Abstract
We utilized a cohort of 828 treatment seeking self-identified white cigarette smokers (50% female) to rank candidate gene single nucleotide polymorphisms (SNPs) associated with the Fagerström Test for Nicotine Dependence (FTND), a measure of nicotine dependence which assesses quantity of cigarettes smoked and time- and place-dependent characteristics of the respondent’s smoking behavior. 1123 SNPs at 55 autosomal candidate genes, nicotinic acetylcholine receptors and genes involved in dopaminergic function, were tested for association to baseline FTND scores adjusted for age, depression, education, sex and study site. SNP P values were adjusted for the number of transmission models, the number of SNPs tested per candidate gene, and their intragenic correlation. DRD2, SLC6A3 and NR4A2 SNPs with adjusted P values < 0.10 were considered sufficiently noteworthy to justify further genetic, bioinformatic and literature analyses. Each independent signal among the top-ranked SNPs accounted for ~1% of the FTND variance in this sample. The DRD2 SNP appears to represent a novel association with nicotine dependence. The SLC6A3 SNPs have previously been shown to be associated with SLC6A3 transcription or dopamine transporter density in vitro, in vivo and ex vivo. Analysis of SLC6A3 and NR4A2 SNPs identified a statistically significant gene-gene interaction (P=0.001), consistent with in vitro evidence that the NR4A2 protein product (NURR1) regulates SLC6A3 transcription. A community cohort of N=175 multiplex ever smoking pedigrees (N=423 ever smokers) provided nominal evidence for association with the FTND at these top ranked SNPs, uncorrected for multiple comparisons.
Keywords: dopamine transporter, Fagerström Test for Nicotine Dependence, single nucleotide polymorphism, candidate gene association scan, gene-gene interaction
INTRODUCTION
Cigarette smoking remains prevalent (Kilmer et al., 2008), a risk factor for more than two dozen diseases (CDC, 2003), and is the largest cause of preventable mortality worldwide (Ezzati and Lopez, 2003). Identifying factors influencing smoking will help us understand the nature of this behavior and potentially guide the development of effective interventions for behavior change.
Tobacco dependence is considered to be a multidimensional construct (Hudmon et al., 2003; Russell et al., 1974) and is measured using a variety of questionnaires, including smoking quantity (cigarettes per day), the Fagerström Tolerance Questionnaire (FTQ) (Fagerstrom, 1978), the Heaviness Smoking Index (HSI), consisting of two items from the FTQ (Heatherton et al., 1989), and the Fagerström Test for Nicotine Dependence (FTND), a revision of the FTQ (Heatherton et al., 1991), among other scales. The FTND is one of the most commonly employed measures of nicotine dependence in smoking intervention trials, epidemiological studies, and genetic studies. Scale items assess current smoking level, as well as other time- and place-dependent characteristics of the respondent’s smoking behavior. Scores on this six-item scale range from 0 to 10, with higher scores considered indicative of greater nicotine dependence. The additive genetic component of nicotine dependence defined by the FTND has been estimated at 67%, 75% and 40% among American, Dutch and Finnish twins, respectively (Broms et al., 2007; Maes et al., 2004; Vink et al., 2005). We chose to use the FTND in this study due to the scale’s established estimates of heritability and its broad and continued use in the field (Piper et al., 2006).
A decade of linkage analyses of smoking behaviors continues to inform and motivate candidate gene studies in the same family based samples (Li, 2008). Through a combination of genome wide association scans (GWAS) and candidate gene studies, nicotinic receptor candidate genes currently are the leading candidates for influencing responses to cigarette smoking, cigarette smoking quantity and nicotine dependence (Li and Burmeister, 2009). Other candidate gene families, e.g., GABAergic (Agrawal et al., 2008; Beuten et al., 2005; Lou et al., 2007b; Saccone et al., 2007), cell adhesion molecule (Bierut et al., 2007; Gelernter et al., 2006; Uhl et al., 2007; Vink et al., 2009), dopaminergic (Caporaso et al., 2009; Li and Burmeister, 2009; McKinney et al., 2000; Morton et al., 2006), transient receptor potential (Bierut et al., 2007; Caporaso et al., 2009; Saccone et al., 2007; Vink et al., 2009) and nicotine metabolic (Caporaso et al., 2009; Malaiyandi et al., 2006; Saccone et al., 2007) candidate gene families, have also been identified in GWAS and candidate gene studies of smoking behaviors.
A candidate gene SNP genotyping panel focused primarily on neuronal pre-synaptic nicotinic receptors and the dopaminergic pathway was developed to test candidate gene association to nicotine addiction and treatment outcomes by the Pharmacogenetics of Nicotine Addiction and Treatment consortium (Conti et al., 2008), a member of the Pharmacogenetics Research Network (Giacomini et al., 2007). We utilized SNP genotypes from this panel and baseline FTND scores from treatment seeking smokers from two randomized clinical trials (RCTs) of smoking cessation pharmacotherapy, one RCT that randomized participants to bupropion and placebo and one RCT that randomized participants to two forms of nicotine replacement therapy (Lerman et al., 2006), to rank SNPs associated with nicotine dependence. Association analyses of SNPs to smoking cessation outcomes have been performed in these two RCTs previously, e.g., significant association of an insertion deletion SNP at the dopamine D2 receptor gene (DRD2) to abstinence has been reported (Lerman et al., 2006). A community ascertained, multiplex ever smoker pedigree cohort was genotyped using the same SNP panel and this genotype data was used to evaluate the significance of association of the top-ranked SNP association findings from the treatment seeking cohort in a second sample. The multiplex ever smoker pedigree cohort consisted of pedigrees with at least three first degree ever-smoking relatives with ever smoking defined as ≥ 100 cigarettes smoked lifetime (Swan et al., 2003). Herein we report the ranking of candidate gene SNP associations to baseline FTND score in a cohort of self-identified white treatment seeking smokers, and the significance of association of top-ranked SNPs to FTND in a second cohort of smokers from a cohort of multiplex ever smoker pedigrees.
PARTICIPANTS and METHODS
Treatment seeking smoker cohort
Current daily smokers were recruited for participation into two RCTs of smoking cessation, one evaluating bupropion versus placebo and another evaluating two modalities of administration of nicotine replacement therapy, as previously described (Lerman et al., 2006). Study sites include Washington, DC, Buffalo, NY and Philadelphia, PA. Inclusion criteria included age ≥ 18 and a recent smoking history of ≥ 10 cigarettes per day, while exclusion criteria included pregnancy or lactation, uncontrolled hypertension, unstable angina, heart attack or stroke within the past 6 months, current treatment or recent diagnosis of cancer, drug or alcohol dependence, current diagnosis or history of a psychiatric disorder, seizure disorder, and current use of bupropion or nicotine-containing products other than cigarettes. Participants were assessed for eligibility, completed a baseline assessment, provided a venous blood sample, were randomized to treatment, and attended at least one clinical visit. In conjunction with the baseline assessment, participants provided demographic information including the Center for Epidemiologic Studies scale for depression symptoms (Radloff, 1977), and completed the FTND. Both genomic DNA and whole genome amplified DNA samples were available from participants in the treatment seeking smoker cohort, and was genotyped using the candidate gene panel as described below and by Conti et al. (Conti et al., 2008).
Multiplex ever smoker pedigree cohort
A community ascertained multiplex ever smoker pedigree cohort was selected to evaluate SNPs that were top-ranked in the treatment seeking smoking cohort. These pedigrees were recruited from a longitudinal study that originally recruited N=734 probands 11 to 15 years of age, their parents and siblings, and assessed longitudinal phenotypes of tobacco use (Hops et al., 2000). Pedigrees with three first degree relatives with a smoking history of ≥ 100 cigarettes smoked were selected for recruitment for an integrated study of the genetics of smoking behavior and nicotine metabolism after administration of a family history of tobacco use questionnaire (Swan et al., 2003). After identification and recruitment of probands and family members, a detailed baseline assessment was performed that included sociodemographics, cigarette smoking history, and the FTND at the period of greatest smoking intensity. This analysis utilized smoking history questionnaire data and genomic DNA from N=175 pedigrees. Genomic DNA was extracted from venous whole blood using a standard salt-based precipitation method (Miller et al., 1988), and DNA was genotyped using the same SNP panel as described (Conti et al., 2008).
Candidate gene and SNP selection
Candidate genes for the association analysis of nicotine dependence and for the pharmacogenetic analysis of smoking cessation trials were selected from the presynaptic neuronal nicotinic acetylcholine receptor genes and dopaminergic receptor genes whose proteins mediate the nicotine-stimulated release of dopamine within the mesolimbic system, corpus striatum and prefrontal cortex of the brain and self-administration in animal models of nicotine addiction (Benowitz, 2008). SNP coverage of candidate genes, i.e., the mean (SD) proportion of HapMap Phase II SNPs tagged at an r2 of > 0.80 by the candidate gene genotyping panel SNPs, is 0.92 (0.10). Additional details on the candidate gene and SNP selection process are provided by Conti et al. (Conti et al., 2008).
