Abstract
MicroRNAs (miRNAs) are short RNA molecules which bind to target mRNAs, resulting in translational repression and gene silencing and are found in all eukaryotic cells. Approximately 2200 miRNA genes have been reported to exist in the mammalian genome, from which over 1000 belong to the human genome. Many major cellular functions such as development, differentiation, growth, and metabolism are known to be regulated by miRNAs. Proximity to other genes in the genome and their locations in introns of coding genes, noncoding genes and exons have been reported to have a major influence on the level of gene expressions in eukaryotic cells. miRNAs are well conserved in eukaryotic system and are believed to be an essential and evolutionary ancient component of gene regulatory networks. Therefore, in recent years miRNAs have been studied as a likely candidate for involvement in most biologic processes and have been implicated in many human diseases.
Keywords: Disease, Human Genome, MicroRNAs, miRNA
Background
Since the first draft of human genome was published in February 2001, many new discoveries have been made which have elucidated the complexity of the human genome and subsequently the human proteome. In the past decade, application of genomics and proteomics technologies for early detection of diseases have demonstrated that many types of diseases can be diagnosed at an early stage which would be helpful in initiation of treatment protocols at an earlier time point in the clinic. We have previously reported the application of proteomics technologies for early detection of ovarian cancer (1, 2, 3), prostate cancer (4), lymphatic vascular system (5) and drug-induced cardiac toxicities (6). Other genomics and proteomics technologies have been employed in a variety of other diseases in the past decade (7, 8).
As a result of human genome studies and the discovery of less than 25,000 genes in the human genome, a shift has occurred in the focus of research from mRNAs to noncoding RNAs as a major regulator of human genome. The presence of noncoding RNAs and its role in many human diseases make these molecules important mediators which have to be understood in medical research. We have recently reviewed the role of noncoding RNAs in cancer (9) and in the present article extend this review in a more comprehensive way to include many other types of diseases that are now known to occur in humans.
In early 90's, it was discovered that two small RNAs (instead of a protein) were the product of lin-4, a gene controlling the timing of larval development of Caenorhabditis elegans (C. elegans) (10). Later studies revealed that the longer RNA (70 nucleotides) was the precursor of the shorter RNA (22 nucleotides) which is now known as a member of the class of microRNA (miRNA) genes (11). By targeting specific mRNAs for degradation and/ or translational repression, microRNAs (miRNAs), a novel class of endo-genous, non-coding RNAs (ncRNAs) plays an important role in controlling gene expression (12). Originally it was thought that lin-4 gene expression was restricted to C. elegans, due to lack of homology with other species. However, in the year 2000 the miRNA gene (let-7) (product of lin-4 gene precursor) was discovered to target lin-41(a protein coding gene) in many species (13). Since then large amounts of microRNAs have been identified in mammals (14).
Members of ncRNAs include microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs) that are known to have well preserved functions in various species. The conservation of ncRNAs’ activities in mediating the binding of RNA-enzymes to target RNA complexes results from their specific hybridization to other nucleic acids in the cell and their targeting ability of different cellular targets (15). Such specific functions are thought to limit sequence co-variation and tendency to evolution (16). As the human genome and its functions are rapidly being deciphered, the roles of miRNAs are becoming more evident in specific cellular functions. For example, miRNAs and a large set of ncRNAs including: Air, H19, Ipw, NTT, Tsix and XIST in mammals (known as ‘‘gene regulators’’) have been proposed to have different functions ranging from imprinting to inactivation of X-chromosome in mammals (17).
