol (MTOL), 5-methoxytryptamine (MOT) (TCI America, Portland, OR), tryptophol (TOL) (Research Organics, Inc., Cleveland, OH), 5-methoxyindole acetic acid (MIAA) (Gold Biotechnology, St. Louis, MO), and 5-hydroxytryptophol (HTOL) (Gold Biotechnology) were dissolved in 2.5 mM citric acid, pH 2.5, + 10% v/v acetone and sonicated on ice for 30–60 min. Standard buffers were prepared by diluting the sheath buffer 1:10 with ultrapure DI water. Fluorescein was prepared in ultrapure DI water. Standard stock solutions were diluted in either 1 mM borate buffer, pH 8.8 (1:50 dilution of stock borate electrophoresis buffer), or in high Ca+2/high Mg+2 modified Gey’s balanced salt solution (high salt mGBSS), pH 7.2, consisting of 3.0 mM CaCl2 (0.44 g), 4.9 mM KCl (0.37 g), 0.2 mM KH2PO4 (0.03 g), 22 mM MgCl2 (4.47 g), 0.6 mM MgSO4 (0.07 g), 138 mM NaCl (8.06 g), 27.7 mM NaHCO3 (2.33 g), 0.8 mM Na2HPO4 (0.11 g), 25 mM HEPES (5.95 g), and 10 mM glucose (1.80 g) dissolved in 1 L of ultrapure DI water. Biological samples were stored in either high salt mGBSS, as described above, or mGBSS, pH 7.2, consisting of 1.5 mM CaCl2 (0.22 g), 4.9 mM KCl (0.37 g), 0.2 mM KH2PO4 (0.03 g), 11 mM MgCl2 (2.24 g), 0.3 mM MgSO4 (0.04 g), 138 mM NaCl (8.06 g), 27.7 mM NaHCO3 (2.33 g), 0.8 mM Na2HPO4 (0.11 g), 25 mM HEPES (5.95 g), and 10 mM glucose (1.80 g) dissolved in 1 L of ultrapure DI water. Buffers were filtered by a 0.45 μm bottle-top filter system (Nalgene, Rochester, NY) and degassed under vacuum with stirring for 30–60 min. NaOH was prepared by dissolving one pellet (~0.002 g) in 0.025 L of ultrapure DI water.
Animals
All experimental procedures were conducted according to protocols approved by the Institutional Animal Care and Use Committee, University of Illinois at Urbana-Champaign. Animal care and experiments were performed in full compliance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Long-Evans/BluGill rats (University of Illinois at Urbana-Champaign), which have been demonstrated to be genetically homogeneous by high density genome scan, were sacrificed by rapid decapitation and the pineal glands isolated from the central nervous system. Males, 6–8 weeks, were sacrificed during the day, between circadian time 3:00 – 5:00, and pineal dissection and preparation was completed within 30 min. Glands were manually triturated for pinealocyte isolation and stored in high salt mGBSS on ice until analysis, typically between 30–60 min.
Biological samples
Rat pinealocyte suspensions were split into two groups. The first was incubated at room temperature for 60 min in 200 μM HTP, dissolved in high salt mGBSS. The second control group had an equal volume of high salt mGBSS and was incubated at room temperature for 100 min before analysis.
Preparation of the CE column inlet
The CE columns, 85–120 cm long, were made from 50 μm inner diameter, 360 μm outer diameter fused silica capillaries (Polymicro Technologies, Phoenix, AZ). The capillary inlets and outlets were etched with hydrofluoric acid (HF) in order to reduce the outer diameters and create sharply tapered tips with a 40° angle (see43 and Supplementary Information for details).
OT design and construction
Unless otherwise noted, all custom-built components were designed and fabricated in-house. The optical axis is parallel to the optical table at a height of 20 cm until the beam is directed 90° vertically by a dichroic mirror. All of the optics are infrared (IR)-coated unless otherwise specified. The near infrared (NIR) beam is viewed with IR viewing cards (VC-1550 and F-IRC-HP, Thorlabs, Newton, New Jersey) and an IR viewer (IRV1-1700, Newport Corp., Irving, CA).
