Abstract
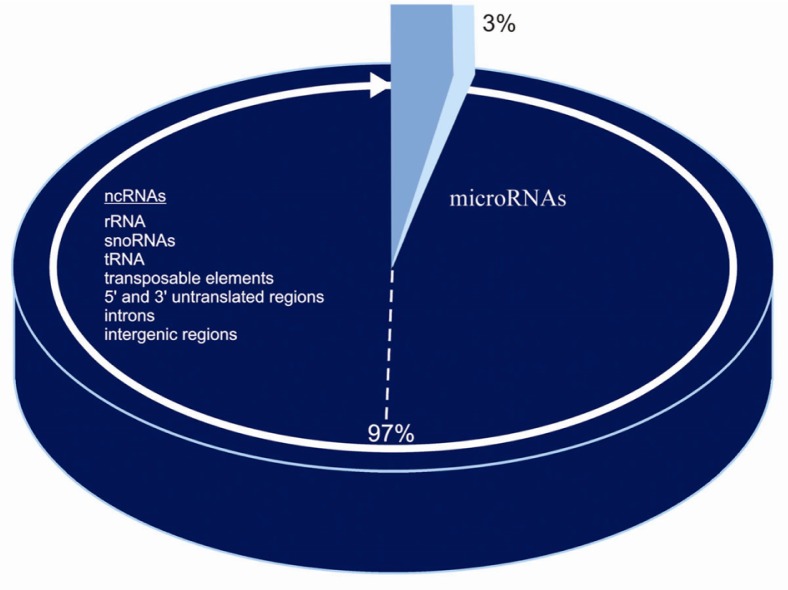
The eukaryotic complexity involves the expression and regulation of genes via RNA-DNA, RNA-RNA, DNA-protein and RNA-protein interactions. Recently, the role of RNA molecules in the regulation of genes in higher organisms has become more evident, especially with the discovery that about 97% of the transcriptional output in higher organisms are represented as noncoding RNAs: rRNA, snoRNAs, tRNA, transposable elements, 5’ and 3’ untranslated regions, introns, intergenic regions and microRNAs. MicroRNAs function by negatively regulating gene expression via degradation or translational inhibition of their target mRNAs and thus participate in a wide variety of physiological and pathological cellular processes including: development, cell proliferation, differentiation, and apoptosis pathways. MicroRNA expression profiles in many types of cancers have been identified. Recent reports have revealed that the expression profiles of microRNAs change in various human cancers and appear to function as oncogenes or tumor suppressors. Abnormal microRNA expression has increasingly become a common feature of human cancers. In this review, we summarize the latest progress on the involvement of microRNAs in different types of cancer and their potential use as potential diagnostic and prognostic tumor biomarkers in the future.
Keywords: Biomarkers, Cellular process, Expression, Micro RNA, Noncoding RNA
Introduction
Ambros et al found lin-4, a gene which controls the timing of larval development of Caenorhabditis elegans (C. elegans) in 1993. But, the product of this gene was not any protein; instead it produced a pair of small RNAs (1). The longer RNA is 70 nucleotides (nt) that can shape a stem-loop structure and is the precursor of the shorter RNA (22 nt) that is now known as a member of the class of microRNA genes (2). MicroRNAs (miRNAs) are a novel class of endogenous small, non-coding RNAs (ncRNAs) that control gene expression by targeting specific mRNAs for degradation and/or translational repression (3). At the time it appeared that lin-4 was restricted to C. elegans because of lack of homology with other species. However, when the second miRNA gene (let-7) with its target miRNA gene (lin-41), was discovered in 2000, it became clear that these miRNA genes are conserved in many species (4). After that, a great number of microRNAs have been identified in mammals (5).
Many members of ncRNAs are part of the functional classes such as miRNAs and small nucleolar RNAs (snoRNAs) that are well conserved in various species. This is because: a) RNAs in both classes function by hybridization to other nucleic acids b) many of the miRNAs have different cellular targets (6) that confine their chance for sequence co-variation and tendency to evolution. Regarding snoRNAs, they encode sequences and structures that mediate binding of compatible RNA-modifying enzymes to the snoRNA target RNA complex, and this further limits sequence change (7).
miRNAs along with a large set of ncRNAs are known as ‘‘gene regulators’’ that include: Air, H19, Ipw, NTT, Tsix and XIST in mammals and have various functions from potential involvement in the imprinting process to X-chromosome inactivation in mammals (8). Multiple steps and specific cellular machinery are involved in the biogenesis of miRNAs (9). The miRNAs are encoded as short inverted repeats having a double-stranded RNA (dsRNA) stem loop about 70 bp long and are found in both introns and intergenic clusters in the genome (9). The introns and exons of both protein-coding and non coding transcripts are synthesized by RNA polymerase II from where miRNAs are derived (10). In the nucleus, miRNAs are transcribed as primary pri-miRNA transcripts, and then they are processed to shape the precursor pre-miRNA stem loop structure and then is transported to the cytoplasm by RanGTP and Exportin 5 where they are cleaved by the Dicer RNAase III endonuclease and produce mature miRNA (21-23 nt) (11).
Human genome and microRNAs
Development, differentiation, growth, and metabolism are regulated by miRNAs and about 500 known miRNA genes are reported to be encoded by the mammalian genome (12). It is estimated that miRNAs regulate about one third of the genes in human genome (13). How the transcription of miRNAs are regulated in the cell is not precisely known However, the transcription of miRNAs are known to depend on their localization within the genome and their proximity to host genes and their locations in introns of coding genes, noncoding genes and exons (14). Recent studies have shown that miRNAs are organized in clusters and share the same transcriptional regulation and if having their own promoters, miRNAs are independently expressed (15, 16).