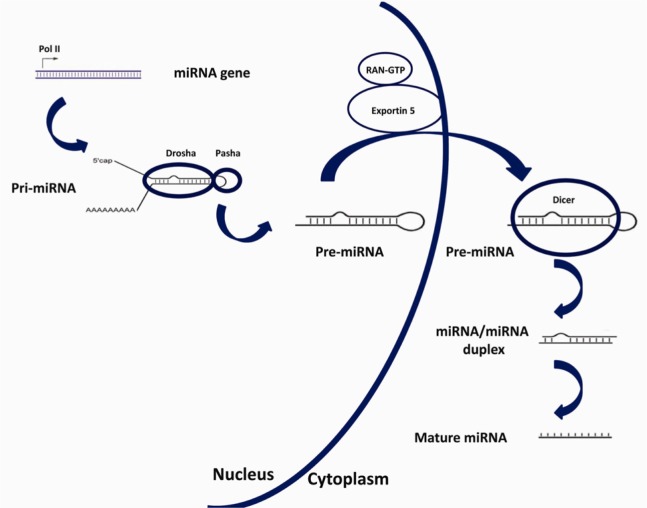
The biogenesis of miRNAs (Figure 1) involves multiple steps and specific cellular machinery (18). miRNAs are encoded as short inverted repeats having a double-stranded RNA (dsRNA) stem loop about 70 bp long and are found in both introns and intergenic clusters in the genome (18). RNA polymerase II is responsible for the synthesis of the introns and exons of both protein-coding and non-coding transcripts from where miRNAs are derived (19). In the nucleus, miRNAs are transcribed as primary pri-miRNA transcripts and then are processed to form the precursor pre-miRNA stem loop structure before transportation into the cytoplasm [where they are cleaved by the Dicer RNAase III endonu-clease and produce mature miRNA (21–23 nucleotides] (20).
Figure 1.
The biosynthesis pathway for miRNAs
MicroRNAs in Human Genome
Many major cellular functions such as development, differentiation, growth, and metabolism are regulated by miRNAs and approximately 2200 miRNA genes have been reported to exist in the mammalian genome (21). One third of the human genome is estimated to be regulated by miRNAs (22). The precise mechanisms involved in the miRNA transcription is not known but proximity to other genes in the genome and their locations in introns of coding genes, noncoding genes and exons are reported to influence their expression (23). In the genome, miRNAs are organized in clusters and share the same transcriptional regulatory units and are independently expressed if they have their own promoters (24, 25). It is estimated that about 50% of miRNAs expressed in the genome are transcribed from non-protein-coding genes and the remaining miRNAs are coded in the introns of coding genes (21).
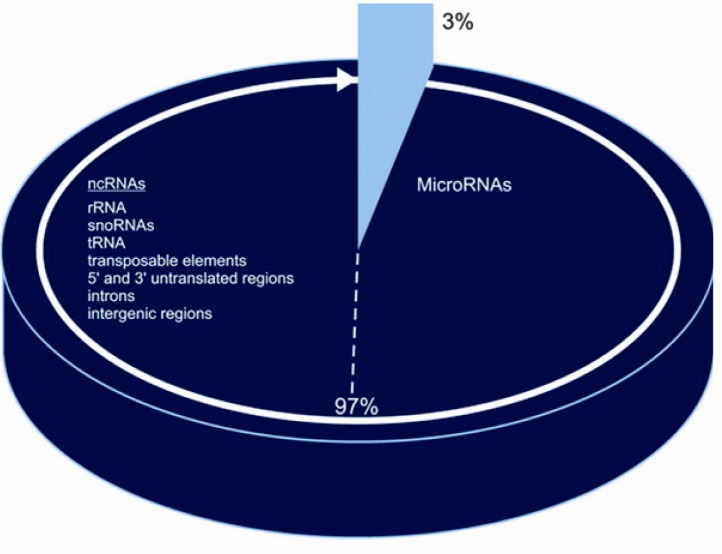
In higher eukaryotic organisms, almost the entire genome (97%) is transcribed as non-codingRNA (ncRNA) which consist of rRNA, tRNA, introns, 5’ and 3’ untranslated regions, transposable elements, intergenic regions, and microRNAs (12, 26). Recently, it has been suggested that mammalian miRNAs are derived from DNA repeats and transposons (27). Such reports have lead the scientific community to re-evaluate the functional role of transposons, especially because it appears that the specific sequences of transposons can play a major role in the developmental processes and epigenetic variations (28, 29). Furthermore, it has been recently demonstrated that miRNAs can be derived from processed pseudogenes (30), which were once believed to have no cellular functions (Figure 2).Most recent sequence analyses of the human genome demonstrates that the protein coding genes may be as low as 25,000 (31). Although the exact number of the protein coding genes in the human genome is not known, the 25,000 figure is at least 3-4 times lower than the figure believed in late 1980's. What these new data reveal is that a large segment of the human genome consists of non-coding protein genes. Further sequence analyses indicate that the Open Reading Frames (ORFs) comprise less than 2%, repetitive sequences around 46% (32, 33) and non-coding parts of protein-coding genes (introns, 5′ and 3′-UTRs) an estimated 25–27% (34) of the 3.2 billion bases in the human genome.