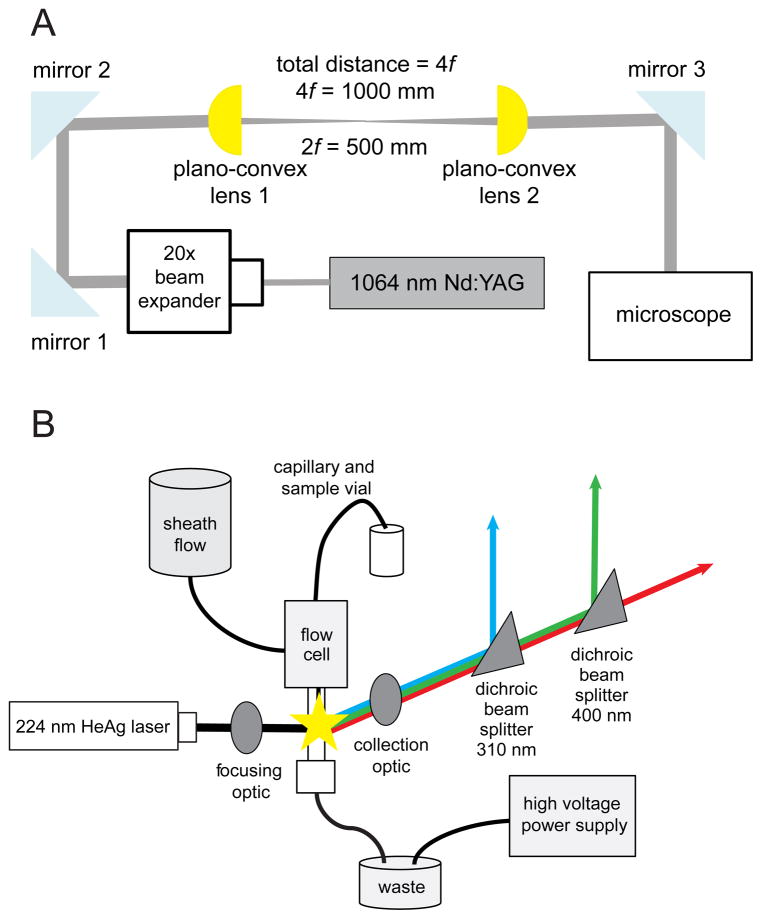
The OT design (Figure 1A) uses a 1064 nm diode-pumped solid state Nd:YAG laser (Compass 1064-2500MN, Coherent Inc., Santa Clara, CA) with a maximum output of 2.5 W. The laser operates in TEM00 mode and has a wavelength stability of < 1 cm−1. The beam has a nominal diameter of 0.4 mm, a divergence of < 3.5 mrad, a pointing stability of < ± 5%, and an ellipticity of < 1.1. It is air-cooled and turn-key operated. The beam is expanded by a 20× high energy beam expander (HB-20X, Newport Corp.) and directed by a pair of gold-coated mirrors (PF10-03-M01, Thorlabs) into a set of two plano-convex lenses (SPX029, Newport Corp.) in a 1:1 telescope configuration, used to steer and parfocalize the beam. The beam expander is housed in a precision gimbal optic mount (605-4, Newport Corp.) and translated in the x-, y-, and z-directions by a translation stage (UMR12.40, Newport Corp.) and heavy duty optical lab jack (L490, Thorlabs). Plano-convex lens 1 is mounted in a 3-axis optical mount (LP-1A-XYZ, Newport Corp.) located 1000 mm from the back aperture of the objective and plano-convex lens 2 is mounted in a 2-axis mount (LP-1A-XY, Newport Corp.) located 500 mm from the back aperture of the objective. This setup can be generalized as the distance between the back aperture of the objective and the steering optic is equal to 4f, where f is the focal length of the lens.44,45 In this case, the focal lengths of both of the plano-convex lenses are 250 mm. The beam is then directed into the epi-fluorescence port of the microscope (AxioObserver A1, Carl Zeiss, Jena, Germany) and directed into the back aperture of the objective (Objective C-Apochromat 63x/1.2 W Corr, 441777-9970-000, Carl Zeiss, Jena, Germany) by a dichroic mirror centered at 1064 nm (950dcsp-laser, Chroma Technology, Rockingham, VT).
Figure 1.
The OT and MC-CE-LINF instrument design (both being interfaced to the same sample stage on the microscope). (A) Schematic of the optical trap. Emission from the Nd:YAG laser is expanded by a 20× beam expander to fill the back aperture of the objective. The beam is directed into a pair of plano-convex lenses, which act to steer and parfocalize the beam. The beam is directed in the epi-fluorescence port of a microscope and a dichroic mirror directs it into the objective. (B) Schematic for the MC-CE-LINF instrument. Emission from the HeAg laser is focused 0.5–1 mm below the capillary outlet. Fluorescence is collected orthogonal to excitation by an objective, which collimates and directs the emission to two dichroic beam splitters. The beam splitters separate the emission into three wavelength ranges: 250–310 nm (PMT blue), 310–400 nm (PMT green), and 400 nm and above (PMT red), and a photomultiplier tube detector quantifies the emission intensity for each wavelength range.
Polystyrene bead stock solutions (1 μm (PS04N/5749) and 10 μm (PS06N/6955) in diameter, Bangs Laboratories, Fishers, IN), were diluted 10-fold to 100-fold in ultrapure DI water and then used to optimize the optical trap. The beads were contained on a coverslip (2735-246, Corning Inc.) by a vacuum grease ring (Silicon High Vacuum Grease, Dow Corning, Midland, MI). Laser power measurements were taken with a PM10 sensor and a LabMax-TOP meter (Coherent Inc.).
MC-CE-LINF instrument design and construction
The injection port for the instrument is housed on a non-conductive breadboard platform on a microscope that is contained in a clear Plexiglas box. The capillary inlet has PEEK fittings (Upchurch Scientific, Oak Harbor, WA) and a FEP sleeve (Upchurch Scientific), attached to allow for fast and easy switching between the syringes and capillary holder.