Analysis of the treatment seeking smoker cohort
As described in Conti et al., genotyping of DNA samples from the RCT participants was performed using the GoldenGate™ assay (Illumina, San Diego, CA), with quality control procedures that included automated sample handling protocols and the inclusion of replicate DNA samples to aid in identifying genotyping errors (Conti et al., 2008). The mean genotype concordance rate across 14 replicate DNA samples was 99.0%. As described in Conti et al., 41 SNPs with a genotype call rate of zero and 57 SNPs with a minor allele frequency < 1% from 1528 SNPs (232 ancestry informative markers on the panel), were removed from the analysis dataset. Of the remaining 1430 SNPs, 95% had genotype completion rates of > 97%. Individual call rates were calculated using the entire multiethnic sample (N=1216) and after stratifying by DNA type, i.e. genomic (gDNA) and whole genome amplified (wgaDNA) DNA. Individuals with a call rate less than 90% were removed from the analysis (N=48) for a total of 1091 individuals with gDNA and 90 individuals with wgaDNA. While there was a substantial difference between the call rate distributions for the gDNA samples (99.6% of individuals with a call rate ≥ 95%) in comparison to the wgaDNA samples (85.3% of individuals with a call rate ≥ 95%), inspection of Illumina data for SNPs with drastically discordant SNP call rates between the two DNA types revealed reliable genotyping calls for SNPs with a SNP call rate calculated using only wgaDNA samples of > 80% (Conti et al., 2008). Thus, for 51 SNPs in which the SNP call rate was < 80% for wgaDNA samples, we only report analyses using the individuals with gDNA samples (Conti et al., 2008). Using ancestry informative markers included in the genotyping panel, Conti et al. demonstrated little individual admixture within each self-identified ethnic group in the treatment seeking smoker cohort (Conti et al., 2008). We report the results of association analysis to covariate adjusted FTND with 1123 SNPs at 55 autosomal candidate genes (51 contiguous candidate gene regions) in N=821 self-identified white treatment seeking smokers. We choose to limit our reporting to results from autosomal genotypes in order to model SNP association to FTND using a single multivariate model, removing 30 SNPs at three X chromosome candidate genes (FLNA, MAOA and MAOB) from the available genotype dataset. We choose to limit our reporting of analyses to self-identified white participants of the RCTs because of the potential for differential linkage disequilibrium across ethnic groups that might lead to heterogeneity of effect estimates (Conti et al., 2008).
FTND SNP association analysis was based on a generalized linear model (GLM), treating FTND as a quantitative variable, with additive or dominant genetic models, and covariates age, self-reported depression symptoms, education, sex, and site. Covariates included in the GLM were significantly associated (univariate P < 0.05) with baseline FTND scores. A GLM including the FTND and covariates was estimated as the base model, and then two GLMs for each SNP using dominant and additive models were estimated. A 1 degree of freedom likelihood ratio test was used to rank SNP GLM results where the P value (PLRT) of the more significant GLM (additive or dominant) is reported. We performed multiple test correction to account for the correlated tests from the estimation of two GLMs (additive and dominant) for each SNP and for the multiple correlated tests from the SNPs within each gene (Conneely and Boehnke, 2007). We report this adjusted P value for correlated tests as PACT, and selected SNPs with PACT < 0.10 as noteworthy SNPs for further genetic, bioinformatic and literature based analyses. We evaluated the distribution of FTND scores by SNP genotype via analysis of variance in post-hoc analyses of noteworthy SNPs. Genetic interaction was modeled using GMDR (Lou et al., 2007a). Power analyses were performed using Quanto (Gauderman, 2002). Statistical analyses were performed using SAS (SAS for Windows, version 9.1, SAS Institute, Cary, NC) and R (Team, 2003).
Analysis of the multiplex ever smoker pedigree cohort
We evaluated association between SNPs and the FTND in the multiplex ever smoker pedigree cohort previously identified as associated with the FTND in the treatment seeking smoker cohort. FTND data were available on N=425 individual ever smokers (≥ 100 cigarettes lifetime) from N=175 nuclear pedigrees. We treated the FTND as an ordinal variable and calculated a covariate-adjusted FTND score using logistic regression (Wang et al., 2006), and utilized this score as the quantitative trait for family based association testing (FBAT) (Schneiter et al., 2005). 16 genotypes inconsistent with Mendelian transmission were deleted prior to FBAT. We performed FBATs using the genetic models suggested by the post-hoc ANOVA analyses of SNP effects on FTND in the treatment seeking smoker cohort, as well as alternative genetic models, i.e., dominant, additive and genotype models, but not recessive models. We did not perform multiple test correction for multiple models or multiple SNPs tested in the FBAT analyses. Genetic interaction of SLC6A3 and NR4A2 SNPs in pedigrees was modeled using PGMDR (Lou et al., 2008).
Genomic annotation and visualization
We evaluated the physical location of SNPs in relationship to NCBI Build 35 genomic annotation with Genewindow (Staats et al., 2005), with the UCSC Genome Browser (Hinrichs et al., 2006) and linkage disequilibrium (LD) among SNPs with Haploview 4.0 (Barrett et al., 2005), using genotype data from the treatment seeking smoker cohort or from the CEU HapMap sample (Consortium, 2005).
RESULTS
FTND, covariates and SNPs in the treatment seeking smoker cohort
The treatment seeking smoker cohort consists of 828 self-identified white treatment seeking smokers, with a mean (SD) baseline FTND score of 5.3 (2.1) (Table 1). Participants are about equally divided between college graduates and non-graduates, women and men, and recruitment site. Older age, a higher depression symptom score, lower educational achievement, male sex and two of three sites are significantly associated with higher FTND scores in this cohort (Table 1). The multivariate model indicates that these covariates account for 10% of the variance of FTND in the treatment seeking smoker cohort (F827=18.01, P<0.0001, r2=0.0987).
Table 1.
Treatment Seeking Smoker Cohort Covariate Associations with the FTND
| Variable | N | FTND M (SD) | r, t or F1 | P2 | t3 | P4 |
|---|---|---|---|---|---|---|
| Age5 | 828 | 5.3 (2.1) | 0.19 | <0.0001 | 5.84 | <0.0001 |
| Depressive symptoms6 | 828 | 5.3 (2.1) | 0.09 | 0.008 | 2.67 | 0.0077 |
| Education | ||||||
| ≥16 years | 410 | 4.9 (2.2) | 5.58 | <0.0001 | 4.37 | <0.0001 |
| <16 years | 418 | 5.7 (2.1) | ||||
| Sex | ||||||
| Female | 420 | 5.1 (2.2) | 3.03 | 0.003 | 3.47 | 0.0006 |
| Male | 408 | 5.6 (2.2) | ||||
| Site | ||||||
| Buffalo | 260 | 5.5 (2.1) | 8.51 | 0.0002 | 2.89 | 0.0039 |
| Philadelphia | 258 | 5.6 (2.1) | ||||
| Washington | 310 | 4.9 (2.2) | ||||
Univariate association estimate.
Univariate P.
Multivariate association estimate.
Multivariate P.
Mean (SD) age is 45.5 (11.5) years.
Mean (SD) CESD score is 11.8 (8.5).
73 SNPs at 22 candidate genes were identified in the treatment seeking smoker cohort with a PLRT value < 0.05 (see Table S1 for all SNP GLM results). Five SNPs at three genes (DRD2, SLC6A3 and NR4A2) were identified with a PACT < 0.10 (Table 2). These SNPs were evaluated in detail using genomic annotation, literature review and genotyping in a second cohort of multiplex ever smoker pedigrees.
Table 2.
Top Ranked SNPs Associated with the FTND in the Treatment Seeking Smoker Cohort
| SNP | Gene | Genomic1 | Major2 | Minor3 | MAF4 | Model5 | Beta6 | SE7 | PLRT8 | PACT9 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs10891552 | DRD2 | IVS1+12127 | A | T | 0.04 | Add | 0.87 | 0.27 | 0.0014 | 0.037 |
| rs2975226 | SLC6A3 | −2303 | T | A | 0.39 | Add | 0.31 | 0.10 | 0.0029 | 0.057 |
| rs2652510 | SLC6A3 | −4547 | T | C | 0.40 | Dom | 0.45 | 0.15 | 0.0026 | 0.081 |
| rs834829 | NR4A2 | −14150 | G | A | 0.29 | Dom | 0.37 | 0.15 | 0.0117 | 0.091 |
| rs2652511 | SLC6A3 | −3076 | G | A | 0.39 | Dom | 0.44 | 0.15 | 0.0033 | 0.097 |
Genomic, location of SNP relative to gene.
Major, major or referent allele.
Minor or variant allele.
MAF, minor allele frequency.
Models are Additive (Add) and Dominant (Dom).
Coefficient of genotype term of GLM model. A positive value implies association of the minor allele with increased FTND.
SE, standard error of Beta.
Likelihood ratio test P value.
P value adjusted for correlated tests.
The top-ranked DRD2 SNP rs10891552 is associated with a ~0.9 unit increase of FTND score and with ~1.3% of FTND variance in an additive or dominant model, as, in this sample, the minor allele homozygote is not observed (Table 3). rs10891552 is located in a linkage disequilibrium (LD) block within the large first intervening sequence (IVS1), but exhibits only weak LD (r2 < 0.23) with SNPs in the local LD block in this sample (Figure S1). There are three additional DRD2 SNPs (rs2440390, rs4586205 and rs1076562) with PLRT values <0.05 (Table S1), located an average of 37 kilo base pairs (kbp) 3’ of rs10891552 within IVS1 of DRD2, and within a LD block that spans IVS1 through the coding region of DRD2, but these SNPs have PACT values ~ 0.6 (Table S1), and rs10891552 has an r2 < 0.01 with these SNPs (Figure S1).
