Abstract
Molecular epidemiology analyses are frequently used in determining epidemiology of tuberculosis. Recently, Mycobacterial Interspersed Repetitive Unit Variable Number Tandem Repeat (MIRU-VNTR) and Spoligotyping has become an important method, as it allows high-through put, discriminatory and reproducible analysis of clinical isolate. The purpose of this study is to compare techniques of “MIRU-VNTR” versus “MIRU-VNTR and Spoligotyping” together for study of genetic pattern of Mycobacterium tuberculosis (M. tuberculosis) strains. Sixty M. tuberculosis (MTB) isolates were selected (30 susceptible, 30 multi-drug resistant) for this study. Thereafter, the “MIRU-VNTR and Spoligotyping” were performed to identify their genetic patterns. The frequency of unknown genetic pattern of MTB was compared using technique of “MIRU-VNTR” alone versus “MIRU-VNTR and Spoligotyping” together. The MIRU-VNTR allelic diversity at each of the loci was calculated by Hunter – Gaston Discriminatory Index (HGDI). Based on differentiation index of all strains 10, 16, 26, 31 and 40 loci were identified as the most distinctive (HGI ≥0.6) and 2, 4, 20 and 24 as the weakest distinctive locus (HGI ≤0.3). By using MIRU-VNTR technique 38% (n = 23) of isolates could not be typed, whereas by applying “MIRU-VNTR and Spoligotyping” together only 15% (n = 9) of isolates remained unknown (p = 0.004). For sensitive strains, the difference was significant (67% vs. 90%, p = 0.028), but only marginally significant for drug resistant strains (57% vs. 80%, p = 0.052). The discrimination power of 12-locus MIRU-VNTR and Spoligotyping was equal to that of MIRU-VNTR analysis. If appropriate loci are added to the standard MIRU analysis, MIRU-VNTR genotyping could be a valuable tool for strain typing and epidemiological research of M. tuberculosis. With this approach a more clear understanding about genetic pattern of MTB can be achieved.
Keywords: Alleles, Genetic Loci, Molecular epidemiology, Mycobacterium tuberculosis, Restriction fragment length polymorphism
Introduction
Tuberculosis is one of the main factors of vital statistic in the third world countries. Ninety percent of all tuberculosis patients live in third world countries (1). In some regions like Saharan desert in Africa 1000 of 100,000 people are infected with tuberculosis and HIV co-infection, among which one out of three dies from HIV virus (2). Recent, spread and rise in multi-drug resistant M. tuberculosis (MTB) isolate had worsened the matter. In addition, the worldwide development of transport and migration contributes to globalize these threats. Therefore, there is an urgent need to understand and estimate the incidence of tuberculosis in large and small biographical ranges (3).
In this context, effective methods for accurate identification and typing is required. However, all the current typing markers suffer from significant drawbacks. For example DNA fingerprinting method based on IS6110 is a powerful tool to study molecular epidemiology of MTB isolates. But RFLP based on IS6110 typing is difficult and time consuming, and cannot be applied to MTB strains with low or no IS6110 banding patterns (4).
Spoligotyping analyzes the Direct Repeat (DR) locus in the genome of M. tuberculosis, which is composed of a cluster of 36 bp repeat sequences interspersed with unique spacers of 35 to 41 bp. By using a reverse Southern blotting technique, the variability in the spacer sequences can be interrogated and recorded in a digital code (5). Although this method provides digital typing data, it is only measuring variability in a single locus and does not generally provide sufficient discrimination for outbreak investigation (6).
Another molecular technique for strain typing of MTB is based on Variable Number Tandem Repeats (VNTRs) of Mycobacterial Interspersed Repetitive Units (MIRUs). The repeated units are 52 to 77 nucleotides in length and the number of repeated units can be determined by the size of the entire locus. Previous studies demonstrated the importance of MIRU-VNTR method for tracking epidemiological key events such as transmission or relapse and provide non-ambiguous data which are highly portable between different laboratories (7).