The capillary is held in place in the instrument by a custom-built acetal resin (Delrin, E. I. duPont de Nemours & Co., Wilmington, DE) sheath flow cell. It enters at the top of the cell and is held in place by liquid-tight fittings (Upchurch Scientific). The sheath buffer enters the cuvette from the right side with respect to the optical table and exits from the bottom of the sheath flow cell. The quartz cuvette (Starna Cells, Atascadero, CA) used for excitation and detection of eluents is open on both ends and is attached to the top and bottom pieces of the sheath flow cell with Tra-Cast 3103 epoxy (Henkel Corp., Billerica, MA).
The instrument optics (Figure 1B) were based on our prior instrument design.46 Deep UV radiation (224.6 nm) from an HeAg hollow cathode ion laser (HeAg70, Photon Systems Inc., Covina, CA) is spectrally filtered using a four-bounce mirror configuration, attached to the front of the laser head. The beam is directed via two UV-coated mirrors (Thorlabs) into a laboratory-built lightproof, non-conductive box and breadboard, which houses the detection optics and protects against spurious arcing. The collimated beam is nominally focused using a plano-convex lens (OptoSigma, Santa Ana, CA) to a 50 μm spot directly below the outlet of the capillary, which has been HF-etched to a cone-shaped tip and is housed in a custom-built sheath flow cell, as described above. As analytes elute from the capillary they are excited by the focused beam and emit fluorescence, which is collected and collimated by a 15× all-reflective objective (13596, Newport Corp.). The fluorescence is directed toward three photomultiplier tube (PMT) detectors (H6780-06, Hamamatsu, Middlesex, NJ) by two dichroic mirrors (310dcxxr-haf #110258 and 400dcxru #111563, Chroma Technology), with transition points at 310 nm and 400 nm, respectively. The first detector (PMT “blue”) measures emission from 250–310 nm, the second detector (PMT “green”) measures emission from 310–400 nm, and the third detector (PMT “red”) measures emission from 400 nm and above. The laser and PMTs are synchronized and controlled by software written in LABView and provided by Photon Systems Inc. Posts, post holders, and other optical mounts were purchased from Newport Corp., Melles Griot (Albuquerque, NM), or custom-built. Optical mounts for the focusing and collection optics are coated in Vinyl Liquid Electric Tape (Star Brite, Ft. Lauderdale, FL) and Scotch Super 88 electrical tape (3M, St. Paul, MN) to reduce arcing from the capillary outlet and tubing to the mounts.
Negative voltage for electrophoresis is applied to the sheath flow waste by a stainless steel cylinder connected to a power supply (PS/MJ30N0400-11, Glassman High Voltage, High Bridge, NJ) and laboratory-built control box. A 10 kΩ resistor and a digital multimeter (Fluke 76, Fluke Corp., Everett, WA) are part of the circuit and are used to measure the current across the capillary.
Sheath buffer is gravity-driven and flow can be adjusted by a right angle switching valve (Upchurch Scientific). High purity Teflon PFA Plus tubing and appropriate fittings were purchased from Upchurch Scientific. All tubing is further encased within FEP-lined polyethylene tubing (McMaster-Carr, Elmhurst, IL) to reduce static attraction and arcing during electrophoresis. Tubing between the optics box and the sheath box is also surrounded by four 16 oz. polyethylene containers and Scotch Super 88 electrical tape.
Interfacing the OT with the MC-CE-LINF instrument
The optical trap and the MC-CE-LINF system are interfaced at the microscope stage (Figure 1S, Supplemental Information). The trap is located at the focal point of the objective, approximately 0.28 mm from the objective surface, including the coverslip thickness (0.13–0.16 mm). The capillary inlet is controlled by a computer-controlled motorized micromanipulator (MP-285, Sutter Instrument Co., Novato, CA), which has 1″ of travel in all three axes, two step sizes—coarse (0.2 μm/step) and fine (0.04 μm/step)—and a maximum speed of 2.9 mm/s. It has a tabletop controller and a rotary optical encoder for manual control; programmable robotic control is also available. The micromanipulator is mounted on a non-conductive optical breadboard, which is stabilized by two 1/4″-28-tapped beams that attach to the microscope stand on either side of the stage. The capillary is held in the micromanipulator by an acetal resin cylinder, which is 6″ long and has a 1/16″ diameter hole drilled in the center. This holder reduces the chance of arcing to the micromanipulator motors.
The sample is held on a coverslip holder, machined out of polycarbonate, with a lip to rest the coverslip edges on, and a 30° angled oval hole for holding the electrophoresis buffer vial, which consists of an Eppendorf tube (Hamburg, Germany) that has its top quarter removed at an angle. A platinum grounding wire (California Fine Wire Co., Grover Beach, CA) is placed in contact with the electrophoresis buffer, completing the circuit.
Trapping, manipulation, and injection of cells and beads are recorded by a monochrome CMOS camera (NT59-365, EO-1312M, Edmund Optics, Barrington, NJ) that is attached to the microscope housing with a 1× C-mount (Carl Zeiss, Jena, Germany).