Table 3.
Unadjusted FTND Distributions by Genotype in the Treatment Seeking Smoker Cohort
| SNP | Major | Mean (SD) | Heterozygote | Mean (SD) | Minor | Mean (SD) | PF1 | r2 |
|---|---|---|---|---|---|---|---|---|
| rs10891552 | 754 | 5.27 (2.18) | 64 | 6.20 (2.10) | - | - | 0.001 | 1.32 |
| rs2975226 | 289 | 5.02 (2.20) | 345 | 5.49 (2.15) | 123 | 5.65 (2.22) | 0.006 | 1.32 |
| rs2652510 | 296 | 5.19 (2.21) | 377 | 5.47 (2.15) | 132 | 5.59 (2.19) | 0.030 | 0.87 |
| rs834829 | 405 | 5.18 (2.12) | 334 | 5.58 (2.23) | 69 | 5.12 (2.29) | 0.010 | 0.82 |
| rs2652511 | 300 | 5.06 (2.21) | 377 | 5.49 (2.13) | 124 | 5.59 (2.20) | 0.016 | 1.04 |
Genetic model of ANOVA is dominant or additive for rs10891552, heterotic for rs834829, and additive for SLC6A3 SNPs. The genotype(s) associated with increased FTND scores can be inferred by inspection.
The top-ranked three SLC6A3 SNPs (rs2975226, rs2652510 and rs2652511) identified as associated with FTND (Table 2) are highly correlated with each other (mean r2 ~ 0.97) in a LD block that extends in this sample from rs12652860, 61 kbp 5’ of SLC6A3 to rs6350 in exon 2 of SLC6A3, and are only modestly correlated (r2~0.2) with SNPs in two 3’ LD blocks extending from IVS1 through IVS7 (Figure 1). SNP FTND association model characteristics are similar for the three SLC6A3 SNPs, i.e., for each of these SNPs, the presence of the minor allele is associated with a ~0.5 unit increase of FTND score and with ~1% of FTND variance in the additive model (Table 3). Inspection of all SLC6A3 SNP FTND association results (Table S1) identifies rs12652860, also in the 5’ linkage block, with a PLRT < 0.01 and a PACT = 0.20, and with similar effects on FTND score and variance (data not shown), while the remaining SLC6A3 SNPs exhibit PLRT values > 0.15 and PACT values > 0.95 (Table S1). Estimation of haplotypes in the 5’ linkage block results in five haplotypes with frequency > 0.01, with the minor allele at each of the three top-ranked SLC6A3 SNPs associated with increased FTND found in three haplotypes with combined frequency 39% (Figure 1).
Figure 1.
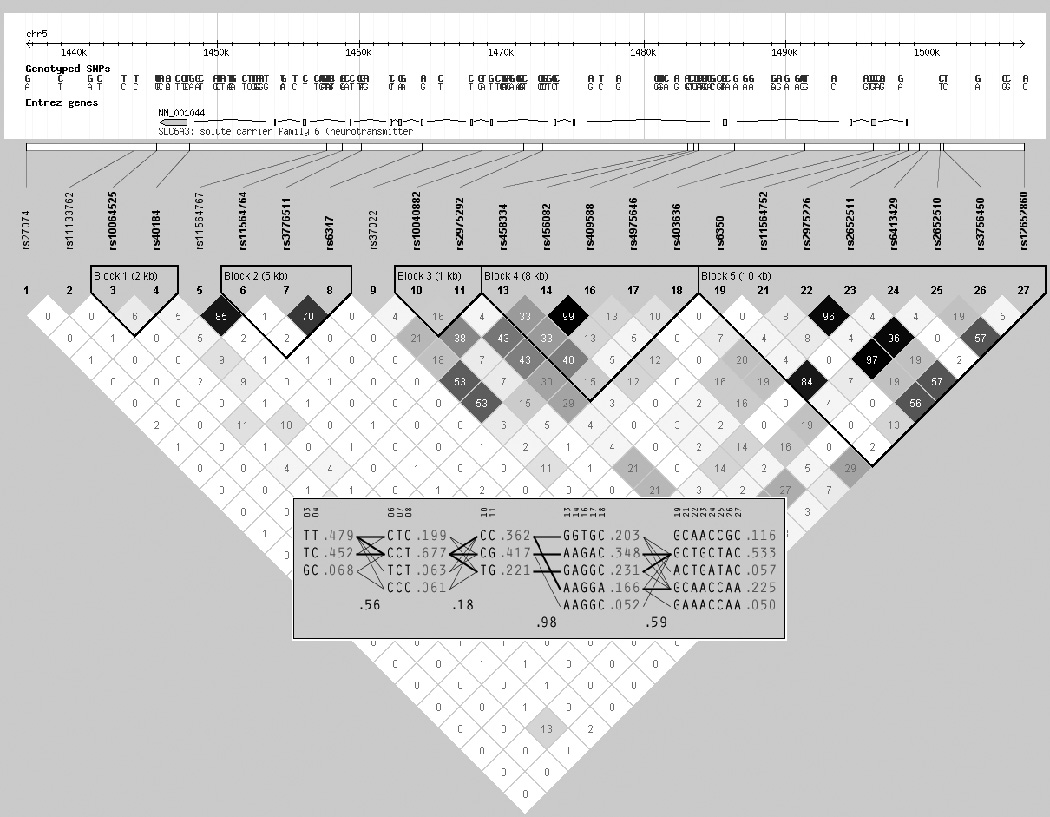
SLC6A3 SNPs, LD and haplotypes, treatment seeking smoker cohort.
The top-ranked NR4A2 SNP rs834829 was identified in the initial association analysis as most significantly associated with a dominant model (Table 2), and while both additive and dominant models are significantly associated with increased FTND score, inspection of FTND distributions by genotype in post-hoc analysis identifies the heterozgote genotype as significantly associated with increased FTND scores, accounting for a ~0.4 unit increase of mean FTND score and ~0.8% of FTND variance (Table 3). The NR4A2 SNP associated with FTND is found ~12 kbp 5’ of the 5’ UTR of the NR4A2 gene, within a LD block that extends from 5’ of the gene, across the entire gene to the 3’ flanking region in this sample (Figure S2). Inspection of all NR4A2 SNP FTND association results (Table S1) identifies rs1150144 and rs1150143, ~2.5 kbp 3’ of the 3’UTR, with PLRT values < 0.05, strong r2 with rs834829 (0.79 and 0.94, respectively) and modest PACT values (0.12 and 0.21, respectively).
In post-hoc power analyses, the sample size of 821 treatment seeking smokers has power of 82% to detect beta coefficients of 78%, 42% and 43% for minor allele frequencies of 4% (DRD2), 29% (NR4A2) and 39% (SLC6A3), respectively, for genetic effects in a dominant model with an r2 of 0.01 and the observed FTND mean and standard deviation (Table 1) at an alpha value of 0.05 with a two-sided test.
SLC6A3 NR4A2 interaction analysis
We evaluated gene-gene interaction between rs2652511 at SLC6A3 and rs834829 at NR4A2, using a continuous model of FTND score, adjusted for age, depressive symptoms, education, sex and site in a sample of N=794 individuals with complete FTND, covariate and genotype data. We chose these two SNPs to perform the interaction analysis because the gene product of NR4A2, NURR1, is known to be required for SLC6A3 gene transcription (see Discussion), and because these two SNPs are highly ranked in our GLM results (Table 2). We evaluated training (prediction) accuracy (TA) of three GMDR analyses to predict the distribution of covariate adjusted normalized FTND score within genotype classes for rs2652511, for rs834829, and for both rs2652511 and rs834829 (Figure S2). While each SNP independently predicted FTND score (for rs2652511, TA=0.56, TA Odds Ratio (OR)=1.71, 95CI 1.37–2.14, P<0.0001, cross validation sign test P=0.02; for rs834829, TA=0.56, TA OR=1.64, 95 CI 1.31–2.04, P<0.0001, cross validation sign test P=0.83), only the analysis with both rs2652511 and rs834829 resulted in a highly significant cross validation sign test, in addition to increased training accuracy and odds ratio (TA=0.59, TA OR=2.15, 95CI 1.72–2.70, P<0.0001, cross validation sign test P=0.001).