Approximately 50% of miRNAs are transcribed from non-protein-coding genes, and the rest are in the introns of coding genes (12). In higher organisms, about 97% of the transcriptional product is non-coding RNA (ncRNA) which consist of rRNA, tRNA, introns, 5’ and 3’ untranslated regions, transposable elements, and intergenic regions, and a large family known as microRNAs, some of which can down-regulate large numbers of target mRNAs (3, 17). Recently, a number of mammalian miRNAs have been reported to be derived from DNA repeats and transposons (18). This finding has lead to a re-evaluation of the functional role of transposons, especially since it appears that their sequences can play a significant role in the developmental processes and epigenetic variations (19, 20). miRNAs have also been shown to be derived from processed pseudogenes (21) (Figure 1).
Figure 1.
Coding and Non-coding DNA in human genome
Analysis of the human genome in the past few years has given an estimated number of the protein coding genes (as low as 25,000) in the human genome (22). Despite a controversy as to a real number of protein coding genes, the 25,000 estimated figures are at least 2-3 times lower than it was previously believed. These new data reveal that large portions of human and other mammalian genomes consists of non-coding proteins genes. The Open Reading Frames (ORFs) in the human genome is estimated to comprise less than 2% of the 3.2 billion bases and 46% of the human genome is composed of repetitive sequences (23, 24). It is also estimated that 25-27% of the genome is non-coding parts of protein-coding genes (introns, 5V- and 3V-UTRs) (25).
The microRNAs and other living organisms
Since the initial discovery of the miRNAs in 1993, it has been discovered that miRNAs play a role in gene regulation in different eukaryotic organisms (26). The miRNA registry (version 1) showed 218 miRNA entries for primates, rodents, birds, fish, worms, flies, plants and viruses by 2002; however, this figure has increased to 6396 as of April 2008 (version 11) (27, 28). In prokaryotes, ORFs account for over 90% of genomic DNA and this is because intergenic and untranslated regions are short and splicing is an exception rather than the rule. Among eukaryotes, simple unicellular organisms have 10-40% of the genome, in invertebrates, 70-90% of the genome, and in mammals 98% of the genome is composed of noncoding DNA regions (25).
The functions of mirRNA vary in different organisms. For example, they control development of leaf and flower in plants (29) or modulate differentiation of hematopoietic cells in mammals (30). Conservation of many mirRNAs sequences among distantly related organisms indicates that they participate in essential processes (31).
Role of microRNAs in cancers
The link between miRNAs and cancer was shown when it was discovered that miR-16 and miR-15 genes are located in a region of chromosome 13 that is deleted in over 65% of Chronic Lymphocytic Leukaemia (CLL) patients and more than half of B-cell chronic lymphocytic leukaemia (B-CLL) (32). Then it was discovered that miRNA genomic positioning seems to be non-random, and that a significant number of miRNA genes are located at fragile sites (unstable regions that have been shown to promote DNA instability in cancer cells) or genomic regions linked to cancers (33, 34).
Deregulation of miRNAs correlates with various human cancers and has a role in its initiation and progression. They can inhibit the expression of cancer-related target genes and promote or suppress tumorigenesis in various tissues by acting as oncogenes or tumor suppressors. Thus identification of miRNAs and their respective targets make them ideal for diagnostic and prognostic tumor biomarkers and new therapeutic strategies to treat cancers. Deletion and down-regulation of miR-15 and miR-16 in B-CLL patients is the first report documenting abnormalities in miRNA expression in tumor samples (32). Currently, it is well documented that up-regulation or down-regulation of miRNAs occurs in various human cancers (35).
Over-expressed miRNAs may play a role as oncogenes by down-regulating tumor suppressor genes and/or genes controlling cell differentiation or apoptosis, whereas the down regulated miRNAs act as tumor suppressor genes by negatively regulating oncogenes and/or genes that control cell differentiation or apoptosis (36, 37). Amplification, deregulation of a transcription factor or demethylation of CpG islands in the promoter regions of a gene in human cancers can result in over expression of miRNAs. Tumor suppressor miRNAs can be down-regulated by deletions, epigenetic silencing, or loss of the expression of trans-cription factors (38). Over-expression or inhibition of a number of miRNAs in cancer cells, have been reported to interfere with cell cycle regulators. Many of which directly interact with critical regulators such as PTEN, Myc, Ras or ABL as well as members of the Rb pathway, Cyclin-CDK complexes or cell cycle inhibitors of INK4 or Cip/Kip families to modulate cell proliferation pathways (39). They can also directly target anti-apoptotic genes and participate in tumorigenesis. Proapoptotic miRNAs such as miR-15 and miR-16 can target and inhibit the pro-survival molecule Bcl-2 to antagonize tumor development by promoting apoptosis through the mitochondrial pathway (40).
Fifty to seventy divisions are finite replicative potentials those human cells normally have, after which cells become unable to continue dividing. This senescence suppresses the proliferation of damaged or stressed cells that are at risk for malignant transformation, thus inhibits the development of cancer (41). Maintaining telomeres by either up-regulating telomerase or alternatively, lengthening telomeres causes immortalization in human cancer cells (42). Recently, some miRNAs have been reported to regulate the limitless replicative potential of cancer cells such as the Dicer-dependent miR-290 cluster which can directly target the Retinoblastoma-like 2 protein (Rbl2) and indirectly affects telomere integrity and telomere-length homeostasis (43).