An ancillary capillary was provided to clear the inlet to the separation capillary (and the trap) as needed by flowing high salt mGBSS at ~1 μL/min. The capillary position was controlled via another micromanipulator (Narishige Scientific Instrument Lab, Tokyo, Japan) placed to the left of the microscope; an HF-etched capillary (50 μm inner diameter, 360 μm outer diameter, and ~50 cm in length) was held by an acetal resin cylinder, similar to the one described above. PEEK fittings and a FEP sleeve were used to connect the capillary outlet to a syringe filled with high salt mGBSS. The pressure on the syringe was controlled by a syringe pump (model 601553, KD Scientific, Holliston, MA), located on top of the injection box.
Coverslip coatings and additives
Glass coverslips were used for all experiments except where noted. Several coatings and additives were tested to reduce adhesion of cells to the coverslip surface (as detailed in the Supplemental Information). Treated coverslips were stored at ambient temperature and humidity in Parafilm M (Pechiney Plastic Packaging Inc., Chicago, IL)-covered glass dishes, unless otherwise noted, until use.
Single-cell injections
Single pinealocytes were injected into the CE column for analysis. A 2.5 μL droplet of sample was pipetted onto the coverslip. The selected cell is held in the OT. The capillary inlet was directed into the cell’s proximity by the micromanipulator, which was programmed to stop near the trap location and further position refinement was performed manually using the rotary optical encoder. Once the capillary was in place, the cell was released from the trap and hydrodynamic injection of the cell was performed by lowering the sheath waste outlet. Once injection was complete, the micromanipulator was used to bring the capillary inlet to the buffer vial, and the voltage and detectors were turned on. Injections were recorded using the CMOS camera on the microscope.
Electrophoresis
The sheath flow buffer was 25 mM citric acid, pH 2.25, and the flow rate was 0.2 mm/s for all experiments. The electrophoresis buffers and sample buffers varied as stated in the text and figure captions and both the capillary zone electrophoresis (CZE) and micellular electrokinetic chromatography (MEKC) modes were used. The voltage for all experiments was −30 kV unless otherwise stated. The injection volume varied as stated, but for bulk injections the volume was 14.7 nL for a 30 s hydrodynamic injection, which was achieved by lowering the sheath flow waste outlet by 32.5 cm. The typical laser pulse energy was between 1.5 μJ/pulse and 2 μJ/pulse.
The capillary was conditioned at the beginning of the day with 0.1 M NaOH for 15–20 min, followed by water for 5 min, and then electrophoresis buffer for a minimum of 5 min.
Data analysis
Data analysis was performed in IgorPro 5.05A (WaveMetrics Inc., Lake Oswego, OR). An automated data analysis script was written that reduces the user input to a single command. Output consists of four tables of calculated values with four corresponding color-coded graphs displaying the raw data, 6-point boxcar averaged data, normalized (with respect to the laser pulse energy) data, and both normalized and boxcar averaged data.
Limits of detection
The baseline range (30 points, 10 s) with the lowest standard deviation was determined and used to calculate the limits of detection (LOD) for each PMT channel. Ratiometric analysis (calculating the intensity ratio between peak maxima in each of the PMT channels) was also automated to aid in analyte identification.
Single-cell analyte concentrations were calculated with an approach that removes background signals from the media and differences in separation conditions, as described here. The ratio of the injection lengths between a sample analyzed under cell lysing conditions and the same sample analyzed under non-cell lysing conditions was used to normalize the analyte concentrations under non-cell lysing conditions. These normalized values were used to calculate equivalent background concentrations, which were subtracted from the concentrations calculated under cell lysing conditions. The analyte amounts were then adjusted based on an assumed cell volume of 4 pL to represent the actual analyte concentration within the cell.
LODs and concentrations of analytes were determined by generating calibration curves for each analyte under the appropriate conditions. Investigated analyte concentrations ranged from the micromolar to the low nanomolar range typical for biological systems. The criterion for calculating the LODs was three times the standard deviation of the baseline.
Results and discussion
Optical trap construction and performance
In order to successfully trap and manipulate objects, the trapping laser should have a symmetric beam profile (usually Gaussian TEM00), low beam divergence, low ellipticity, and high pointing stability. Also, the NIR wavelength region is typically used for biological manipulations, as the NIR as a whole (from ~ 700 nm to 1300 nm) produces little damage to biological structures compared with visible and UV light. While water does not absorb NIR radiation, absorption of NIR radiation by endogenous chromophores makes some wavelength ranges better suited to these measurements than others.47,48 An Nd:YAG laser with 1064 nm emission was selected. No beam correction was needed, and the beam was expanded (by a factor of 20) to 8 mm to fill the back aperture of the objective, ensuring a steeper gradient and hence a stronger trap. The 1:1 telescope configuration was set up with the steering optic (plano-convex lens 1) located at a distance of 4f (1000 mm) from the back aperture. Translations in this lens correspond to movements of the trap, and were used to direct trapped objects to specific locations. The optical throughput of this system was experimentally determined to be 32% of the initial power, with the highest loss occurring at the objective. The trap size was ~1 μm, based on the numerical aperture of the objective and the wavelength of the laser. Polystyrene beads suspended in water were used to align the trap. A minimum initial power of 90 mW (power density ~6.5 × 106 W/cm2) was used to determine optimal conditions for the manipulation and translation of trapped beads.