Multiplex smoking pedigree cohort analysis
FTND score, age, and the proportion of individuals with 16 or more years of education were significantly lower in the multiplex smoking pedigree cohort than in the treatment seeking smoker cohort (all P < 0.0001). Age, depressive symptoms, education and sex exhibited significant association with FTND in the multiplex smoking pedigree cohort in the same direction as in the treatment seeking smoker cohort (Table 4). The multivariate model of covariate association with FTND indicates that these covariates account for 15% of the variance of FTND in the multiplex smoking pedigree cohort (F405=18.21, P<0.0001, r2=0.1537). With one exception (rs834829), each of the five top-ranked SNPs in the treatment seeking smoker cohort (Table 2) exhibited nominally significant (P < 0.05) association with FTND in the multiplex smoking pedigree cohort in FBAT analyses uncorrected for multiple testing (Table 5). For rs10891552 (DRD2), the minor allele (in additive, dominant or genotype models) was associated with increased FTND. For the SLC6A3 SNPs, the minor allele (in the genotype model) was associated with increased FTND. For rs834829, the FBAT with the smallest P value was the heterozygote genotype model (P = 0.0511). We evaluated statistical interaction between rs2652511 at SLC6A3 and rs834829 at NR4A2 in 411 individuals with FTND scores from N=175 pedigrees using PGMDR (Figure S3). The PGMDR analysis with both rs2652511 and rs834829 had the greatest training (prediction) accuracy, but did not exhibit a significant cross validation sign test P value (TA=0.56, TA OR=1.65, 95 CI 1.24, 2.21, P=0.0006, cross validation sign test P=0.82). The increase in TA over the PGMDR analysis of rs2652511 (TA=0.54, TA OR= 1.66, 95 CI 1.15, 2.40, P=0.0063, cross validation sign test P=0.38) was modest. The PGMDR analysis of rs834829 had a non-significant training accuracy estimate and OR (data not shown).
Table 4.
Multiplex Ever Smoker Pedigree Cohort Covariate Associations with the FTND
| Variable | N | FTND M (SD) | r or t1 | P2 | t3 | P4 |
|---|---|---|---|---|---|---|
| Age5 | 423 | 3.9 (2.7) | 0.24 | <0.001 | 6.87 | <0.0001 |
| Depressive symptoms6 | 410 | 3.9 (2.7) | 0.11 | 0.022 | 3.65 | 0.0003 |
| Education | ||||||
| ≥16 years | 110 | 3.3 (3.0) | 3.07 | 0.002 | 4.93 | <0.0001 |
| <16 years | 312 | 4.2 (2.5) | ||||
| Sex | ||||||
| Female | 219 | 3.6 (2.7) | 2.43 | 0.015 | 2.83 | 0.0048 |
| Male | 206 | 4.3 (2.6) | ||||
Univariate association estimate.
Univariate P.
Multivariate association estimate.
Multivariate P.
Mean (SD) 39.4 (13.6).
Mean (SD) 11.9 (9.4)
Table 5.
FBAT of the FTND in the Multiplex Ever Smoker Pedigree Cohort
| SNP | Gene | Modell | Frequency2 | Pedigrees3 | Z4 | PFBAT5 |
|---|---|---|---|---|---|---|
| rs10891552 | DRD2 | A/D & G | 0.11 & 0.21 | 15 | 2.301 | 0.0214 |
| rs2975226 | SLC6A3 | G | 0.17 | 42 | 3.065 | 0.0022 |
| rs2652510 | SLC6A3 | G | 0.11 | 47 | 3.059 | 0.0022 |
| rs834829 | NR4A2 | G | 0.32 | 67 | 1.951 | 0.0511 |
| rs2652511 | SLC6A3 | G | 0.10 | 42 | 2.856 | 0.0043 |
FBAT genetic model: A=Additive, D=Dominant, G=Genotype;
allele or genotype frequency of minor allele model tested;
number of informative pedigrees;
normalized Z score, (S-E(S))/Var(S)), where a positive Z implies association of the genotype with a higher FTND score;
P value of FBAT.
DISCUSSION
This analysis ranked 1123 SNPs at 55 autosomal candidate genes for significance of association to baseline FTND score in 821 self-identified white treatment seeking smokers. We observed five SNPs assigned to three candidate genes with P values adjusted for correlated tests of < 0.10. We focused our subsequent analyses on these five SNPs to avoid the post-hoc exercise of going deeper into the list of ranked SNPs in search of biological coherence. These top-ranked SNPs were found within dopamine candidate genes, with a single association finding each at a dopamine receptor involved in intracellular signaling through multiple pathways (Bonci and Hopf, 2005), at the dopamine transporter, which regulates extracellular dopamine concentration by reuptake of dopamine (Sotnikova et al., 2006), and at an essential regulator of dopaminergic gene transcription and dopaminergic neuron development (Jankovic et al., 2005). The panel of candidate genes was developed using nicotinic acetylcholine receptor and dopaminergic candidate genes, and thus observing top-ranked SNPs at dopaminergic candidate genes is not unexpected.
However, our analysis results do raise the question of why a nicotinic acetylcholine receptor (nACHR) SNP was not identified within the small group of SNPs that met our threshold for follow-up analyses. In our analyses of the treatment seeking smoker cohort, we tested 121 SNPs at nine nACHR genes for association with FTND (Table S1). Three of these nACHR SNPs (CHRNB4 rs3971872 and CHRNA2 rs748283 and rs4733065, all several kbp 3’ of their assigned gene) exhibited unadjusted P values < 0.05, but adjusted P values were > 0.10. We did not observe significant association of two CHRNB2 SNPs with FTND in this analysis that were previously identified by Conti et al. in an age, gender and FTND score adjusted analysis of associated with abstinence in ~ half of the treatment seeking smoker cohort investigated here (Conti et al., 2008). FTND is associated with abstinence in the cohort of treatment seeking smokers analyzed here (data not shown), however, this analysis is focused on baseline nicotine dependence as measured by the FTND in treatment seeking smokers of two RCTs (Lerman et al., 2006).
Based on recent association studies of nicotinic receptor SNPs and nicotine dependence, the lack of nACHR SNPs among the top-ranked SNPs in this study may be assumed to be the result of a lack of statistical power and/or differences between the treatment seeking smokers in our study and the smokers in other recent population based and clinically based studies. Our treatment seeking smoker cohort was ~19 and ~13 fold smaller than two GWAS studies that identified the chr15q25.1 nACHR locus as associated with heavy smoking (≥ 25 cigarettes per day (CPD) versus < 5 CPD) in population based and clinical based samples originally recruited for other genetic studies (Berrettini et al., 2008), and with cigarettes smoked per day in smokers originally recruited for other genetic studies (Thorgeirsson et al., 2008). Our treatment seeking smoker cohort was ~3.4 and ~2.3 fold smaller than two candidate gene association studies of the chr15q25.1 nACHR locus and FTND that characterized risk and protective haplotypes in three cohorts of current and former smokers, one of which was composed of treatment seeking smokers (Weiss et al., 2008), and a study that evaluated association of SNPs at 348 candidate genes, including 40 of the candidate genes investigated in this study, to a population based sample of non-dependent and dependent ever smokers (Saccone et al., 2007). While our treatment seeking smoker sample was much smaller than prior studies, our noteworthy SNPs at dopaminergic candidate genes accounted for ~1% of FTND variance. Thorgeirsson et al. (Thorgeirsson et al., 2008) estimated that ~0.7% of the variance of cigarettes smoked per day is explained by the chr15q25.1 variant (rs1051730) identified in the Icelandic population sample of 10,995 smokers used in their study. rs1051730 was tested in our treatment seeking smoker cohort for association with FTND with non-significant unadjusted and adjusted P values (0.47 and 0.99, respectively). Finally, our sample of treatment seeking smokers exhibited a normal distribution of FTND scores, rather than a dichotomized distribution utilized by some investigators, i.e., selected for extremes of the CPD or nicotine dependence distributions from a larger population or clinically based sample. Our results and the ascertainment differences between our treatment seeking cohort and previously studied cohorts raise the possibility that dopaminergic candidate genes may exert a greater influence on nicotine dependence among treatment seeking smokers with average nicotine dependence scores than do the chr15q25.1 nicotinic receptor candidate genes. Additional recent GWAS in population based and clinically ascertained populations have identified a wide variety of candidate genes as significantly associated with smoking behaviors in addition to nicotinic receptor candidates (Bierut et al., 2007; Caporaso et al., 2009; Uhl et al., 2007; Uhl et al., 2008; Vink et al., 2009), suggesting that many factors, including sample ascertainment, sample size, genotyping platforms and analyses approaches, may influence the ranking of specific candidate genes or candidate gene families in association analyses of smoking behaviors.
We evaluated association of dopaminergic SNPs identified in the treatment seeking smoker cohort in an independent cohort of non-treatment seeking ever smokers from multiplex ever smoking pedigrees that were significantly younger, with significantly less education and with significantly lower FTND scores than the individuals in the treatment seeking smoker cohort. We also evaluated the biological coherence of these findings by performing interaction analyses among SNPs from two genes with a priori evidence for biological interaction, which revealed a significant SLC6A3 NR4A2 interaction in the treatment seeking smoker cohort, although we failed to observe a significant gene-gene interaction in the multiplex smoker pedigree cohort. The sample size of the multiplex ever smoker pedigree cohort, about half that of the treatment seeking smoker cohort, may have limited our ability to observe statistically significant interaction in these pedigrees. Below, we attempt to place our top-ranked association findings of DRD2, SLC6A3 and NR4A2 SNPs to the FTND in the context of the nicotine dependence susceptibility candidate gene association literature.