A recent study has revealed oncogenic function of miR-378 which can enhance tumor cell survival, blood vessel expansion and tumor growth by repressing the expression of two tumor suppressors, Sufu and Fus-1(44). Several miRNAs have been also identified that mediate metastasis (35). miR-21 has been shown to promote tumorigenesis and metastasis through suppressing expression of PTEN, PDCD4, TPM1 and Maspin, being all negative regulators of both growth and migration and invasion (45–48). miR-373 and miR-520c have also involvement in tumor migration and invasion by down-regulating metastasis suppressor, CD44 (49). miR-335 has been reported to target a set of genes which are required for tumor metastasis such as SOX4 and TNC (50).
A multistep process of sequential alternations of several, often many, oncogenes, tumor suppressor genes, and miRNA genes result in cancer (51). The miR-17 and miR-20a, two C-Myc-regulated miRNAs, have been reported to govern the transition through G1 checkpoint in normal human cells. Inactivation of these miRNAs causes a G1 checkpoint due to accumulation of DNA double-strand breaks as a result of accumulation of the E2F1 transcription factor (52). Current evidence suggest that more than half of miRNAs are clustered in regions of genomic instability or fragile sites of chromosome areas associated with various human cancers (33, 53). Unique miRNA expression profiles have been demonstrated in a number of cancers including breast, ovary, colon, blood, lung, esophageal, prostate, bladder and thyroid cancers. In the following pages the involvement of mirRNAs in variety of cancers will be reviewed.
The microRNAs in breast cancer
Breast carcinoma, is the second most prevalent cancer in women and fatal disease when not detected at early stages. Breast tumors are grouped into four subtypes (luminal A, luminal B, HER2 over expressing, and basal) according to their difference in gene expression, phenotypes, prognosis, and susceptibility to specific treatments (54). Another more traditional classification is based on the status of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-like 2 (HER2/neu) (55).
It has been shown that multiple miRNAs are associated with a given cancer signature. In breast cancer, up-regulation of miR-21 has been reported across tumor specimens whereas other miRNAs (miR-141, miR-200b, miR- 200c, miR-214, miR-221, and miR-222) exhibited irregular pattern of expression (56). In two of the four ER-, PR-, HER2- tumor specimens, miR-205 was highly expressed, which may be indicative of an association between miR-205 expression with this aggressive tumor subtype (56). The specificity of miRNA expression at different stages of the breast tumor is such that the level of miRNA expressions can be used to distinguish breast cell lines according to their malignancy status. For example, the expressions of several miRNAs have been observed to be elevated in tumorigenic cell lines compared with nontumorigenic cell lines (57). In the same study, it was shown that in the breast tumor tissue, miR-21 was frequently having higher levels in carcinoma cells than in matching normal tissue.
In another study (57) whose results were confirmed by microarray and Northern blot analyses, the differentially expressed miRNAs, miR-10b, miR-125b, miR145, miR-21, and miR-155 were demonstrated to be among the most consistently deregulated in breast cancer. The miR-10b, miR-125b, and miR-145 were down-regulated and miRNA-21 and miR-155 were up-regulated. The miRNA genes have been reported to be frequently deleted in human cancer and interestingly, miR-125b which is down regulated in breast cancer is located at chromosome 11q23-24, one of the most frequently deleted regions in breast cancer (58).
The results of these studies indicate that the differential expressions of miRNAs and their correlation with specific breast cancer bio-pathological features such as tumor stage, vascular invasion, estrogen and progesterone receptor expressions may in the future be used in the clinic as biomarkers to specific stages of breast cancer.
The microRNAs in ovarian cancer
The most common gynecologic malignancy and the sixth most common cancer in women worldwide is Epithelial Ovarian Cancer (EOC), causing almost 125,000 deaths yearly (59). Even with advances in detection and cytotoxic therapies, only 30% of patients with advanced-stage ovarian cancer have 5 years survival after initial diagnosis (60). Late-stage diagnosis for >70% of ovarian cancers is the dominant reason for high mortality of this disease.
Four major histological subtypes for ovarian adenocarcinomas are serous, (being the most common), mucinous, endometrioid and clear cell. Many studies have demonstrated that each of these histologic types is associated with specific morphologic and molecular genetic changes including miRNA gene expressions. For example, in ovarian adenocarcinomas miR-200a and miR-141 were shown to be significantly up-modulated and miR-199a, miR-140, miR-145, and miR-125b1 were most significantly down-modulated. In all the three histotypes considered (serous, endometrioid and clear cell) miR-200a and miR-200c were found up-modulated. Up-modulation of three additional miRNAs, miR-21, miR-203, and miR-205 were detected in the endometrioid histotype (61). In a recent study (62), high frequency copy number abnormalities of Dicer1, Argonaute2, and other miRNA-associated genes in breast and ovarian cancers have been found. These findings suggest that copy number alterations of miRNAs and their regulatory genes are prevalent in cancer.
In the three studies (62–64) reported on miRNAs in ovarian cancer, more than 136 miRNAs have been reported to be deregulated. Fifty six new miRNAs which were not previously reported have been recently demonstrated to be deregulated in ovarian cancer (65). In this study, the most consistently and highly up-regulated miRNA in both ovarian tumor tissues and ovarian cancer cell lines was reported to be miR-221. The oncogene KIT (66) and the tumor suppressor p27kip1 (67) have been shown to be targeted by miR-221.