MC-CE-LINF instrument performance
The MC-CE-LINF instrument has several unique features; notable aspects include deep UV excitation, synchronization between the laser and PMTs, multiple detectors, a tapered outlet, post-column detection, and the ability to independently optimize the separation and detection conditions.
This excitation and detection system is controlled via the program provided by Photon Systems Inc. The laser pulse length and the PMT on-time are synchronized, resulting in reduced dark noise and low background signal. The net intensity for a given peak is determined in each channel and used to calculate three different ratios (green/blue or G/B, blue/red or B/R, and green/red or G/R). These ratios provide information on the unique spectral characteristics of the analytes and, when combined with migration time information, can allow for identification without using a complimentary technique or sample spiking, as would be required with a traditional single-channel system.
Post-column detection has been used for CE-LIF since the 1980s.49 For deep UV excitation, post-column detection is important because light scattering is proportional to λ−4. Another benefit of post-column detection is the ability to independently optimize the separation and detection conditions.50 This is useful because the maximal fluorescence for biogenic amines is achieved under acidic conditions but the best analyte separations use basic electrolytes. The HF-etched capillary outlet reduces background signal from scattering off of the edge of the capillary wall and changes the sheath flow profile around the outlet, allowing the excitation beam to be focused closer to the outlet.
To demonstrate the instrument’s performance, the LODs for 13 indolamines known to be present in the pineal gland have been determined for both MEKC and CZE (Table 1) modes. The LODs are appropriate for measuring the physiological concentrations of the indoles present in pinealocytes.
Table 1.
Limits of detection for indolamines and tyrosine in MEKC and CZE modes.
| Analyte | MEKC (nM)1 | CZE (nM)2 |
|---|---|---|
| 5-hydroxyindole-3-acetic acid, HIAA | 3.5 | 2.8 |
| 5-hydroxytryptophan, HTP | 5.4 | 1.6 |
| 5-hydroxtryptophol, HTOL | 2.6 | 6.7 |
| 5-methoxyindole-3-acetic acid, MIAA | 3.4 | 2.0 |
| 5-methoxytryptamine, MOT | 14 | 9.5 |
| 5-methoxytryptophan, MTrp | NA | 1.3 |
| 5-methoxytryptophol, MTOL | 3.5 | 3.0 |
| melatonin, MT | 14 | 2.8 |
| N-acetylserotonin, NAS | 5.1 | 3.7 |
| serotonin, 5-HT | 14 | 14 |
| tyrosine, Tyr | 39 | 17 |
| tryptophan, Trp | 6.0 | 1.6 |
| tryptophol, TOL | 2.5 | 2.6 |
MEKC conditions: 20 mM borate buffer, pH 8.8, + 50 mM SDS or 15 mM borate buffer, pH 8.8, + 37.7 mM SDS.
CZE conditions: 40 mM or 30 mM borate buffer, pH 8.8.
Optimizing conditions for single-cell sampling
Challenges that arose during single pinealocyte trapping and injection were adhesion to the coverslip and multiple cells and/or debris entering the capillary simultaneously. Pinealocytes can adhere to the surface of the glass coverslip within minutes after sample deposition, which makes selecting and trapping a cell for injection time-sensitive and can waste valuable sample. Once the cells are adhered, trapping is ineffective and moving the cells is difficult, even when using the capillary inlet to “push” them around the surface. Pinealocytes are robust and adhesion is strong; typically, pinealocytes do not lyse when they forcefully come into contact with the capillary inlet. Several coverslip coatings and solution additives were tested with pinealocytes to determine the best method to reduce adhesion, as detailed in the Supplemental Information. The efficacy of the coating was determined by three figures of merit: how easily a cell could be trapped and manipulated, and how long it took for cells to adhere to the coverslip. Based on the results of these experiments, poly(2-hydroxyethyl methacrylate)-coated coverslips were subsequently used for these studies.
Another issue is that multiple cells or cellular/tissue debris can enter the capillary during injection. We tried several approaches to minimize this from occurring. While one can prevent this is by using capillary out-flow (to push the untrapped objects away from the capillary inlet), this is effective only when the buffer is compatible with the sample. Here the electrophoresis buffer used for these experiments contained a surfactant to lyse the trapped cell once it entered the capillary. Using capillary out-flow under these conditions lysed the cells in the vicinity, including the trapped cell. We successfully used a second capillary, housed in and controlled by a second micromanipulator, to flush the area before injection of the undesired cells and debris. The second micromanipulator was manually adjusted and the second capillary’s outlet was fitted to a syringe filled with high salt mGBSS with flow controlled via a syringe pump.