The purpose of this study was to compare the frequency of unknown genetic pattern of MTB isolates by using single technique of “MIRU-VNTR” versus the combined techniques of “MIRU-VNTR and Spoligotyping”.
Materials and Methods
Design and setting
The study was conducted in Mycobacteriology Research Center, NRITLD, Shahid Beheshti University of Medical Sciences, Tehran, Iran (2009).
Participants and sampling
Sixty MTB isolate along with their clinical files (30 sensitive and 30 multi-drug resistant MTB from Mycobacteriology bank in MRC) were used for this study.
Extraction DNA
DNA from M. tuberculosis isolates was extracted from growth on LJ slants by a well standized method. In brief, fifty µl of Lysozyme (Merck, Germany) was added to the mycobacterial suspension in 400 µl of water, followed by stirring and incubation at 37 °C. Ten µl with 10 mg/ml of proteinase K (Merck, Germany) was added to the sample, which was incubated for 10 min at 65°C, and then heated at 100°C for 30 min. One volume of phenol/ chloroform/ isoamyl-alcohol 25: 24: 1 was added to the supernatant, mixed by inversion and centrifuged at 12,000 xg for 10 min. The aqueous phase was transferred to other tubs, and the extraction procedure was repeated. A volume of chloroform/ isoamyl-alcohol 24: 1 was added and mixed by inversion followed by centrifugation for 10 min at 12,000 xg. One hundred µl of 5 M Nacl and two volumes of absolute ethanol were added to the supernatant. Samples were incubated for 60 min at −20°C and centrifuged for 15 min at 12,000 xg. The pellet was washed twice with 1 ml of 70% ethanol and re suspended in 200 µl of 0.1 x TE buffer (1 mM Tris- Hcl, 0.1 mM EDTA, Ph 8). Four µl of this sample was used for PCR (8).
MIRU – VNTR typing
PCR was performed in 25 µl volume that contained 5 to 50 ng of DNA, 0.5 µm of specific primers (Table 1) in the presence of 1.5 m MgCl2, 100 µm of each d NTP, 70 m PCR buffer and 1.25 u recombinant DNA polymerase (Cinagen Co. Iran) (9). DNA was amplified by general PCR. All PCRs were initiated by a 10 min denaturizing step at 72 °C. The temperature cycles for different types of PCRs were as follow: 35 cycles of 1 min at 94 °C, annealing temperatures were used as follow: 61, 60, 64, 67, 65, 64, 61, 64, 58, 63, 62 and 64 for MIRU loci 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39 and 40, respectively. Also the MIRU copy numbers in the 12 MIRU-VNTR loci are shown in Table 2, (5).
Table 1.
Primer sequence of the MIRU-VNTR loci and Spoligotyping in this study
| Locus of MIRU | Primer sequence (5′→3′) |
|---|---|
| 2 | TGGACTTGCAGCAATGGACCAACT F |
| TACTCGGACGCCGGCTCAAAA‘ R | |
| 4 | GCGCGAGAGCCCGAACTGC F |
| GCGCAGCAGAAACGTCAGC R | |
| 10 | GTTCTTGACCAACTGCAGTCGTCC F |
| GCCACCTTGGTGATCAGCTACCT R | |
| 16 | TCGGTGATCGGGTCCAGTCCAAGTA F |
| CCCGTCGTGCAGCCCTGGTAC R | |
| 20 | TCGGAGAGATGCCCTTCGAGTTAG F |
| GGAGACCGCGACCAGGTACTTGTA R | |
| 23 | CAGCGAAACGAACTGTGCTATCAC F |
| CGTGTCCGAGCAGAAAAGGGTAT R | |
| 24 | CGACCAAGATGTGCAGGAATACAT F |
| GGGCGAGTTGAGCTCACAGAA R | |
| 26 | CCCGCCTTCGAAACGTCGCT F |
| TGGACATAGGCGACCAGGCGAATA R | |
| 27 | TCGAAAGCCTCTGCGTGCCAGTAA F |
| GCGATGTGAGCGTGCCACTCAA R | |
| 31 | ACTGATTGGCTTCATACGGCTTTA F |
| GTGCCGACGTGGTCTTGAT R | |
| 39 | CGCATCGACAAACTGGAGCCAAAC F |
| CGGAAACGTCTACGCCCCACACAT R | |
| 40 | GGGTTGCTGGATGACAACGTGT F |
| GGGTGATCTCGGCGAAATCAGATA R | |
|
| |
| Locus of Spoligotyping | Primer sequence (5′→3′) |
|
| |
| DR a | Biotin- GGTTTTGGGTCTGACGAC |
| DRb | CCGAGAGGGGACGGAAAC |
MIRU-VNTR: Mycobacterial Interspersed Repetitive Unit Variable Number Tandem Repeat
Table 2.