Microarray studies, validated by Northern blots of the differentially expressed miRNAs have shown that many miRNAs are up- regulated and/or down-regulated in serous ovarian cancer tissues (68). In this study several miRNAs were demonstrated to be differentially expressed in more than 16 patients compared with normal ovarian tissues, including miR-21, miR-99a, miR-125a, miR-125b, miR-100, miR-145, and miR-16. An interesting finding of this study was the correlation found between the expression levels of some microRNAs with the survival in patients with serous ovarian carcinoma. Specifically, the higher expression of miR-200, miR-141, miR-18a, miR-93, and miR-429 and lower expression of let-7b, and miR-199a were significantly correlated with a poor prognosis.
The microRNAs in colorectal cancer
Experimental studies on colorectal cancer (CRC), which is the most common cancer site and the second most common cause of death, has shed light on miRNA-mediated regulatory links to the oncogenic and tumor suppressor signalling pathways. Currently, the connection between miRNA and CRC is investigated by two different approaches. On the one hand, many known oncogenic and tumor suppressor pathways involved in the pathogenesis of CRC seems to be regulated by miRNAs. This is very interesting because in colorectal neo-plasms, miRNA regulation affects many proteins such as p53, RAS and Epithelial-Mesenchymal Transition (EMT) transcription factors and members of the PI-3-K and the Wnt/β-catenin pathway (69). On the other hand, the expressions of hundreds of different miRNAs have been shown to have a higher potential as biomarkers when compared to mRNAs for prediction of prognosis and diagnosis of specific stages in colorectal cancer (69).
Experimental data performed on colorectal cancer and CRC cell lines have demonstrated several miRNAs to function as Oncomirs (refers to tumor suppressor and oncogenic effects of mirRNAs). Several miRNAs (miR-17-92, miR-21, miR-135) are identified as oncogenic miRs and others (miR-34, miR-126, miR-143) are identified as tumor suppressor miRs because they are involved in myc, PTEN, PDCD4, APC pathways and p53, CDK, p85β, KRAS pathways, respectively (70).
The Wnt/β-catenin pathway plays a central role in early colorectal tumor development. More than 60% of all colorectal adenomas and carcinomas have mutation in the APC gene, leading to stimulation of the Wnt pathway via free β-catenin (71). Recently (72), miR-135 family has been shown to regulate the adenomatous polyposis coli gene in colorectal cancer. The miR-135a and miR-135-b target the 3′-untranslated region and decrease the translation of the APC transcript in vitro. Interestingly, up-regulation of miR- 135 was also found in vivo in colorectal adenomas and carcinomas and correlated with low APC levels (72). These results suggest that the mir-135 family is deregulated in neo-plastic colorectal tissues.
The down-regulation of Ecadherin and the successive loss of cell-cell adhesion are described by EMT, thus leading to a mesen-chymal phenotype which contributes to the invasiveness and dissemination of epithelial tumor cells in several carcinomas including colorectal cancer (73). The members of miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) that are suppressed by TGF-β-signaling have been shown to have functional link to EMT (74). Thus, the miR-200 family members might act as important regulators of epithelial phenotype in colorectal cancer.
Reduced levels of miR-143 and miR-145 in colonic adenomas and carcinomas are demonstrated by investigations on paired colorectal neoplasias and normal mucosal samples (75). The reduction of these two miRNAs in CRC and additional two miRNAs (miR-126 and miR-133b) has been confirmed by others (76, 77). Although a number of studies have shown increased or reduced levels of miRNAs in colon adenocarcinomas, however, very few studies have evaluated the association between mirRNA expression patterns and colon cancer prognosis or therapeutic outcome.
In a recent report (78), this question was addressed by a study involving microRNA microarray expression profiling of tumors and paired nontumor tissues on a cohort of 84 patients with incident colon adeno-carcinoma. In this study, thirty-seven mirRNAs were reported to be differentially expressed in tumors from the test cohort. Expression patterns of mirRNAs were shown to be systematically altered in colon adeno-carcinomas and a high miR-21 expression was demonstrated to be associated with poor survival and poor therapeutic outcome. In another study (79), forty-eight clinical colorectal samples (24 samples with 24 paired normal samples) were evaluated for the presence of miRNAs and their significance as markers for disease prognosis. Among the ten miRNAs, miR-15b, miR-181b, miR-191, and miR-200c were highly expressed and miR-200c was significantly associated with patient survival.
The microRNAs in hematological cancers
A. Acute myeloid leukemia
Clonal expansion of hematopoietic stem cells blocked at different stages of erythroid, granulocytic, monocytic or megakaryocytic differentiation is characterized as Acute Myeloid Leukemia (AML) (80). miRNA signatures in bone marrow disorders have been found by an increasing number of studies. Recently, it has been shown that miRNA expression profiles are linked to the karyotype in AML, the most common acute leukemia in adults (81) and expression of a specific miRNA (hsa-miR-181a) is correlated with AML morphological subtype (82). In other studies, association of miRNA signatures and cytogenetic abnormalities in AML were found and the high expression of hsa-miR-191 and hsamiR-199a were correlated with patients having poor prognosis (83, 84).