Single pinealocyte analysis
We used the OT-MC-CE-LINF system for single pinealocyte analyses, and bulk cell suspensions were analyzed with the MC-CE-LINF system. Two different CE conditions were used for these analyses: MEKC51,52 and CZE. Several of the indolamines of interest co-eluted without MEKC. The use of such distinct separation mechanisms allowed confirmation of analyte identity as it is less likely that the same analytes would coelute in both systems.
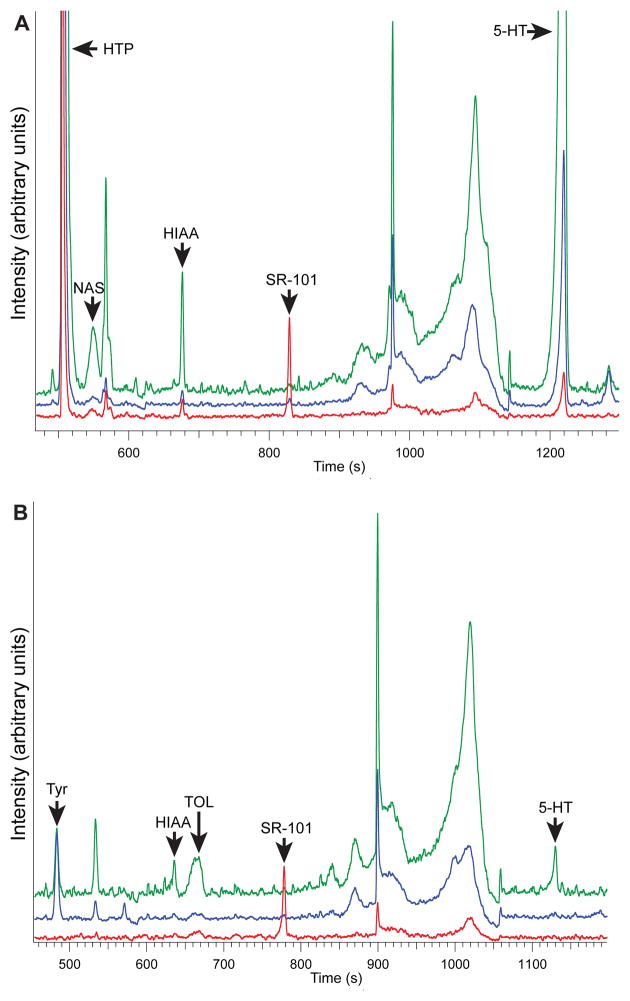
Prior to analysis, pinealocyte suspensions were split into two aliquots, referred to as incubated and untreated samples. Incubated samples were exposed to HTP to increase production of indolamines within the cells. This also tests for cell viability as only viable cells synthesize analytes of interest. The untreated samples were not incubated with HTP but rather an equal amount of high salt mGBSS was added to the suspension. Representative electropherograms from single pinealocyte injections under both incubation (Figure 2A) and untreated conditions (Figure 2B), respectively, from pineal samples acquired from the same animal are shown. Several analytes of interest have been detected and identified, such as serotonin 5-HT, NAS, HIAA, and TOL. In the HTP-incubated sample, elevated levels of 5-HT and HIAA are present compared to native levels (Figure 2A), as expected; a roughly 50-fold difference in 5-HT concentration is observed. HIAA is an enzymatic degradation product of 5-HT, so an increase in its concentration is also expected when elevated 5-HT levels are present. NAS is a product of 5-HT, the precursor to melatonin, and is present in low amounts relative to the other indolamines detected. The amounts of analytes present within an individual HTP-incubated pinealocyte correspond to 10−14 mol to 10−16 mol (mM to μM levels in a 10 μm diameter pinealocyte), an order of magnitude higher than the native levels present in an untreated pinealocyte. Two unknown indolamines (based on fluorescence properties) were also detected in the incubated sample (data not shown); work is ongoing to identify them.
Figure 2.
Electropherograms of a treated and untreated single pinealocyte from the same animal. The green trace represents the signal from PMT green (310–400 nm), the blue trace represents the signal from PMT blue (250–310 nm), and the red trace represents the signal from PMT red (400+ nm). (A) A pinealocyte incubated in 5-hydroxytryptophan. (B) An untreated pinealocyte, with native levels of analytes such as indolamines and Tyr detected. Recordings from all three detection channels are shown. 5-HT = serotonin, HIAA = 5-hydroxyindole-3-acetic acid, HTP = 5-hydroxytryptophan, NAS = N-acetylserotonin, SR-101 = sulforhodamine-101 (internal standard), TOL = tryptophol, Tyr = tyrosine. Conditions are: 15 mM borate buffer, pH 8.8, + 37.5 mM SDS (electrophoresis buffer), high salt mGBSS (sample buffer), 25 mM citric acid buffer, pH 2.25 (sheath buffer), −30 kV (separation voltage), 3 Hz (laser repetition rate), 100 μs (laser pulse length), 8 A (laser current), 420 V (laser BUSS voltage), 470 pF (PMT gain), 64% (gain voltage), 110 μs (PMT integration time).