MIRU locus information for M. tuberculosis H37RV
| MIRU alias | Locus name | Location on H37RV genome | Expected length in H37RV | No. of repeat and size repeat | Size of repeat (bp) |
|---|---|---|---|---|---|
| MIRU 02 | H37RV0154-53 bp | 154111 | 508 | 2 (53) | |
| MIRU 04 | H37RV0580-77 bp | 580546 | 353 | 3(77) | |
| MIRU 10 | H37RV0959-53 bp | 959868 | 643 | 3(53) | |
| MIRU16 | H37RV1644-53 bp | 1644026 | 671 | 2(53) | |
| MIRU20 | H37RV2059-77 bp | 2059429 | 591 | 2(77) | |
| MIRU23 | H37RV2531-53 bp | 2531560 | 873 | 6(53) | |
| MIRU24 | H37RV2387-54 bp | 2684427 | 447 | 1(52) | |
| MIRU26 | H37RV2996-51 bp | 2996002 | 614 | 3(51) | |
| MIRU27 | H37RV3006-53 bp | 3006875 | 657 | 3(53) | |
| MIRU31 | H37RV3192-53 bp | 3192168 | 651 | 3(53) | |
| MIRU39 | H37RV4348-53 bp | 4348401 | 646 | 2(53) | |
| MIRU40 | H37RV0802-54 bp | 802194 | 408 | 1(54) |
MIRU-VNTR: Mycobacterial Interspersed Repetitive Unit Variable Number Tandem Repeat
Analysis
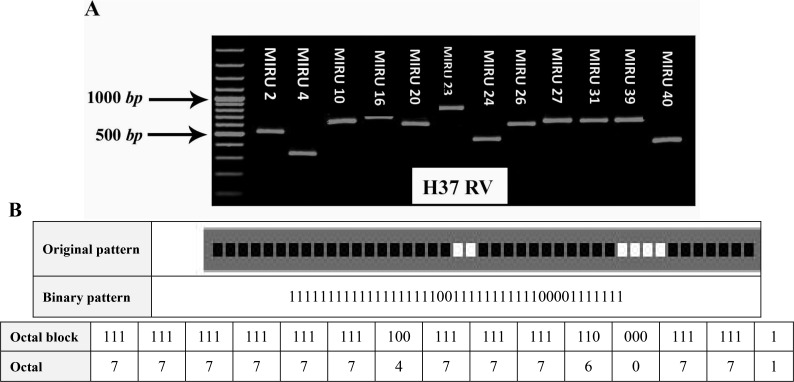
Genetic analysis patterns MIRU-VNTR for each strain to a twenty digit profile from right to left indicate loci 2, 4, 10, 16, 20, 23, 24, 26, 27, 31, 39 and 40, respectively. For example pattern of genetic strains H37RV (M. tuberculosis standard strain) a 2, 3, 3, 2, 2, 6, 1, 3, 3, 3, 2 and 1 is the genetic pattern that can be seen in this model in Figure 1A (10).
Figure 1.