Figure 2.
Coding and non-coding DNA in human genome
MicroRNAs in Other Genomes
In addition to their major presence in the human genome, microRNAs have been shown to be involved in regulation of genes in higher eukaryotes (35). The rapid growth of research in the field of miRNAs is observed in the number of entries in the miRNA registry (version 1), with only 218 entries in 2002 and about 6500 entries in 2008 (version 11) (36, 37). The sequence analyses of genomes in eukaryotes indicate that simple unicellular organisms, invertebrates and mammals have 10-40%, 70–90% and 98% of their genomes composed of noncoding DNA regions, respectively (34).
The cellular functions of miRNAs appear to vary in eukaryotes, including regulation of leaf and flower development in plants (38) and modulation of differentiation of hematopoietic cells in mammals (39). The fact that many micro RNAs’ sequences are conserved among distantly related organisms indicates that miRNAs are involved in basic cellular processes (40).
MicroRNAs in Human Diseases
MicroRNAs have been been demonstrated to play a major role in a wide range of developmental processes including metabolism, cell proliferation, apoptosis, developmental timing, and neuronal cell fate (15, 41–44). Other regulatory roles include neuronal gene expression (45), brain morphogenesis (46), muscle differentiation (47), and stem cell division (48).
The role of miRNAs as a major source in the development of cancer is still very much unappreciated (49). But altered patterns of miRNAs in cells have been shown to be responsible for changes that cause cells to make a decision to turn malignant (50).
MicroRNAs are now recognized to play a pivotal role in the regulation of certain processes related to development in all eukaryotes and because of their potential role as agents controlling cell growth and differentiation, they have been proposed to be good candidates for cancer therapy (51, 52).
MicroRNAs’ deficiencies or excesses have been linked to a number of other clinically important diseases ranging from myocardial infarction to autoimmune disease. Single point mutations in miRNA or its target or epigenetic silencing of miRNA transcription units is a mechanism by which the functions of miRNA in cell are affected (53). Great discoveries and rapid progress in the past few years on miRNAs provide the hope that miRNAs will in the near future have a great potential in the diagnosis and treatment of many diseases. In the following pages, the role of miRNAs as important new regulatory molecules in different human diseases will be reviewed (54).
MicroRNAs in Cancer
It is now well documented that upregulation or down-regulation of miRNAs occurs in various human cancers (9). Over-expressed miRNAs may function as both oncogenes (through down-regulation of tumor-suppressor genes) and/ or regulator of cellular processes such as cell differentiation or apoptosis (55, 56).
Unique miRNA expression profilings have been demonstrated for many types of cancer. A list of such profiles for reproductive cancers (breast, ovary, and endometrioid adenocarcinoma) and colon, hematological cancers (AML, ALL, CML, CLL), esophagus, gastro-intestinal, lung, bladder, and thyroid tumors are shown in Tables 1 and 2, respectively (9). It has been predicted that microRNAs will have a great potential to be used in diagnosis and treatment of cancer in the near future (Tables 1–3).