Conclusions
Optical trapping is a versatile approach for manipulation of small samples such as individual cells and is well suited for positioning live cells adjacent to a CE capillary inlet. One can time cell injection with the physiological state of the cell under investigation. The current system works well when injecting and analyzing analytes from smaller mammalian cells such as the pinealocytes studied here. Future work will include investigating day- and night-time differences in pinealocyte content and chemical profile, as well as understanding the surprisingly complex indole chemistry within these cells.
Several modifications can be made to improve the performance and ease of use of the hyphenated instrument, including using smaller inner diameter CE capillaries and refining the second micromanipulator/capillary combination for improved injections. Using smaller inner diameter capillaries will reduce the likelihood of trapping and injecting multiple cells and reduce analyte dilution after cell lysis, enabling lower concentrations analytes within the cells to be characterized.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the useful advice and assistance of Jennifer M. Arnold concerning the animal experiments. The help provided by Bill Hug, Ray Reid, and Prashant Oswal (at Photon Systems Inc.) was essential during the building of the analytical system. This work was supported by the National Institute of Neurological Disorders and Stroke under award number R01 NS031609 and by the National Institute of Dental and Craniofacial Research under award number R01 DE018866 and the National Heart, Lung, and Blood Institute under award number R01 HL086870. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Lapainis T, Sweedler JV. J Chromatogr. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 2.Li G-W, Xie XS. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krylov SN, Dovichi NJ. Electrophoresis. 2000;21:767–773. doi: 10.1002/(SICI)1522-2683(20000301)21:4<767::AID-ELPS767>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 4.Lin Y, Trouillon Rl, Safina G, Ewing AG. Anal Chem. 2011;83:4369–4392. doi: 10.1021/ac2009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stuart JN, Sweedler JV. LC-GC Europe. 2003:427–429. [Google Scholar]
- 6.White HS, editor. Single, Analyst. 2012. in press. [DOI] [PubMed] [Google Scholar]
- 7.Anderson BB, Ewing AG. J Pharm Biomed Anal. 1999;19:15–32. doi: 10.1016/s0731-7085(98)00088-0. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RD, Navratil M, Poe BG, Xiong G, Olson KJ, Ahmadzadeh H, Andreyev D, Duffy CF, Arriaga EA. Anal Bioanal Chem. 2007;387:107–118. doi: 10.1007/s00216-006-0689-6. [DOI] [PubMed] [Google Scholar]
- 9.Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV. Anal Chem. 2011;83:6810–6817. doi: 10.1021/ac2015855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boardman A, Chang T, Folch A, Dovichi NJ. Anal Chem. 2010;82:9959–9961. doi: 10.1021/ac1022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Anal Chem. 2010;82:967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chueh B-h, Li C-W, Wu H, Davison M, Wei H, Bhaya D, Zare RN. Anal Biochem. 2011;411:64–70. doi: 10.1016/j.ab.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Omiatek DM, Santillo MF, Heien ML, Ewing AG. Anal Chem. 2009;81:2294–2302. doi: 10.1021/ac802466g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen PB, Doepker BR, Chiu DT. Anal Chem. 2009;81:3784–3791. doi: 10.1021/ac900099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D, Sims CE, Allbritton NL. Electrophoresis. 2010;31:2558–2565. doi: 10.1002/elps.201000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando J, Bautista G, Smith N, Fujita K, Daria VR. Rev Sci Instrum. 2008;79:103705. doi: 10.1063/1.2999542. [DOI] [PubMed] [Google Scholar]
- 17.Hellmich W, Pelargus C, Leffhalm K, Ros A, Anselmetti D. Electrophoresis. 2005;26:3689–3696. doi: 10.1002/elps.200500185. [DOI] [PubMed] [Google Scholar]
- 18.Maghelli N, Tolić-N⊘rrelykke IM. Meth Cell Biol. 2010;97:173–183. doi: 10.1016/S0091-679X(10)97010-6. [DOI] [PubMed] [Google Scholar]
- 19.Ashkin A. Phys Rev Lett. 1970;24:156–159. [Google Scholar]
- 20.Ashkin A, Dziedzic JM. Science. 1987;235:1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- 21.Ashkin A, Dziedzic JM, Yamane T. Nature. 1987;330:769–771. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- 22.Block SM. Nature. 1992;360:493–495. doi: 10.1038/360493a0. [DOI] [PubMed] [Google Scholar]