B. Acute lymphocytic leukemia
Either T or B lymphocyte precursors cause Acute Lymphocytic Leukemia (ALL). However, B-ALL is the most common type (85). The predominant cancer in the childhood which has a favorable prognosis compared to AML is ALL. An up-regulation of miR-155 has been shown in a subset of AML patients, which may repress genes implicated in hematopoietic development and disease (86). miR-155 was demonstrated to have an important role in the mammalian immune system, specifically in regulating T cell differentiation (87). Defective miR-155 cells were shown to induce alteration in PU.1 transcription factor which is critical in early B cell commitment (84, 87).For differential diagnosis of AML and ALL at the genetic level, recently (88) an additional method has been developed to discriminate between AML and ALL by bead-based miRNA expression profiling assay. The results of this study demonstrated that several miRNAs were differentially expressed between AML and ALL where miR-128a, miR-128b, miR-223 and let-7b were the most significant and discriminatory. The authors found that to discriminate between AML and ALL diseases, a signature of only two miRNAs was sufficient (84).
C. Chronic myeloid leukemia
A multi-step chronic bone marrow disorder involving progression from chronic phase to an accelerated phase which ends up in a blast crisis is characterized as Chronic Myeloid Leukemia (CML) (89). A translocation involving chromosomes 9 and 22, generating the Philadelphia chromosome (Ph), characterizes CML (90). In a recent miRNA study, CML-CD34+ cells from patients with chronic phase have been isolated and shown to have seven miRNAs (miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-19b-1, miR- 20a and miR-92a-1) up-regulated as compared to blast phase. Since expression of this miRNA cluster appears to be BCR-ABL and c-Myc dependent, this observation from microarray analysis (miCHIP) may have implications in CML pathogenesis (91).
D. Chronic lymphocytic leukemia
Chronic Lymphocytic Leukemia (CLL) is characterized by elevated number of clonal B lymphocytes in circulation usually arrested in G0/G1 phase (92). In a recent study (93), micro-array analysis identified differentially expressed miRNAs, including miR-15a and miR-16, which might have the potential for CLL prognosis. CLL Patients with good prognosis have been associated with down-regulation of miR-15a and miR-16, whereas bad prognosis was associated with down-regulation of miR-29. Interestingly, in CLL patients the location of 2 miRNAs, miR-15a and miR- 16 are in 13q14.3, a chromosome region frequently deleted (32, 40).
In a recent study (94), it was shown that the expression of miR-143 and miR-145, the levels of which were previously shown to be reduced in variety of cancers was also decreased in CLLand B-cell lymphomas. In another study (95), a profile of the expression of miRNAs in patients with primary CLL cells showed the over-expression of two miRNAs (miR-21 and miR-155). The pattern of miRNA expression in CLL cells was recently analyzed (96) and a global reduction in miRNA expression levels in CLL cells was found associated to a consistent under expression of miR-181a, let-7a and miR-30d. An interesting and surprising finding in this study was that the predicted mRNA targets for these novel miRNAs were located on a small region of chromosome 1 which is frequently altered in human cancer. A number of miRNAs have been found to be relevant to diagnosis and prognosis of CLL. Recently (97), thirteen miRNAs were reported to be associated with prognostic factors and specifically two of these miRNAs, miR-15 and miR-16 were found to be located on chromosome 13q14, a region deleted in more than half of B-CLL patients and absent or down-regulated in the majority of CLL patients.
The microRNAs in endometrioid adenocarcinoma
The most common pelvic malignancy in China and the fourth most common cancer in women worldwide (98) is endometrial carcinoma. Endometrioid adenocarcinoma is the major form of endometrial carcinoma, comprising 75-80% of the cases (99). The miRNAs may play an important role in the carcinogenesis of endometrial carcinoma although it has received very little attention in the miRNA expression profiling.
In a recent study on expression profile of mammalian miroRNAs in endometrioid adenocarcinoma (100), it was reported that seventeen human miRNAs exhibited higher expression and six miRNAs exhibited lower expression in endometrioid adenocarcinoma samples. Among those miRNAs there were three miRNAs (miR-205, miR-449, and miR-429) up-regulated in tumor samples with more than fifteen-fold change and there were three miRNAs (miR-204, miR-99b, and miR-193b) down-regulated with more than five-fold change. Interestingly, miR-205, miR-449 and miR-204 have been shown to regulate the estrogen receptor gene expression which is considered to be involved in endometrioid adenocarcinoma (100)(Table 1).
Table 1.
Cancer-related miRNAs
| Cancer Type | miRNA | Up/Down Regulation | References |
|---|---|---|---|
| Breast | miR-21 ,miR-155, miR-23, and miR-191 | Up | (57,58) |
| miR-205,miR-145,miR-10b,and miR-125b | Down | ||
| Ovary | miR-200a,miR-200c, and miR-141 | Up | (61) |
| miR-199a,miR-140,miR-145,and miR125b1 | Down | ||
| Colon | miR-135,miR-21,miR-15b,miR-181b,miR-191,miR-200c | Up | (72,79) |
| miR143,miR145,miR-133b,and miR-126 | Down | (75, 76, 77) | |
| AML | Has-miR-191,199a,miR155 | Up | (83,86) |
| CML | miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-19b-1, | Up | (91) |
| miR- 20a, and miR-92a-1 | |||
| CLL | miR-21,miR-155 | Up | (96) |
| miR-15a,miR16,miR-29, miR143,miR-45,miR-30d,miR-let7a,miR-181a | Down | (32, 40, 94, 96) | |
| Endometrioid adenocarcinoma | miR-205,miR-449, miR-429 | Up | (100) |
| miR-193a,miR-204, miR-99b | Down |
The microRNAs in esophageal cancer
Esophageal Adenocarcinoma (AC) has the fastest rising incidence of any solid tumor (>300% in the past 30 years and continues to rise) in the United States (101) and is among the high-incidence cancers in Iran (102). Chronic Gastroesophageal Reflux Disease (GERD) which is associated with an approximately 16-fold increased risk of AC and occurs in up to 60% of patients diagnosed with this tumor, is the most common risk factor for AC (103).