DRD2
This study has identified one DRD2 SNP (rs10891552) among 52 SNPs tested at the ANKK1 and DRD2 loci as worthy of followup evaluation (SNPs from the two regions were tested together because SNPs at these genes are in LD). There are three extant studies that have evaluated association with nicotine dependence measures and DRD2 SNPs (Gelernter et al., 2006; Huang et al., 2008; Saccone et al., 2007). Saccone et al. tested 41 SNPs at the two genes for association with nicotine dependence and rs4245150, 69 kbp 5’ of DRD2, was identified as significant with a FDR < 0.40 level (Saccone et al., 2007). Huang et al., 2008 (Huang et al., 2008) tested 23 SNPs at the two genes for association with nicotine dependence in the Mid South Tobacco Family sample and identified one ANKK1 SNP (rs2734849, Ex8+475, H490R) associated with nicotine dependence in African-American pedigrees after multiple test correction. Gelernter et al. (Gelernter et al., 2006) tested 43 SNPs at these two genes and two additional flanking genes (TTC12 and NCAM1) for association to both DSM-IV and FTND nicotine dependence and identified one, four and five SNPs that exhibited significant association to DSM-IV nicotine dependence in African-American pedigrees, European-American pedigrees and both sets of pedigrees combined, however, no SNPs were considered to be significantly associated to FTND after multiple test correction. Inspection of r2 in the HapMap CEU sample between the SNP associated with FTND in this study and SNPs found associated with nicotine dependence in Saccone et al., Huang et al., and Gelernter et al., identified no substantial LD (r2≤ 0.01 for six such SNPs). PACT values for SNPs declared significantly associated with nicotine dependence by Huang et al. (Huang et al., 2008) and Gelernter et al. (Gelernter et al., 2006) were non-significant (for five such SNPs, PACT values > 0.98). rs10891552 does not appear to be in LD with flanking DRD2 SNPs (Figure 1), or with DRD2 SNPs associated with nicotine dependence in other studies (Gelernter et al., 2006; Huang et al., 2008; Saccone et al., 2007), and those other SNPs do not exhibit association in this study. The association of rs10891552 in the cohort of treatment seeking smokers thus represents a novel finding of association of a DRD2 SNP to nicotine dependence and the cohort of multiplex smoking pedigrees provides nominal support for this association in a second sample.
SLC6A3
There are three published studies investigating SLC6A3 SNPs for association with nicotine dependence (Ling et al., 2004; Saccone et al., 2007; Segman et al., 2007). Ling et al. (Ling et al., 2004) evaluated association of a 3’ UTR SNP (rs27072) with nicotine dependence and age of smoking onset in a sample (668 individuals, 99% male, from 253 sibships) of severely nicotine dependent smokers (mean (SD) FTND = 8.1 (2.1)). Ling et al. did not identify a significant association with nicotine dependence, but did identify significant association with smoking onset ≤ 18 years of age, in the subsample of severely dependent smokers with a FTND score ≥ 8 (Ling et al., 2004). Segman et al. evaluated association of four SLC6A3 SNPs (located in the promoter (rs2652510), IVS1, Exon 9, and Exon 15) and the 3’ VNTR polymorphism, located in exon 15, to smoking initiation and to dichotomized nicotine dependence in a sample of 390 female undergraduate students, 242 of whom who had smoked daily for at least one year and 148 of whom had never smoked (Segman et al., 2007). The 242 daily smokers were dichotomized into a group with FTQ scores of ≥ 6 (N=127) and a group with scores ≤ 4 (N=115). After multiple test correction, Segman et al. did not observe significant association between any SNP and either smoking initiation or nicotine dependence. Segman et al. did identify significant association to both phenotypes with a haplotype composed of the 3’ VNTR and a SNP in the same (15th) exon (Segman et al., 2007). Saccone et al. tested five SLC6A3 SNPs located in the 3’UTR (3 SNPs), exon 12, and exon 13; none of these SNPs were associated with dichotomized FTND (Saccone et al., 2007).
In contrast to these two studies evaluating association between SLC6A3 SNPs and nicotine dependence, there have been many studies focused on assessing association between a 3’ polymorphic variable number of tandem repeat (VNTR) locus found in exon 15 of the SLC6A3 gene (Vandenbergh et al., 1992) and smoking behaviors, including smoking status (Jorm et al., 2000; Lerman et al., 1999; Sabol et al., 1999; Segman et al., 2007; Timberlake et al., 2006; Vandenbergh et al., 2002), adolescent smoking progression (Audrain-McGovern et al., 2004; Laucht et al., 2008), smoking cessation (David et al., 2007; David and Munafo, 2008; Han et al., 2008; Lerman et al., 2003; O'Gara et al., 2007; Styn et al., 2008; Swan et al., 2007; Ton et al., 2007), and affective, behavioral and brain responses to smoking (Brody et al., 2006; Erblich et al., 2004; Erblich et al., 2005; Franklin et al., 2008; Newberg et al., 2007; Perkins et al., 2008; Yang et al., 2008). Only one of these studies has evaluated a sample of smokers for potential association of the SLC6A3 3’ VNTR with nicotine dependence (Lerman et al., 1999). Lerman et al. evaluated four smoking behaviors in a sample of 289 smokers and 233 controls; association of 3’ VNTR genotypes with FTND in the sample of 289 smokers was not observed (Lerman et al., 1999). Lerman et al. did identify significant association with smoking status in the entire sample, and with age of initiation and quitting history in the subsample of smokers (Lerman et al., 1999).
The SLC6A3 3’ VNTR polymorphism has been associated with dopamine transporter expression differences in vivo via single photon emission computed tomography of the radioligand 123I-2β-carbomethoxy-3β-4(iodophenyl)tropane (β-CIT) for measurement of striatal dopamine transporter binding potential (Heinz et al., 2000; Martinez et al., 2001; van Dyck et al., 2005), and with differences in SLC6A3 gene expression via quantitative polymerase-chain reaction analysis of RNA ex vivo in lymphocytes (Mill et al., 2002) and in post-mortem human brain (Brookes et al., 2007; Mill et al., 2002). The presence of the 9 repeat allele of the 3’ VNTR has generally been associated with reduced dopamine transporter binding potential or reduced levels of SLC6A3 transcripts. However, the SLC6A3 promoter and 5’ UTR region and the 3’ VNTR region (exon 15) are in very modest linkage disequilibrium in the sample investigated in this study (Figure 1), and in other samples (Greenwood and Kelsoe, 2003; Kelada et al., 2005).
SLC6A3 promoter SNPs have been associated with dopamine transporter expression in vivo via [11C] cocaine positron emission tomography (PET) (Drgon et al., 2006), in vitro via expression constructs (Greenwood and Kelsoe, 2003; Kelada et al., 2005; Sacchetti et al., 1999) and ex vivo in post mortem human brain via [3H] carboxyfluorotropane binding studies (Drgon et al., 2006). In studies of promoter sequence constructs, Sacchetti et al. found that a proximal promoter construct that includes rs2975226 drives expression up to 150 times greater than an expression construct without this proximal promoter sequence, while SLC6A4 promoter sequences further 5’ serve to repress transcription (Sacchetti et al., 1999). Greenwood and Kelsoe evaluated promoter constructs that differed by promoter sequence and by allelic content (Greenwood and Kelsoe, 2003). Promoter expression constructs that include the very proximal promoter and 5’UTR sequence, including rs2975226, have greater reporter activity than expression constructs that include more 5’ promoter and/or more 3’ sequence, as observed by Sacchetti et al. (Sacchetti et al., 1999). However, promoter constructs that included the minor allele of common promoter SNPs, e.g., at rs2652510, within the core or extended promoter constructs, exhibited reduced reporter activity in vitro(Greenwood and Kelsoe, 2003). Another analysis of expression constructs of the SLC6A3 promoter and more 3’ sequence spanning 7391 basepairs evaluated transcription levels for six haplotypes that included rs2975226, rs2652511 and rs2652510, as well as other SLC6A3 promoter SNPs (Kelada et al., 2005). Expression constructs containing the minor alleles at these three SNPs in the promoter haplotype exhibited significantly reduced transcription in vitro (Kelada et al., 2005). in vivo [11C] cocaine PET and ex vivo [3H] carboxyfluorotropane binding studies indicate that the minor allele at rs2975226 is associated with reduced dopamine transporter binding (Drgon et al., 2006). Thus, four studies of SLC6A3 promoter SNPs including in vitro, in vivo and ex vivo designs showed significantly reduced expression with the minor allele of rs2975226, rs2652510 and/or rs2652511 (Drgon et al., 2006; Greenwood and Kelsoe, 2003; Kelada et al., 2005; Sacchetti et al., 1999).
NR4A2
There are three published reports associating NR4A2 polymorphisms with substance dependence (Ishiguro et al., 2002; Nielsen et al., 2008; Saccone et al., 2007). Ishiguro et al. identified rs34884856 in the promoter (-2920), and a biallelic CA dinucleotide repeat sequence in the 3’ UTR of the NR4A2 gene and identified a significant haplotypic association with alcohol dependence in a sample of 171 individuals with a DSM-IV diagnosis of alcohol dependence and 161 unscreened individuals (Ishiguro et al., 2002). Nielsen et al. identified rs1405735, 45kbp 5’ of NR4A2, as the fourth ranked candidate gene SNP from an a priori candidate gene genotypic association analysis nested within a case:control GWAS of heroin dependence with 99 individuals meeting Federal criteria for methadone maintenance and 99 individuals screened for past and current history of drug use genotyped for this SNP (Nielsen et al., 2008). Information on the smoking status of cases and controls was not provided by Nielsen et al. and Ishiguro et al. (Ishiguro et al., 2002; Nielsen et al., 2008). Saccone et al. tested two SNPs at NR4A2 (rs12803 and rs834835, located in the 3’ UTR and IVS1, respectively) for association with dichotomized FTND, however, neither exhibited significant association with nicotine dependence (Saccone et al., 2007). The level of LD (r2) in the HapMap CEU sample between rs834829 and rs1405735, rs12803 and rs834835 is 0.12, 0.49 and 1.0, respectively. The NR4A2 SNP (rs34884856) tested by Ishiguro et al. (Ishiguro et al., 2002) was not genotyped in the HapMap project.