- 23.Dholakia K, Čižmár T. Nat Photonics. 2011;5:335–342. [Google Scholar]
- 24.Shi L, Shao B, Chen T, Berns M. J Biophotonics. 2009;2:167–177. doi: 10.1002/jbio.200810053. [DOI] [PubMed] [Google Scholar]
- 25.Lautenschlaeger F, Guck J. International Conference on Optomechatronic Technologies. 2009:419–422. [Google Scholar]
- 26.Chiu DT, Wilson CF, Ryttsen F, Stromberg A, Farre C, Karlsson A, Nordholm S, Gaggar A, Modi BP, Moscho A, Garza-Lopez RA, Orwar O, Zare RN. Science. 1999;283:1892–1895. doi: 10.1126/science.283.5409.1892. [DOI] [PubMed] [Google Scholar]
- 27.Applegate R, Squier J, Vestad T, Oakey J, Marr D. Opt Express. 2004;12:4390–4398. doi: 10.1364/opex.12.004390. [DOI] [PubMed] [Google Scholar]
- 28.Murata M, Okamoto Y, Park Y-S, Kaji N, Tokeshi M, Baba Y. Anal Bioanal Chem. 2009:1–7. doi: 10.1007/s00216-009-2648-5. [DOI] [PubMed] [Google Scholar]
- 29.Nève N, Kohles SS, Winn SR, Tretheway DC. Cell Mol Bioeng. 2010;3:213–228. doi: 10.1007/s12195-010-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu DT, Hsiao A, Gaggar A, Garza-Lopez R, Orwar O, Zare RN. Anal Chem. 1997;69:1801–1807. doi: 10.1021/ac961226g. [DOI] [PubMed] [Google Scholar]
- 31.Chiu DT, Lillard SJ, Scheller RH, Zare RN, Rodriguez-Cruz SE, Williams ER, Orwar O, Sandberg M, Lundquist JA. Science. 1998;279:1190–1193. doi: 10.1126/science.279.5354.1190. [DOI] [PubMed] [Google Scholar]
- 32.Alexander T, Pellegrino P, Gillespie J. Appl Spectrosc. 2003;57:1340–1345. doi: 10.1366/000370203322554482. [DOI] [PubMed] [Google Scholar]
- 33.Bankapur A, Zachariah E, Chidangil S, Valiathan M, Mathur D. PLoS ONE. 2010;5:e10427. doi: 10.1371/journal.pone.0010427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kühn S, Phillips BS, Lunt EJ, Hawkins AR, Schmidt H. Lab Chip. 2009;10:189. doi: 10.1039/b915750f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snijder-Van As MI, Rieger B, Joosten B, Subramaniam V, Figdor CG, Kanger JS. J Microsc. 2009;233:84–92. doi: 10.1111/j.1365-2818.2008.03099.x. [DOI] [PubMed] [Google Scholar]
- 36.Ganguly S, Coon S, Klein D. Cell Tissue Res. 2002;309:127–137. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- 37.Goto M, Oshima I, Tomita T, Ebihara S. J Pineal Res. 1989;7:195–204. doi: 10.1111/j.1600-079x.1989.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 38.Bondarenko L. Bull Exp Biol Med. 1985;100:1001–1003. [Google Scholar]
- 39.Mills MH, Finlay DC, Haddad PR. J Chromatogr. 1991;564:93–102. doi: 10.1016/0378-4347(91)80072-k. [DOI] [PubMed] [Google Scholar]
- 40.Fuller RR, Moroz LL, Gillette R, Sweedler JV. Neuron. 1998;20:173–181. doi: 10.1016/s0896-6273(00)80446-8. [DOI] [PubMed] [Google Scholar]
- 41.Squires LN, Talbot KN, Rubakhin SS, Sweedler JV. J Neurochem. 2007;103:174–180. doi: 10.1111/j.1471-4159.2007.04739.x. [DOI] [PubMed] [Google Scholar]
- 42.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Nat Methods. 2011;8:S20–29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann P, Dutoit B, Salathe RP. Ultramicroscopy. 1995;61:165–170. [Google Scholar]
- 44.Fallman E, Andersson M, Axner O. Proc of SPIE (Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues IV) 2006;6088:1–12. [Google Scholar]
- 45.Fallman E, Axner O. Appl Opt. 1997;36:2107–2113. doi: 10.1364/ao.36.002107. [DOI] [PubMed] [Google Scholar]
- 46.Lapainis T, Scanlan C, Rubahkin SS, Sweedler JV. Anal Bioanal Chem. 2007;387:97–105. doi: 10.1007/s00216-006-0775-9. [DOI] [PubMed] [Google Scholar]
- 47.Liang H, Vu KT, Krishnan P, Trang TC, Shin D, Kimel S, Berns MW. Biophys J. 2005;70:1529–1533. doi: 10.1016/S0006-3495(96)79716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman KC, Liou GF, Block SM, Bergman K. Lasers and Electro-Optics, CLEO’98. 1998:203–204. [Google Scholar]
- 49.Zarrin F, Dovichi NJ. Anal Chem. 1985;57:2690–2692. doi: 10.1021/ac00286a010. [DOI] [PubMed] [Google Scholar]
- 50.Park YH, Zhang X, Rubakhin SS, Sweedler JV. Anal Chem. 1999;71:4997–5002. doi: 10.1021/ac990659r. [DOI] [PubMed] [Google Scholar]
- 51.Otsuka K, Terabe S, Ando T. J Chromatogr. 1985;332:219–226. [Google Scholar]
- 52.Quirino JP, Terabe S, Bocek P. Anal Chem. 2000;72:1934–1940. doi: 10.1021/ac990566+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.