miRNAs that were differentially expressed among different histological groups of patients with AC were recently analyzed (104). The results of this study demonstrated that thirteen miRNAs from three histological groups were differentially expressed. The miR-194, miR-192 and miR-200c were significantly up-regulated in AC, but not in Squamous Cell Carcinomas (SCC). The miR-342 was found differentially expressed in SCC but not in AC in comparison with normal epithelium. The significant finding of this study was that different esophageal tissue types and malignant tissues could be distinguished from normal esophagus by miRNA expression profiles.
The microRNAs in gastrointestinal cancer
The most recent study (105) on the role of miRNAs in gastrointestinal cancers has shown that down-regulation and up-regulation of some miRNAs occur which may function as tumor suppressors and oncogenes, respectively. Extensive lists of miRNAs functioning as tumor suppressors or oncogenes in gastrointestinal cancers are provided (105).Recent studies have demonstrated that certain miRNAs are down-regulated in gastrointestinal cancers and are involved in the regulation of cellular activities. For example, miR-15b and miR-16 which are down regulated in human gastric cancer cells participate in the development of multidrug resistance by targeting BCL2 (106). In another study miR-106b-25 cluster which is over-expressed in human gastric cancers has been shown to impair the TGF-β tumor suppressor pathway by regulating the expression of p21 (107). These findings suggest that miRNAs play an important role in the mechanism underlying human carcinogenesis and thus their abnormal expressions may contribute to the initiation and progression of gastrointestinal cancers in human.
The microRNAs in lung cancer
The leading cause of cancer deaths in the world is lung cancer, etiology of which is primarily genetic and epigenetic damage is caused by tobacco smoke (108). To define the molecular network of lung carcinogenesis, systematic analysis of mRNA and protein expression levels of thousands of genes have been accomplished and among them, p53 and RB/p16 defective pathways and expression of several genes such as K-ras, PTEN, FHIT, and MYO18B, have been demonstrated to be altered (109).
Focusing on known genes and proteins have yielded a lot of new information however, the newly discovered micRNAs may help further to understand the molecular mechanisms of lung cancer. Recently, it was reported that DICER expression levels were reduced in lung cancer and this was reported to be associated with poor prognosis in these patients (110). Recently, it was identified that six miRNAs (hsa-mir-205, hsa-mir-99b, hsa-mir-203, hsa-mir-202, hsa-mir-102 and hsa-mir-204-prec) were expressed differently in the two most common histological types of Non-Small Cell Lung Carcinoma (NSCLC), adenocarcinoma and SCC (109).
The potential involvement of reduced miRNA let-7 type expression in lung cancers has been reported (111). Recently, the over-expression of mir-17-92 cluster was reported in small cell lung cancer which is the most aggressive form of lung cancer (112). Altered expression of miRNAs and their molecular mechanisms in lung cancers are not completely understood. However, alteration of the expression of five miRNAs (hsa-mir-155, hsamir- 17-3p, hsa-let-7a-2, hsa-mir-145 and hsa-mir-21) in lung cancers indicate that miRNAs may potentially have a prognostic impact on the survival of patients with lung cancers (109).
The microRNAs in bladder cancer
The five most common malignancies worldwide, and the second most common tumor of the genitourinary tract and cause of death in patients with genitourinary tract malignancies is Bladder Cancer (BC). An invasive Transitional Cell Carcinoma (TCC) of the urinary bladder aggressively progresses to metastatic disease with a poor prognosis (113, 114). Few studies have been reported on the regulatory effects of miRNAs in bladder cancer. In a recent study (115), the role of mirRNAs in bladder primary tumors was investigated and it was found that different mirRNAs are deregulated in bladder cancer. In another study, where a vast number of miRNAs in bladder tumors were analyzed, interesting pattern of expressions was observed (116). These changes included: 18-50 miRNAs up-regulated whereas 9-89 miRNAs were down-regulated when a total of 464 miRNAs in bladder tumors were compared with those in normal tissue in each patient. The results of this study showed that out of 464 different miRNAs, four miRNAs (miR-29c, miR-26a, miR-30c and miR-30e-5p) were consistently down-regulated in all the bladder tissue samples irrespective of tumor grade or stage. These observations suggest that loss of miRNAs may be required for development of bladder cancer.
In a very recent study on mirRNA signatures in bladder cancer, novel mirRNA targets were reported (117). In this study, subsets of seven miRNAs (miR-145, miR-30a-3p, miR-133a, miR-133b, miR-195, miR-125b and miR-199a) were demonstrated to be signifycantly down-regulated in bladder tumor samples. The target search algorithm and gene-expression profiling from this study revealed that Keratin7 (KRT7) mRNA was a common target of the down-regulated miRNAs. The mRNA expression levels of KRT7 were significantly higher in bladder cancer than in normal bladder epithelium. Furthermore, in this study the gain-of-function analysis showed that KRT7 mRNA was significantly reduced by transfection of miR-30a-3p, miR-133a and miR-199a in the bladder cancer cell line (KK47). These results suggest that miRNAs may have a tumor suppressive function.