A) PCR products of the various M. tuberculosis H37RV isolates with using primer that amplify 12 locus MIRU-VNTR. Lane M, 100-bp molecular marker. B) Genotypic of M. tuberculosis H37RV. Spoligotyping is from the MIRU-VNTR plus database MIRU-VNTR: Mycobacterial Interspersed Repetitive Units Variable Number Tandem Repeat
Spoligotyping
Spoligotyping was performed as previously described by Kamerbeek et al (11). The DR region was amplified by PCR using primers derived from the DR sequence. Fifty µl of the following reaction mixture was used for the PCR: 10 ng of DNA, 20 pmol each of primers DRa and DRb, each deoxynucleoside triphosphate at 200 mM, PCR buffer, and 0.5 u of taq polymerase (Cinagen Co. Iran). The mixture was heated for 2 min at 94 °C and subjected to 30 cycles of 30 sec at 94 °C, 30 sec at 55 °C, and 1 min at 72 °C. The amplified DNA was hybridized to a set of 43 immobilized oligo-nucleotides, each corresponding to one of the unique spacer DNA sequence within the DR locus.
The sequence of the oligonucleotides used is given in Table 2. These oligonucleotides were covalently bound to a hybridization membrane for hybridization. Twenty µl of the amplified PCR product was diluted in 150 µl of 2 Saline – Sodium Phosphate – EDTA (SSPE), supplemented with 0.1% sodium dodecyl sulfate, and heat denatured. The diluted samples (130 µl) were pipetted in to the parallel channels in such a way that the channels of the miniblotter apparatus were perpendicular to the rows of oligonucleotides deposited previously. Hybridization was done for 60 min at 60 °C.
After hybridization, the membrane was washed as previously described. Detection of hybridizing DNA was done using a (chemilu-minescent ammersham ECL direct nucleotide acid) detection kit. The 4 digit binary result was converted in to a 15 digit octal designa-tion as previously described (12).
Analysis
These rational systems (Figure 1B) are based on the identification of the spoligotype pattern as a 43- digit binary number (taking a negative as 0 and positive as 1), which can be presented in a shorter format by converting to either octal or hexadecimal formats. For the octal system, the pattern is read in groups of 3, as 3 – digit binary number can be read as a single digit octal number. In this way the binary series 000, 001, 010, 011, 100, 101, 110, 111 is read respectively as 0, 1, 2, 3, 4, 5, 6, 7. Following 14 groups of 3, the 43rd spacer is read on its own, simply as 0 or 1 (13).
MIRU-VNTR plus
The information was analysed by MIRU-VNTR plus website. MIRU-VNTR plus software compares and analyses the received information with the information bank in its own software.
Codes of ethics
This study was short term research project and information regarding each patient kept confidential. The study was approved by the medical board of NRITLD.
Statistical analysis
The Hunter – Gaston Discriminatory Index (HGDI) described by Hunter and Gaston (Hunter and Gaston, 2003) was used as a numerical index for MIRU-VNTR discriminatory power. HGDI was calculated by using the following formula: (14)
Where, N stands for the total number of strains in the typing scheme, S stands for the total number of different MIRU-VNTR patterns, and xj stands for the number of strains belonging to the Jth pattern.
P less than 0.05 was considered as significant. Statistical package SPSS (15) was used for analysis of data. Chi square was used to determine possible difference by means of unknown MTB strains between the single and combined techniques.
Results
As it shown in Table 3 34% of susceptible strains could not identified by MIRU-VNTR, and the remaining 66% were identified as Haarlem family (17%), Dehli/ CAS (17%), NEW1 (14%), and LAM, Uganda and M.X families were 7%. Similarly, 44% drug resistant strains could not be identified by MIRU-VNTR technique. Using both “MIRU-VNTR and Spoligotyping” techniques together significantly reduced the percentage of unknown strains (Table 3).
Table 3.