Table 1.
miRNAs in reproductive cancers (9)
| Cancer type | miRNA | Up/Down Regulation |
|---|---|---|
| Breast | ||
| miR-21, miR-155, miR-23, and miR-191 | Up | |
| miR-205, miR- 145, miR-10b, and miR-125b | Down | |
| Ovary | ||
| miR-200a, miR-200c, and miR-141 | Up | |
| miR-199a, miR-140, miR-145, and miR125bl | Down | |
| Endometrioid adenocarcinoma | ||
| miR-205, miR155 miR 200a, 200b, 200c | Up | |
| miR-193a, 193b | Down | |
Table 2.
miRNAs in cancer (9)
| Cancer type | miRNA | Up/Down Regulation |
|---|---|---|
| Colon | ||
| miR-let 7g, miR-21, miR-20a, miR-17- 19 family, miR 31, miR 135, miR-181b, and miR 200c | Up | |
| miR-34, miR-let7, miR 143, miR 145, miR-133b, and miR- 126 | Down | |
| AML | ||
| Has- miR- 191, 199a, miR 155 | Up | |
| CML | ||
| miR-17-5p, miR-173p, miR- 18a, miR-19a, miR-19b-1, miR- 20a and miR- 92a-1 | Up | |
| CLL | ||
| miR-21, miR 150, miR-155 | Up | |
| miR- 15a, miR16, miR-29, miR143, miR-45, miR-30d, miR- let 7a, miR-181a | Down | |
Table 3.
miRNAs in cancer (9)
| Cancer type | miRNA | Up/Down Regulation |
|---|---|---|
| Esophagus | ||
| miR-194, miR- 192, miR-200c | Up | |
| miR- 203 | Down | |
| Gastrointestinal | ||
| miR-106b- 25 | Up | |
| miR-15b, miR- 16 | Down | |
| Lung | ||
| has-mir-21 and has-mir- 205, miR- 17-92 | Up | |
| has-mir-126*, miR-let 7, hsa-let-7a-2, let-7f-1 | Down | |
| Bladder | ||
| miR-2 23, miR- 26b, miR- 221, miR- 103-1, miR-185, miR-23 b, miR- 203,miR 17-5p, miR-23, miR- 205 | Up | |
| miR-29c, miR-26a, miR-30c, miR- 30e-5p | Down | |
| Thyroid tumors | ||
| PC | miR- 146 b, miR- 221, miR- 222, miR- 181b, miR- 155, miR- 224 | Up |
| AC | miR-30d, miR-125b, miR- 26a, miR- 30a- 5p | Down |
Endothelial cells are known to play a major role in the angiogenesis process. In recent studies (57–59) the mirRNA expression pattern (known for its tissue and cell type specificity) in endothelial cells has been demonstrated to include let-7b, miR-16, miR-21, miR-23a, miR-29, miR-100, miR-221, and miR-222. The regulation of miRNAs during patho-physiological processes has been suggested to help uncovering the role of miRNAs in vascular cells. For example in two recent reports (60, 61), the regulation of miRNAs in vascular cells in response to serum and hypoxia have been studied. In one study (60) the pro-angiogenic miR-130a is expressed at low levels in quiescent HUVEC and is upregulated in response to foetal bovine serum. In another study (61) hypoxia was shown to induce miR-210 expression in endothelial cells. The overexpression of miR-210 was reported to be associated with the enhanced formation of capillary-like structures whereas inhibition of miR-210 expression was linked to decreased tube formation and migration (62).
miR-221 and miR-222 are among the highly expressed miRNAs in HUVEC that exhibit anti-angiogenic effects (59). Similar observations were reported (57) for the anti-angiogenic function of miR-221 miR-222 in endothelial cells. This study also showed that the overexpression of miR-221 and miR-222 also indirectly reduces the expression of the endothelial Nitric Oxide Synthase (eNOS) (57) Nitric oxide (NO) is an important molecule in regulating endothelial cell growth (63), migration (64), vascular remodeling (65), and angiogenesis (66) and its impaired bioavailability is the cause of diseases including: atherosclerosis and ischemic cardiomyopathy (67). Recently it was demonstrated that eNOS also plays a major role in the mobilization and functional activity of stem cells (68–70); therefore, designing miRNAs to target eNOS is thought to regulate vasculogenesis (62).
MicroRNAs in Cardiovascular Disease
The homoeostasis of the vascular system depends on the functionality of endothelial cells and coordinated regulation of angiogenesis, vasculogenesis, and vessel regression. Little is known about the regulatory machinery at the gene expression level during neovascularization and vascular remodeling (62). However, the discovery of microRNAs in recent years has made it evident that these RNA molecules have an important function in regulation of heart function (71) and mammalian cardiovascular system in general (72).