The microRNAs in thyroid tumors
Thyroid carcinomas mostly originate from thyroid follicular cells. Papillary Carcinoma (PC) and Follicular Carcinoma (FC) are the two most common types of cancer types. Both PCs and FCs may progress to Poorly Differentiated Carcinoma (PDC) or transform to Anaplastic Carcinoma (AC) by complete loss of differentiation. The Medullary Carcinoma (MC) accounts for less than 5% of C thyroid tumors and originate from the thyroid cells (118).
MicroRNA expression profiling in thyroid tumors have identified several miRNAs (miR-146b, miR-221, miR-222, miR-181b, miR-155 and miR-224) up-regulated in PCs when comparison was made to normal thyroid cells (119). An interesting observation in this study was that the miRNA expression levels were correlated with the mutations of genes such as BRAF, RET/PTC and RAS. Specifically, the miR-222 and the miR-221 were the most consistently up-regulated in PC. In the same study, miRNA analysis of ACs demonstrated up-regulation of several miRNAs that exhibited over-expression in well-differentiated tumors driving from follicular cells. Many miRNAs was shown to be down-regulated in ACs and in particular a signifycant decrease was observed in expression of miR-30d, miR-125b, miR-26a, and miR-30a-5p (120). Over-expression of miR-125b and miR-26a was demonstrated to be able to reduce cell growth and proliferation of human AC-derived cell lines, suggesting that miRNAs may have a role in thyroid cancer development (120).
The miRNAs, are an important regulators of single genes and whole genome and have a great potential for therapeutic use in humans (121). The results presented in this review indicate that miRNA deregulation is common in human cancers (35). Specifically, miRNAs as inhibitors can be used to over-express or inhibit miRNAs for treatment of cancers and the expression of miRNAs can be artificially regulated by modified antisense oligonucleotides to control the growth of cancer cells (122, 123) (Table 2).
Table 2.
Cancer-related miRNAs
| Cancer Type | miRNA | Up/Down regulation | References |
|---|---|---|---|
| Esophagus | miR_194, miR_192,miR_200c | Up | (104) |
| Gastrointestinal | miR-106b-25 | Up | (107) |
| miR-15b,miR-16 | Down | (106) | |
| Lung | miR-17-92 | Up | (112) |
| let- 7 | Down | (111) | |
| Bladder | miR-29c, miR-26a, miR-30c, miR-30e-5p,miR-145, miR-30a-3p, miR-133a, miR-133b, miR-195, miR-125b and miR-199a | Down | (117) |
| Thyroid tumors | |||
| -PC | miR-146b, miR-221, miR-222, miR-181b, miR-155,miR-224 | Up | (119) |
| -AC | miR-30d, miR-125b, miR-26a, miR-30a-5p | Down | (120) |
Perspectives
The microRNAs (miRNAs) as a special class of noncoding RNAs have been discovered in most organisms although, the precise biological effects of miRNAs are not yet known. In this review article, an attempt was made to present in a comprehensive manner, the most recent findings in this exciting new area of research with emphasis on the involvement of miRNAs in cancer development. As the results of many studies demonstrate in the above pages, the carcinogenic processes in many different types of cancer are associated with alterations in the expression of miRNAs, suggesting oncogenic or tumor suppressor roles for microRNAs in a variety of cancer types. To establish other roles, further studies are needed to decipher the molecular and biological functions of miRNAs in cancer development.
Better understanding of the role of microRNAs in cancer development at the molecular level should provide a basis for novel strategies for cancer diagnosis and therapy in the near future. Because mirRNAs are a special class of noncoding RNAs that post-transcriptionally regulate gene expression in a negative manner, a prediction is made that mirRNAs will have a great potential in diagnosis and treatment of cancer in medicine. Currently, several techniques such as miRNA silencing, antisense blocking, and miRNA modification are under investigation for possible use as potential therapeutic agents for treatment of cancers.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Cinzia Sevignani, George A. Calin, Linda D. Siracusa, Carlo M. Croce. Mammalian micro RNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17(3):189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C. MicroRNAs: role in cardiovascular boilogy and disease. Clin Sci. 2008;114(12):699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 6.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 7.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22(1):1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 9.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13(3):302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 10.Morey C, Avner P. Employment opportunities for non-coding RNAs. FEBS Lett. 2004;567(1):27–34. doi: 10.1016/j.febslet.2004.03.117. [DOI] [PubMed] [Google Scholar]
- 11.Bilen J, Liu N, Bonini NM. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5(24):2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- 12.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 14.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Smalheiser NR, Torvik VI. Mammalian miRNAs derived from genomic repeats. Trends Genet. 2005;21(6):322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27(4):361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 20.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, et al. Retro-transposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Devor EJ. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J Hered. 2006;97(2):186–190. doi: 10.1093/jhered/esj022. [DOI] [PubMed] [Google Scholar]
- 22.Herbert. A. The four Rs of RNA-directed evolutions. Nat Genet. 2004;36(1):19–25. doi: 10.1038/ng1275. [DOI] [PubMed] [Google Scholar]
- 23.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 25.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochim Biophys Acta. 2005;1756(1):65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erson AE, Petty. EM. MicroRNAs in development and disease. Clin Genet. 2008;74(4):296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 29.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynam-Lennon N, Maher SG, Reynolds JV. Reynolds. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84(1):55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 35.Ruan K, Fang X, Ouyang G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.04.031. (Epub ahead of print-available online). [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Pan X, Cobb GP, Anderson TA. Micro RNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 37.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumor suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–511. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 39.Bueno MJ, de Castro IP, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7(20):3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 40.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2(5):331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 43.Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15(3):268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 46.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 over-expression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 48.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 49.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumor invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 50.Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 52.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 check point to regulate cell cycle progression. Oncogene. 2009;28(1):140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sevignani C, Calin GA, Nnadi SC, Shimizu M, Davuluri RV, Hyslop T, et al. MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA. 2007;104(19):8017–8022. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer. 2004;40(18):2667–2675. doi: 10.1016/j.ejca.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 55.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 56.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered micro RNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67(24):11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 57.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 58.Negrini M, Rasio D, Hampton GM, Sabbioni S, Rattan S, Carter SL, et al. Definition and refinement of chromosome 11 regions of LOH in breast cancer: identification of a new region at 11q23-q24. Cancer Res. 1995;55(14):3003–3007. [PubMed] [Google Scholar]