Frequency of sensitive and drug resistance strains of MTB by “MIRU-VNTR” assay versus “MIRU-VNTR and Spoligotyping” together
| Families | MIRU-VNTR No. (%) | MIRU-VNTR+ Spoligotyping No. (%) |
|---|---|---|
| Sensitive strains | ||
| Known | 20 (67) Case | 27 (90) Case |
| Haarlem | 5 (16) Case | 7 (23) Case |
| Dehli/CAS | 5 (16) Case | 8 (26) Case |
| NEW1 | 4 (14) Case | 6 (20) Case |
| LAM | 2 (7) Case | 2 (7) Case |
| Uganda1 | 2 (7) Case | 2 (7) Case |
| M.X | 2 (7) Case | 2 (7) Case |
| Unknown | 10 (33) Case | 3 (10) Case |
| Multi-drug resistant strains | ||
| Known | 17 (57) Case | 24 (80) Case |
| Haarlem | 4 (14) Case | 6 (20) Case |
| Dehli/CAS | 4 (14) Case | 5 (16) Case |
| LAM | 3 (10) Case | 4 (14) Case |
| Uganda 1 | 2 (7) Case | 2 (7) Case |
| Cameron | 1(3) Case | 2 (7) Case |
| Beijing | 1 (3) Case | 1(3) Case |
| Ghana | 0 (0) Case | 1(3) Case |
| Bovis | 1 (3) Case | 1(3) Case |
| Caparie | 1 (3) Case | 1(3) Case |
| M.X | 0 (0) Case | 1(3) Case |
| Unknown | 13 (43) Case | 6(20) Case |
| All strains | ||
| Known | 37 (62) Case | 51(85) Case |
| Unknown | 23 (38) Case | 9(15) Case |
MIRU-VNTR: Mycobacterial Interspersed Repetitive Units Variable Number Tandem Repeat
As shown in Table 3, thirteen strains with unknown sample (by “MIRU-VNTR” technique) decreased to 6 strains by using “MIRU-VNTR and Spoligotyping” technique. On the other hand strains (samples specified) increased by using the two techniques; because of the setting of the family samples for some of the unknown strains (Table 3).
Overall by using “MIRU-VNTR” technique, 38% of isolates could not be identified whereas by applying “MIRU-VNTR and Spoligotyping” together only 15% of the isolates remained unknown (p < 0.05).
In Table 4, the allelic differences for MIRU loci had accounted based on HGDI statistic formula; that is loci 20, 24 and 27 had lower allele difference (HGDI ≤0.3), loci 31, 39 and 40 were in middle (0.3 ≤HGDI ≤0.6) and loci 10, 16, 23 and 26 had the most allele difference among the locus MIRU (HGDI ≥0.6).
Table 4.
Allellic diversity of each MIRU-VNTR locus
| Locus | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | HGDI | |
| 2 | M | 1 | 60 | – | – | – | – | – | – | – | 0 |
| S | – | 33 | – | – | – | – | – | – | – | 0 | |
| 4 | M | – | 61 | – | – | – | – | – | – | – | 0 |
| S | – | 31 | – | – | – | – | – | – | – | 0 | |
| 10 | M | 1 | 18 | 14 | 11 | 13 | 4 | – | – | – | 70% |
| S | – | 16 | 5 | 8 | 3 | 1 | – | – | 66% | ||
| 16 | M | 4 | 16 | 32 | 8 | 1 | – | – | – | – | 64% |
| S | 2 | 2 | 18 | 10 | 1 | – | – | – | – | 59% | |
| 20 | M | 2 | 59 | – | – | – | – | – | – | – | 10% |
| S | 3 | 30 | – | – | – | – | – | – | – | 14% | |
| 23 | M | – | – | – | – | 6 | 55 | – | – | – | 16% |
| S | – | – | – | 1 | – | 32 | – | – | – | 10% | |
| 24 | M | 61 | – | – | – | – | – | – | – | – | 0.02% |
| S | 33 | – | – | – | – | – | – | – | – | 0 | |
| 26 | M | – | 2 | 2 | 6 | 24 | 13 | 8 | 5 | 1 | 78% |
| S | – | – | – | 7 | 10 | 1 | 9 | 6 | – | 76% | |
| 27 | M | – | 3 | 3 | 55 | – | – | – | – | – | 17% |
| S | – | – | 1 | 30 | 2 | – | – | – | – | 14% | |
| 31 | M | 1 | 1 | 40 | 8 | 11 | – | – | – | 51% | |
| S | – | – | – | 20 | 6 | 7 | – | – | – | 45% | |
| 39 | M | – | – | 44 | 17 | – | – | – | – | – | 42% |
| S | – | 1 | 20 | 12 | – | – | – | – | – | 52% | |
| 40 | M | 3 | 5 | 41 | 12 | – | – | – | – | – | 50% |
| S | 1 | 4 | 25 | 3 | – | – | – | – | – | 38% | |
MIRU-VNTR: Mycobacterial Interspersed Repetitive Units Variable Number Tandem Repeat; S strain (Susceptible), M strain (Resistant)
Based on Table 4 in both sensitive and resistant drug loci 10, 16 and 26 had the most allelic profile between the studied loci. Also loci 2, 4, 20 and 24 had the least allelic difference between all loci.