NURR1, the protein product of the NR4A2 gene, is an orphan nuclear receptor transcription factor (Law et al., 1992) expressed in dopaminergic neurons (Zetterstrom et al., 1996), required for the development and maintenance of dopaminergic neurons (Saucedo-Cardenas et al., 1998; Zetterstrom et al., 1997), and coexpressed with tyrosine hydroxlase and the dopamine transporter (DAT) within striatal neurons (Cossette et al., 2005). Together with DAT, NURR1 is decreased in expression in dopamine neurons of cocaine (Bannon et al., 2002) and heroin (Horvath et al., 2007) abusers, suggesting that DAT and NURR1 are involved in the adaptation of the striatal dopaminergic system in addiction, as well as in normal development. NURR1 regulates DAT expression using sites in the promoter region ~1000 basespairs 5’ of the transcription start site of SLC6A3 (Sacchetti et al., 1999; Sacchetti et al., 2001). The finding that NR4A2 variation is associated with nicotine, heroin and alcohol dependence is of great interest because it suggests that genetic variation at NURR1 may influence the development and course of substance dependence generally and not just for a single substance (Goldman and Bergen, 1998). The SLC6A3-NR4A2 interaction identified in this study is consistent with what is known regarding the development and maintenance of dopaminergic neurons by NURR1 (Zetterstrom et al., 1997) and the regulation of the SLC6A3 gene by NURR1 (Sacchetti et al., 2001). A challenge now will be to identify the specific biological interaction between the NURR1 protein and the SLC6A3 promoter that increases the risk for nicotine dependence.
Conclusion
In this study, the minor alleles at DRD2 SNP rs10891552, SLC6A3 SNPs rs2975226, rs2652510 and rs2652511, and NR4A2 SNP rs834829 are top-ranked in association with increased FTND scores among both treatment seeking and community smokers. The DRD2 SNP appears to be a novel association with nicotine dependence. There is abundant existing information on the three SLC6A3 promoter SNP SNPs identified in this study demonstrating association with SLC6A3 expression. One hypothesis generated by this study and the extant literature is that the minor alleles of these SLC6A3 promoter SNPs result in increased nicotine dependence severity among smokers via reduced expression of the SLC6A3 gene. Hypotheses on whether SLC6A3 promoter variation influences risk for nicotine dependence among individuals naïve to tobacco smoke, or whether the primary influence is on DAT expression during smoking initiation or during regular smoking, are not addressed by this analysis. Finally, evidence from this study and the existing drug dependence literature suggests that NR4A2 is a candidate susceptibility gene influencing nicotine, alcohol and opioid dependence. Our interaction analysis suggests that some component of this influence occurs through interactive effects with variation at SLC6A3.
The primary limitation of this candidate gene association study with the FTND is the modest sample size of treatment seeking smokers. A larger sample or a similarly sized sample oversampled for extreme FTND scores from a larger sample would have had greater power to detect SNPs associated with FTND (Schork et al., 2000). To limit the reporting of false positives, we used adjustment for testing of additive and dominant models and for testing of multiple SNPs at a candidate gene to rank SNPs in the treatment seeking smoker cohort. This adjustment does not adjust for the number of genes evaluated. We used a second multiplex ever smoker pedigree cohort to evaluate the top-ranked SNPs, although this cohort had substantial demographic and behavioral differences from the treatment seeking smoker cohort, suggesting that its value in this analysis is supportive in nature. With these limitations, noteworthy SNPs associated with FTND in the treatment seeking smoker cohort at three dopminergic genes were associated with FTND in a second multiplex ever smoker pedigree cohort. For SLC6A3 and NR4A2 SNPs, supportive evidence for association to substance dependence is available in the genetic epidemiological literature (Li and Burmeister, 2009; Nielsen et al., 2008), and for the SLC6A3 SNPs, there is abundant evidence for functional effects of these noteworthy SNPs.
Acknowledgements
Supported by U01 DA020830-03, R01 CA63562, P50 CA84718, P50 CA084735, R01 GM069890, R01 DA03706 and 7PT2000-2004 from UC TRDRP. We thank Drs. Faith Allen and Judy Andrews for their contribution to PNAT data curation and to the collection of the multiplex ever smoker pedigree cohort, respectively, and we thank the participants in the treatment seeking smoker and multiplex ever smoker pedigree cohorts.
Footnotes
Disclosure/Conflict of Interest
Related to THIS WORK. Authors Conti, Van Den Berg, Lee, Liu, Li, Guo, Mi, Thomas, Krasnow, He, Nishita, Jiang, McClure, Tildesley, Hops, Tyndale, Benowitz, Lerman, and Swan declare that all financial and material support for this work was provided by their primary employer. In the past three years, Dr. Bergen has been an employee of the National Cancer Institute. In the past three years, Dr. Lessov-Schlaggar has received compensation for research and professional services from SRI International. Related to ALL OTHER COMPENSATION. Authors Van Den Berg, Lee, Liu, Li, Guo, Mi, Thomas, Krasnow, He, Nishita, Jiang, McClure, and Tildesley declare that, except for income provided from their primary employer, no financial support or compensation has been received from any individual or corporate entity over the past three years for research or professional services and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest. In the past three years, Dr. Bergen has been an employee of the National Cancer Institute and has received compensation for professional services from the National Institutes of Health, and from the Price Foundation Ltd. In the past three years, Dr. Conti has been a paid consultant to Pfizer, Inc. In the past three years, Dr. Lessov-Schlaggar has received compensation for research and professional services from SRI International. In the past three years, Dr. Hops has received compensation from the University of Washington for consulting on federally funded projects in adolescent drug use and abuse. In the past three years, Dr. Tyndale has consulted for Novartis, a company that develops and/or markets nicotine dependence medications. Dr. Tyndale is also a shareholder and scientific officer in Nicogen, a company focused on the development of novel smoking cessation treatments. Over the past 3 years Dr. Benowitz has received compensation from pharmaceutical companies marketing or developing smoking cessation medications, including Pfizer, GlaxoSmithKline, Novartis and Aradigm. He has also served as a paid expert witness in litigation against tobacco companies. In the past three years, Dr. Lerman has received compensation and/or research support from GlaxoSmithKline, Pfizer, Astra Zeneca, and Novartis, companies that develop and/or market nicotine dependence medications. In the past three years, Dr. Swan has received compensation from Pfizer.
Supplementary information is available at the Neuropsychopharmacology website.