- 59.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 60.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics. CA Cancer J Clin. 2001;51(1):15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 61.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 64.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 65.Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih IeM, Zhang Y, Wood W, 3rd, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PloS One. 2008;3(6):e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 67.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27 (Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 69.Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454(4):359–367. doi: 10.1007/s00428-009-0751-9. [DOI] [PubMed] [Google Scholar]
- 70.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12(5):414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 71.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 72.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68(14):5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 73.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131(3):830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 74.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal represssion between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2006;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891. [PubMed] [Google Scholar]
- 76.Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR- 126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes chromosomes Cancer. 2008;47(11):939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–121. [PMC free article] [PubMed] [Google Scholar]
- 80.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3(2):89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 81.Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukemia. PloS One. 2008;3(5):e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 83.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105(10):3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Costa A, Osório C, Dias S. MicroRNA expression profiling in bone marrow: Implications in hemato-logical malignancies. Biotechnol J. 2009;4(1):88–97. doi: 10.1002/biot.200800194. [DOI] [PubMed] [Google Scholar]
- 85.Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukemia. Nat Rev Drug Discov. 2007;6(2):149–165. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 86.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. Exp Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 88.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104(50):19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melo JV, Barnes DJ. Chronic myeloid leukemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 90.Rowley JD. Letter: Deletions of chromosome 7 in haematological disorders. Lancet. 1973;2(7842):1385–1386. doi: 10.1016/s0140-6736(73)93347-3. [DOI] [PubMed] [Google Scholar]
- 91.Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 92.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 93.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and 145 in B-cell malignancies. Cancer Sci. 2007;98(12):1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109(11):4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 96.Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, et al. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22(2):330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 97.Zhang H, Chen Y. New insight into the role of miRNAs in leukemia. Sci China C Life Sci. 2009;52(3):224–231. doi: 10.1007/s11427-009-0036-1. [DOI] [PubMed] [Google Scholar]
- 98.Yang L, Parkin DM, Whelan S, Zhang S, Chen Y, Lu F, et al. Statistics on cancer in China: cancer registration in 2002. Eur J Cancer Prev. 2005;14(4):329–335. doi: 10.1097/00008469-200508000-00004. [DOI] [PubMed] [Google Scholar]
- 99.Pecorelli S, Pasinetti B, Angioli R, Favalli G, Odicino F. Systemic therapy for gynecological neoplasms: ovary, cervix and endometrium. Cancer Chemother Bio Response Modif. 2005;22:515–544. [PubMed] [Google Scholar]
- 100.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18(1):50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 101.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97(2):142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 102.Kamangar F, Malekzadeh R, Dawsey SM, Saidi F. Esophageal cancer in northeastern Iran. Arch Iran Med. 2007;10(1):70–82. [PubMed] [Google Scholar]
- 103.Lagergren J, Bergstrom R, Lindgren A, Nyren O. Symptomatic gastroesophageal refluxes as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340(11):825–31. doi: 10.1056/NEJM199903183401101. [DOI] [PubMed] [Google Scholar]
- 104.Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–260. doi: 10.1016/j.jtcvs.2007.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saito Y, Suzuki H, Hibi T. The role of microRNAs in gastrointestinal cancers. J Gastro-enterol. 2009;44(Suppl 19):18–22. doi: 10.1007/s00535-008-2285-3. [DOI] [PubMed] [Google Scholar]
- 106.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123(2):372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 107.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGF-beta dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 108.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Lyon: IARC Press; 2004. World Health Organization Classification of Tumors, Pathology and Genetics: Tumors of the Lung, Pleura, Thymus and Heart; pp. 12–15. [Google Scholar]
- 109.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 110.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 112.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 113.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 114.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 115.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25(5):387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 116.Wang G, Zhan H, He H, Tong W, Wang B, Liao G, et al. Up-regulation of microRNA in bladder tumor tissue is not common. Int Urol Nephrol. 2009 doi: 10.1007/s11255-009-9584-3. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 117.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125(2):345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 118.Nikiforova MN, Chiosea SI, Nikiforov YE. MicroRNA expression profiles in thyroid tumors. Endocr Pathol. 2009;20(2):85–91. doi: 10.1007/s12022-009-9069-z. [DOI] [PubMed] [Google Scholar]
- 119.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 121.Thum T, Catalucci D, Bauersachs J. Micro RNAs: novel regulators in cardiac development and disease. Cardiovas Res. 2008;79(4):562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 122.Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence specific inhibition of small RNA function. PLoS Biol. 2004;2(4):E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA and siRNA-induced RNA silencing. RNA. 2004;10(3):544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]