Discussion
This study showed that “MIRU-VNTR” had low discriminatory power when it is used alone in comparison to when it is used together with Spoligotyping. Using both techniques together decreased the unknown genetic strains to about 10% in sensitive strains and 20% in drug resistant strains. Therefore, for epidemiological studies we suggest to use “MIRU-VNTR and Spoligotyping” techniques together.
DNA fingerprinting of M. tuberculosis isolates is useful for determining the extent of recent transmission in a community and the potential risk factors for recent transmission for identifying previously unsuspected transmission, monitoring the transmission of drug-resistant strains, and confirming laboratory cross contamination (16).
In this study, based on MIRU-VNTR analysis the obtained genetic patterns of sensitive and resistance strains were as follows: 13.3% and 16.6% of them belonged to Haarlem, 10% and 16.6% of the second group belonged to Dehli/ CAS (Figure 2A). When both “MIRU-VNTR and Spoligotyping” techniques were used together, the patterns of Haarlem family reduced about 23% and Dehli/CAS was increased about 26% (Figure 2B). These strains are the most frequent patterns in Iranian population, as shown by Farnia et al in 2004 (17).
Figure 2.
A) Genotypic and characteristic of resistance and sensitive strains. MIRU-VNTR are from the MIRU-VNTR plus database, MIRU-VNTR; Mycobacterial Interspersed Repetitive Units Variable Number Tandem Repeat; B) Genotypic and other characteristics of resistant and sensitivity strains. Spoligotyping and MIRU-VNTR from the MIRU-VNTR plus database, MIRU-VNTR; Mycobacterial Interspersed Repetitive Units Variable Number Tandem Repeat; RIF: Rifampicin, INH: Isoniazid
In another study by Dou et al in 2002, out of 356 isolates, 66 could not be identified by Spoligotyping, but when he applied both “MIRU-VNTR and Spoligotyping”, the number reduced to 30 isolates (18). In a study conducted by Goyal and co-workers (on 167 patients), Spoligotyping technique identified only genetic pattern of 67 strains but when they used RFLP and Spoligotyping together, the genetic patterns of 135 strains were identified (19).
In this study the allele differences were accounted too. According to Sola et al loci 10, 16, 23, 26 and 40 were introduced as loci with the most allele polymorphism and loci 4, 20, 24 and 27 as the most poorly discriminated loci (20). Another study conducted by Roring et al in 2002 demonstrated that by using another locus [(VNTR) which their polymorphism is more], we can identify genetic patterns of strains and their families more carefully. In a study by Romano et al, RFLP and Spoligotyping were used to determine the molecular epidemiology of the strains (21). In the research performed by Skuce et al it was shown that techniques like MIRU-VNTR can be replaced by IS6110 in molecular epidemiology studies (22).
Conclusion
The results showed that in molecular epidemiologic studies of MTB, using the two techniques of “MIRU-VNTR and Spoligotyping” simultaneously could disseminate the genetic patterns of MTB strains in a much better manner as compared to using only “MIRU-VNTR” technique.
Acknowledgement
This study was supported by the Mycobacteriology Research Center of Shahid Beheshti University of Medical Sciences.