REFERENCES
- Agrawal A, Pergadia ML, Saccone SF, Hinrichs AL, Lessov-Schlaggar CN, Saccone NL, et al. Gamma-aminobutyric acid receptor genes and nicotine dependence: evidence for association from a case-control study. Addiction. 2008;103:1027–1038. doi: 10.1111/j.1360-0443.2008.02236.x. [DOI] [PubMed] [Google Scholar]
- Audrain-McGovern J, Lerman C, Wileyto EP, Rodriguez D, Shields PG. Interacting effects of genetic predisposition and depression on adolescent smoking progression. Am J Psychiatry. 2004;161:1224–1230. doi: 10.1176/appi.ajp.161.7.1224. [DOI] [PubMed] [Google Scholar]
- Bannon MJ, Pruetz B, Manning-Bog AB, Whitty CJ, Michelhaugh SK, Sacchetti P, et al. Decreased expression of the transcription factor NURR1 in dopamine neurons of cocaine abusers. Proc Natl Acad Sci U S A. 2002;99:6382–6385. doi: 10.1073/pnas.092654299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Clinical pharmacology of nicotine: implications for understanding, preventing, and treating tobacco addiction. Clin Pharmacol Ther. 2008;83:531–541. doi: 10.1038/clpt.2008.3. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, et al. Single- and multilocus allelic variants within the GABA(B) receptor subunit 2 (GABAB2) gene are significantly associated with nicotine dependence. Am J Hum Genet. 2005;76:859–864. doi: 10.1086/429839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci A, Hopf FW. The dopamine D2 receptor: new surprises from an old friend. Neuron. 2005;47:335–338. doi: 10.1016/j.neuron.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Brody AL, Mandelkern MA, Olmstead RE, Scheibal D, Hahn E, Shiraga S, et al. Gene variants of brain dopamine pathways and smoking-induced dopamine release in the ventral caudate/nucleus accumbens. Arch Gen Psychiatry. 2006;63:808–816. doi: 10.1001/archpsyc.63.7.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Madden PA, Heath AC, Pergadia ML, Shiffman S, Kaprio J. The Nicotine Dependence Syndrome Scale in Finnish smokers. Drug Alcohol Depend. 2007;89:42–51. doi: 10.1016/j.drugalcdep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes KJ, Neale BM, Sugden K, Khan N, Asherson P, D'Souza UM. Relationship between VNTR polymorphisms of the human dopamine transporter gene and expression in post-mortem midbrain tissue. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:1070–1078. doi: 10.1002/ajmg.b.30572. [DOI] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS ONE. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Cigarette smoking-attributable morbidity---United States, 2000. MMWR Morb Mortal Wkly Rep. 2003;52:842–844. [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. 2007;81 doi: 10.1086/522036. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, et al. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17:2834–2848. doi: 10.1093/hmg/ddn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossette M, Levesque D, Parent A. Neurochemical characterization of dopaminergic neurons in human striatum. Parkinsonism Relat Disord. 2005;11:277–286. doi: 10.1016/j.parkreldis.2005.02.008. [DOI] [PubMed] [Google Scholar]
- David SP, Brown RA, Papandonatos GD, Kahler CW, Lloyd-Richardson EE, Munafo MR, et al. Pharmacogenetic clinical trial of sustained-release bupropion for smoking cessation. Nicotine Tob Res. 2007;9:821–833. doi: 10.1080/14622200701382033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SP, Munafo MR. Genetic variation in the dopamine pathway and smoking cessation. Pharmacogenomics. 2008;9:1307–1321. doi: 10.2217/14622416.9.9.1307. [DOI] [PubMed] [Google Scholar]
- Drgon T, Lin Z, Wang GJ, Fowler J, Pablo J, Mash DC, et al. Common human 5' dopamine transporter (SLC6A3) haplotypes yield varying expression levels in vivo. Cell Mol Neurobiol. 2006;26:875–889. doi: 10.1007/s10571-006-9014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich J, Lerman C, Self DW, Diaz GA, Bovbjerg DH. Stress-induced cigarette craving: effects of the DRD2 TaqI RFLP and SLC6A3 VNTR polymorphisms. Pharmacogenomics J. 2004;4:102–109. doi: 10.1038/sj.tpj.6500227. [DOI] [PubMed] [Google Scholar]
- Erblich J, Lerman C, Self DW, Diaz GA, Bovbjerg DH. Effects of dopamine D2 receptor (DRD2) and transporter (SLC6A3) polymorphisms on smoking cue-induced cigarette craving among African-American smokers. Mol Psychiatry. 2005;10:407–414. doi: 10.1038/sj.mp.4001588. [DOI] [PubMed] [Google Scholar]
- Ezzati M, Lopez AD. Estimates of global mortality attributable to smoking in 2000. Lancet. 2003;362:847–852. doi: 10.1016/S0140-6736(03)14338-3. [DOI] [PubMed] [Google Scholar]
- Fagerström KO. Measuring degree of physical dependence to tobacco smoking with reference to individualization of treatment. Addict Behav. 1978;3:235–241. doi: 10.1016/0306-4603(78)90024-2. [DOI] [PubMed] [Google Scholar]
- Franklin TR, Lohoff FW, Wang Z, Sciortino N, Harper D, Li Y, et al. DAT genotype modulates brain and behavioral responses elicited by cigarette cues. Neuropsychopharmacology. 2008;34:717–728. doi: 10.1038/npp.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Bergen A. General and specific inheritance of substance abuse and alcoholism. Arch Gen Psychiatry. 1998;55:964–965. doi: 10.1001/archpsyc.55.11.964. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Han DH, Joe KH, Na C, Lee YS. Effect of genetic polymorphisms on smoking cessation: a trial of bupropion in Korean male smokers. Psychiatr Genet. 2008;18:11–16. doi: 10.1097/YPG.0b013e3282df0939. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Rickert W, Robinson J. Measuring the heaviness of smoking: using self-reported time to the first cigarette of the day and number of cigarettes smoked per day. Br J Addict. 1989;84:791–799. doi: 10.1111/j.1360-0443.1989.tb03059.x. [DOI] [PubMed] [Google Scholar]
- Heinz A, Goldman D, Jones DW, Palmour R, Hommer D, Gorey JG, et al. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology. 2000;22:133–139. doi: 10.1016/S0893-133X(99)00099-8. [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hops H, Andrews JA, Duncan SC, Duncan TE, Tildesley E. Adolescent Drug Use Development, A Social Interactional and Contextual Perspective. In: Arnold J, Sameroff ML, Suzanne M, Miller, editors. Handbook of Developmental Psychopathology. Second edn. New York: Kluwer Academic/Plenum Publishers; 2000. pp. 589–605. [Google Scholar]
- Horvath MC, Kovacs GG, Kovari V, Majtenyi K, Hurd YL, Keller E. Heroin abuse is characterized by discrete mesolimbic dopamine and opioid abnormalities and exaggerated nuclear receptor-related 1 transcriptional decline with age. J Neurosci. 2007;27:13371–13375. doi: 10.1523/JNEUROSCI.2398-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, et al. Significant Association of ANKK1 and Detection of a Functional Polymorphism with Nicotine Dependence in an African-American Sample. Neuropsychopharmacology. 2008;34:319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- Hudmon KS, Marks JL, Pomerleau CS, Bolt DM, Brigham J, Swan GE. A multidimensional model for characterizing tobacco dependence. Nicotine Tob Res. 2003;5:655–664. doi: 10.1080/1462220031000158672. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Okubo Y, Ohtsuki T, Yamakawa-Kobayashi K, Arinami T. Mutation analysis of the retinoid X receptor beta, nuclear-related receptor 1, and peroxisome proliferator-activated receptor alpha genes in schizophrenia and alcohol dependence: possible haplotype association of nuclear-related receptor 1 gene to alcohol dependence. Am J Med Genet. 2002;114:15–23. doi: 10.1002/ajmg.1620. [DOI] [PubMed] [Google Scholar]
- Jankovic J, Chen S, Le WD. The role of Nurr1 in the development of dopaminergic neurons and Parkinson's disease. Prog Neurobiol. 2005;77:128–138. doi: 10.1016/j.pneurobio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Jorm AF, Henderson AS, Jacomb PA, Christensen H, Korten AE, Rodgers B, et al. Association of smoking and personality with a polymorphism of the dopamine transporter gene: results from a community survey. Am J Med Genet. 2000;96:331–334. doi: 10.1002/1096-8628(20000612)96:3<331::aid-ajmg19>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Kelada SN, Costa-Mallen P, Checkoway H, Carlson CS, Weller TS, Swanson PD, et al. Dopamine transporter (SLC6A3) 5' region haplotypes significantly affect transcriptional activity in vitro but are not associated with Parkinson's disease. Pharmacogenet Genomics. 2005;15:659–668. doi: 10.1097/01.fpc.0000170917.04275.d6. [DOI] [PubMed] [Google Scholar]
- Kilmer G, Roberts H, Hughes E, Li Y, Valluru B, Fan A, et al. Surveillance of certain health behaviors and conditions among states and selected local areas--Behavioral Risk Factor Surveillance System (BRFSS), United States, 2006. MMWR Surveill Summ. 2008;57:1–188. [PubMed] [Google Scholar]
- Laucht M, Becker K, Frank J, Schmidt MH, Esser G, Treutlein J, et al. Genetic variation in dopamine pathways differentially associated with smoking progression in adolescence. J Am Acad Child Adolesc Psychiatry. 2008;47:673–681. doi: 10.1097/CHI.0b013e31816bff77. [DOI] [PubMed] [Google Scholar]
- Law SW, Conneely OM, DeMayo FJ, O'Malley BW. Identification of a new brain-specific transcription factor, NURR1. Mol Endocrinol. 1992;6:2129–2135. doi: 10.1210/mend.6.12.1491694. [DOI] [PubMed] [Google Scholar]
- Lerman C, Caporaso NE, Audrain J, Main D, Bowman ED, Lockshin B, et al. Evidence suggesting the role of specific genetic factors in cigarette smoking. Health Psychol. 1999;18:14–20. doi: 10.1037//0278-6133.18.1.14. [DOI] [PubMed] [Google Scholar]
- Lerman C, Jepson C, Wileyto EP, Epstein LH, Rukstalis M, Patterson F, et al. Role of functional genetic variation in the dopamine D2 receptor (DRD2) in response to bupropion and nicotine replacement therapy for tobacco dependence: results of two randomized clinical trials. Neuropsychopharmacology. 2006;31:231–242. doi: 10.1038/sj.npp.1300861. [DOI] [PubMed] [Google Scholar]