References
- 1.World Health Organization. Global tuberculosis control. WHO report 2001. WHO/CDS/TB/ 2001.287; Geneva, Switzerland: World Health Organization; 2001. [Google Scholar]
- 2.Raviglione MC, Snider DE, Kochi A. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA. 1995;273(3):220–226. [PubMed] [Google Scholar]
- 3.Drobniewski F, Balabanova Y, Ruddy M, Weldon L, Jeltkova K, Brown T, et al. Rifampin- and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg Infect Dis. 2002;8(11):1320–1326. doi: 10.3201/eid0811.020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Supply P, Lesjean E, Savine K, Kremer D, van Soolingen J, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39(10):3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groenen PM, Bunschoten EA, van Soolingen D, van Errtbden JDA. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10(5):1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 6.Kremer K, van Soolingen D, Forthingham R, Haas WH, Hermans PW, Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37(8):2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazors E, Lesjean S, Banuls L, Gilbert M, Vincent V, Gicquel B, et al. High resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. PNAS. 2001;98(4):1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive unit in a mycobacterial two-component system operon. Mol Microbiol. 1997;26(5):991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 9.Hawkey PM, Smith EG, Evan JS, Philip M, Bryan G, Mohamed HH, et al. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110 –based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J Clin Microbiol. 2003;41(8):3514–3520. doi: 10.1128/JCM.41.8.3514-3520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human mini satellite-like regions in the mycobacterium tuberculosis genome. Mol Microbiol. 2000;36(3):762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 11.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscohl J, et al. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendation for standardized nomenclature. Tuber Lung Dis. 2001;5(3):216–219. [PubMed] [Google Scholar]
- 13.Sola C, Filliol I, Legrand E, Mokrousov I, Rastogi N. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J Molecular Evol. 2001;53(6):680–689. doi: 10.1007/s002390010255. [DOI] [PubMed] [Google Scholar]
- 14.Mokrousov I, Narvskaya O, Limeschenko E, Vyazovaya A, Otten T, Vyshnevskiy B. Analysis of the allelic diversity of the mycobacterial interspersed repetitive units in Mycobacterium tuberculosis strains of the Beijing family: practical implications and evolutionary considerations. J Clin Microbiol. 2004;42(6):2438–2444. doi: 10.1128/JCM.42.6.2438-2444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer K, Arnold C, Cataldi A, Gutierez MC, Haas WH, Panaiotov S, et al. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J Clin Microbiol. 2005;43(11):5628–5638. doi: 10.1128/JCM.43.11.5628-5638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan LS, Mosher L, Diem L, Massey JP, Crawford JT. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J Clin Microbiol. 2002;40(5):1592–1602. doi: 10.1128/JCM.40.5.1592-1602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farnia P, Mohammadi F, Masjedi MR, Varnerot A, Zarifi A, Tabatabee J, et al. Evaluation of tuberculosis transmission in Tehran: using RFLP and spoligotyping methods. J Infect. 2004;49(2):94–101. doi: 10.1016/j.jinf.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Dou HY, Tseng FC, Lin CW, Chang JR, Sun JR, Tsai WS, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect Dis. 2008;8:170. doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goyal M, Saunders NA, van Embedn JD, Young DB, Shaw RJ. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35(3):647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003;3(2):125–133. doi: 10.1016/s1567-1348(03)00011-x. [DOI] [PubMed] [Google Scholar]
- 21.Romano MI, Amadio A, Bigi F, Klepp L, Etchechoury I, Nato Liana M, Morsella C, et al. Further analysis of VNTR and MIRU in the genome of Mycobacterium avium complex, and application to molecular epidemiology of isolates from South America. Vet Microbiol. 2005;110(3-4):221–237. doi: 10.1016/j.vetmic.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Skuce RA, McCorry TP, McCarroll JF, Roring M, Scott AN, Brittain D, et al. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology. 2002;148:519–528. doi: 10.1099/00221287-148-2-519. [DOI] [PubMed] [Google Scholar]