- Lerman C, Shields PG, Wileyto EP, Audrain J, Hawk LH, Jr, Pinto A, et al. Effects of dopamine transporter and receptor polymorphisms on smoking cessation in a bupropion clinical trial. Health Psychol. 2003;22:541–548. doi: 10.1037/0278-6133.22.5.541. [DOI] [PubMed] [Google Scholar]
- Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10:225–231. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D, Niu T, Feng Y, Xing H, Xu X. Association between polymorphism of the dopamine transporter gene and early smoking onset: an interaction risk on nicotine dependence. J Hum Genet. 2004;49:35–39. doi: 10.1007/s10038-003-0104-5. [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Mangold JE, Zhu J, et al. A combinatorial approach to detecting gene-gene and gene-environment interactions in family studies. Am J Hum Genet. 2008;83:457–467. doi: 10.1016/j.ajhg.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007a;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Ma JZ, Sun D, Payne TJ, Li MD. Fine mapping of a linkage region on chromosome 17p13 reveals that GABARAP and DLG4 are associated with vulnerability to nicotine dependence in European-Americans. Hum Mol Genet. 2007b;16:142–153. doi: 10.1093/hmg/ddl450. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34:1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11:400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- Martinez D, Gelernter J, Abi-Dargham A, van Dyck CH, Kegeles L, Innis RB, et al. The variable number of tandem repeats polymorphism of the dopamine transporter gene is not associated with significant change in dopamine transporter phenotype in humans. Neuropsychopharmacology. 2001;24:553–560. doi: 10.1016/S0893-133X(00)00216-5. [DOI] [PubMed] [Google Scholar]
- McKinney EF, Walton RT, Yudkin P, Fuller A, Haldar NA, Mant D, et al. Association between polymorphisms in dopamine metabolic enzymes and tobacco consumption in smokers. Pharmacogenetics. 2000;10:483–491. doi: 10.1097/00008571-200008000-00001. [DOI] [PubMed] [Google Scholar]
- Mill J, Asherson P, Browes C, D'Souza U, Craig I. Expression of the dopamine transporter gene is regulated by the 3' UTR VNTR: Evidence from brain and lymphocytes using quantitative RT-PCR. Am J Med Genet. 2002;114:975–979. doi: 10.1002/ajmg.b.10948. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton LM, Wang SS, Bergen AW, Chatterjee N, Kvale P, Welch R, et al. DRD2 genetic variation in relation to smoking and obesity in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Pharmacogenet Genomics. 2006;16:901–910. doi: 10.1097/01.fpc.0000230417.20468.d0. [DOI] [PubMed] [Google Scholar]
- Newberg A, Lerman C, Wintering N, Ploessl K, Mozley PD. Dopamine transporter binding in smokers and nonsmokers. Clin Nucl Med. 2007;32:452–455. doi: 10.1097/01.rlu.0000262980.98342.dd. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Ji F, Yuferov V, Ho A, Chen A, Levran O, et al. Genotype patterns that contribute to increased risk for or protection from developing heroin addiction. Mol Psychiatry. 2008;13:417–428. doi: 10.1038/sj.mp.4002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gara C, Stapleton J, Sutherland G, Guindalini C, Neale B, Breen G, et al. Dopamine transporter polymorphisms are associated with short-term response to smoking cessation treatment. Pharmacogenet Genomics. 2007;17:61–67. doi: 10.1097/01.fpc.0000236328.18928.4c. [DOI] [PubMed] [Google Scholar]
- Perkins KA, Lerman C, Grottenthaler A, Ciccocioppo MM, Milanak M, Conklin CA, et al. Dopamine and opioid gene variants are associated with increased smoking reward and reinforcement owing to negative mood. Behav Pharmacol. 2008;19:641–649. doi: 10.1097/FBP.0b013e32830c367c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper ME, McCarthy DE, Baker TB. Assessing tobacco dependence: a guide to measure evaluation and selection. Nicotine Tob Res. 2006;8:339–351. doi: 10.1080/14622200600672765. [DOI] [PubMed] [Google Scholar]
- Radloff L. The CES-D Scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- Russell MAH, Peto J, Patel UA. The classification of smoking by factorial structure of motives. J R Statist Soc A. 1974;137:313–346. [Google Scholar]
- Sabol SZ, Nelson ML, Fisher C, Gunzerath L, Brody CL, Hu S, et al. A genetic association for cigarette smoking behavior. Health Psychol. 1999;18:7–13. doi: 10.1037//0278-6133.18.1.7. [DOI] [PubMed] [Google Scholar]
- Sacchetti P, Brownschidle LA, Granneman JG, Bannon MJ. Characterization of the 5'-flanking region of the human dopamine transporter gene. Brain Res Mol Brain Res. 1999;74:167–174. doi: 10.1016/s0169-328x(99)00275-2. [DOI] [PubMed] [Google Scholar]
- Sacchetti P, Mitchell TR, Granneman JG, Bannon MJ. Nurr1 enhances transcription of the human dopamine transporter gene through a novel mechanism. J Neurochem. 2001;76:1565–1572. doi: 10.1046/j.1471-4159.2001.00181.x. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, et al. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter K, Laird N, Corcoran C. Exact family-based association tests for biallelic data. Genet Epidemiol. 2005;29:185–194. doi: 10.1002/gepi.20088. [DOI] [PubMed] [Google Scholar]
- Schork NJ, Nath SK, Fallin D, Chakravarti A. Linkage disequilibrium analysis of biallelic DNA markers, human quantitative trait loci, and threshold-defined case and control subjects. Am J Hum Genet. 2000;67:1208–1218. doi: 10.1086/321201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segman RH, Kanyas K, Karni O, Lerer E, Goltser-Dubner T, Pavlov V, et al. Why do young women smoke? IV. Role of genetic variation in the dopamine transporter and lifetime traumatic experience. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:533–540. doi: 10.1002/ajmg.b.30507. [DOI] [PubMed] [Google Scholar]
- Sotnikova TD, Beaulieu JM, Gainetdinov RR, Caron MG. Molecular biology, pharmacology and functional role of the plasma membrane dopamine transporter. CNS Neurol Disord Drug Targets. 2006;5:45–56. doi: 10.2174/187152706784111579. [DOI] [PubMed] [Google Scholar]
- Staats B, Qi L, Beerman M, Sicotte H, Burdett LA, Packer B, et al. Genewindow: an interactive tool for visualization of genomic variation. Nat Genet. 2005;37:109–110. doi: 10.1038/ng0205-109. [DOI] [PubMed] [Google Scholar]
- Styn MA, Nukui T, Romkes M, Perkins K, Land SR, Weissfeld JL. The impact of genetic variation in DRD2 and SLC6A3 on smoking cessation in a cohort of participants 1 year after enrollment in a lung cancer screening study. Am J Med Genet B Neuropsychiatr Genet. 2008;150B:254–261. doi: 10.1002/ajmg.b.30801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Hudmon KS, Jack LM, Hemberger K, Carmelli D, Khroyan TV, et al. Environmental and genetic determinants of tobacco use: methodology for a multidisciplinary, longitudinal family-based investigation. Cancer Epidemiol Biomarkers Prev. 2003;12:994–1005. [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Jack LM, Valdes AM, Ring HZ, Ton CC, Curry SJ, et al. Joint effect of dopaminergic genes on likelihood of smoking following treatment with bupropion SR. Health Psychol. 2007;26:361–368. doi: 10.1037/0278-6133.26.3.361. [DOI] [PubMed] [Google Scholar]
- Team RDC. R Foundation for Statistical Computing. Vienna, Austria: 2003. [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake DS, Haberstick BC, Lessem JM, Smolen A, Ehringer M, Hewitt JK, et al. An association between the DAT1 polymorphism and smoking behavior in young adults from the National Longitudinal Study of Adolescent Health. Health Psychol. 2006;25:190–197. doi: 10.1037/0278-6133.25.2.190. [DOI] [PubMed] [Google Scholar]
- Ton TG, Rossing MA, Bowen DJ, Srinouanprachan S, Wicklund K, Farin FM. Genetic polymorphisms in dopamine-related genes and smoking cessation in women: a prospective cohort study. Behav Brain Funct. 2007;3:22. doi: 10.1186/1744-9081-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genet. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65:683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dyck CH, Malison RT, Jacobsen LK, Seibyl JP, Staley JK, Laruelle M, et al. Increased dopamine transporter availability associated with the 9-repeat allele of the SLC6A3 gene. J Nucl Med. 2005;46:745–751. [PubMed] [Google Scholar]
- Vandenbergh DJ, Bennett CJ, Grant MD, Strasser AA, O'Connor R, Stauffer RL, et al. Smoking status and the human dopamine transporter variable number of tandem repeats (VNTR) polymorphism: failure to replicate and finding that never-smokers may be different. Nicotine Tob Res. 2002;4:333–340. doi: 10.1080/14622200210142689. [DOI] [PubMed] [Google Scholar]
- Vandenbergh DJ, Persico AM, Uhl GR. A human dopamine transporter cDNA predicts reduced glycosylation, displays a novel repetitive element and provides racially-dimorphic TaqI RFLPs. Brain Res Mol Brain Res. 1992;15:161–166. doi: 10.1016/0169-328x(92)90165-8. [DOI] [PubMed] [Google Scholar]
- Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, et al. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84:367–379. doi: 10.1016/j.ajhg.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Beem AL, Boomsma DI. The Fagerstrom Test for Nicotine Dependence in a Dutch sample of daily smokers and ex-smokers. Addict Behav. 2005;30:575–579. doi: 10.1016/j.addbeh.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Wang X, Ye Y, Zhang H. Family-based association tests for ordinal traits adjusting for covariates. Genet Epidemiol. 2006;30:728–736. doi: 10.1002/gepi.20184. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YK, Yao WJ, Yeh TL, Lee IH, Chen PS, Lu RB, et al. Decreased dopamine transporter availability in male smokers -- a dual isotope SPECT study. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:274–279. doi: 10.1016/j.pnpbp.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Mitsiadis T, Olson L, Perlmann T. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656–1666. doi: 10.1210/mend.10.12.8961274. [DOI] [PubMed] [Google Scholar]