The miRNA expression levels have been linked to deregulation of developmental processes and disease states, such as cardiac hypertrophy and failure. Many miRNAs are expressed in a tissue-cell-specific manner (73) and in adult cardiac tissue, miR-1, miR-16, miR-27b, miR-30d, miR-126, miR-133, miR-143, and the let-7 family are abundantly expressed (74). Studies have shown that three mirRNAs (miR-1, miR-133, and miR-208) are highly expressed in the heart (23, 75) and are important regulators of heart development and myocyte differentiation (72, 76–78). Recently, deregulated expression of miR-1 and miR-133 were reported in human heart failure (71, 79, 80).
Cardiac Failure
Severe cardiac remodeling with detrimental outcomes can result from disturbances in the physiological stimuli to the heart (81). One of the major responses of the heart to such stimuli is an extensive tissue remodeling known as pathological hypertrophic growth. Although the precise mechanisms involved in cardiovascular biological functions are not known, an increasing number of studies suggest that miRNAs are important regulators of cardiovascular growth, proliferation, cell differentiation, and apoptosis.
Cardiac hypertrophy in humans is a major determinant of mortality and morbidity in cardiovascular diseases. Because miRNAs are important regulators for the differentiation and growth of cardiac cells, they are hypothesized to have an important role in cardiac hypertrophy and heart failure (82). Indeed, several recent reports have found aberrant expression of miRNAs in diseased hearts and vessels. miR-23a, miR-23b, miR-24, miR-195, miR-199a, and miR-214 were upregulated during cardiac hypertrophy (Table 4), and their over-expression in cardiomyocytes in vitro caused an induction of hypertrophic growth. Interestingly, miR-24, miR-125b, miR-195, miR-199a, and miR-214 were similarly upregulated in the tissue of patients with end-stage failing human hearts (74).
Table 4.
miRNAs in human diseases
| Disease type | miRNA | Up/Down Regulation | References |
|---|---|---|---|
| Cardiac hypertrophy | |||
| miR-23a, miR-23b, miR-24, miR-195, miR-199a, and miR-214 | Up | 74 | |
| Down syndrome | |||
| miR-99a, let-7c, miR-125b-2, miR-155 and miR-802 | Up | 133 | |
| Alzheimer | |||
| miR-9, miR-128a, miR-125b | Up | 140 | |
| Rheumatic arthritis | |||
| miR-155, miR-146 | Up | 156 | |
| Systemic lupus erythematosus | |||
| miR-189, miR-61, miR-78, miR-21, miR-142-3p, miR 342, miR-299-3p, miR-198 and miR-298 | Up | 165 | |
| miR-196a, miR-17-5p, miR- 409-3p, miR-141, miR-383, miR- 112, and miR-184 | Down | ||
| Psoriasis | |||
| miR-203 | Up | 183 | |
Cardiac arrhythmias are still considered a serious health problem because of their sudden, unpredictable and potentially fatal nature. In human hearts with coronary heart disease and the rat model of myocardial infarction, it has been shown that the muscle-specific miRNA (miR-1) is upregulated in ischaemic heart tissue (61). Furthermore, the results of this study show that the injection of mature miR-1 exacerbates arrhythmogenesis, whereas inhibition of miR-1 by an antisense inhibitor suppresses arrhythmias. When the genes for the ion channels GJA1 and KCNJ2 were silenced, these proteins were shown to be important in mediating the miR-1-induced arrhythmogenic effect (61). The investigation into the role of miRNAs as a novel class of gene regulators in cardiovascular disease is a new frontier for research and it is hoped that the next decade will bring a greater understanding to their functions in cardiovascular biology (82).
MicroRNAs in Inflammatory Disease
Inflammation is an essential component of host defense system and a major response to infection and injury, which is believed to contribute to multiple acute and chronic diseases (83, 84). Recently a number of studies have investigated the role of miRNAs in vascular inflammation and leukocyte activation and their infiltration into the vascular wall. Indeed, a recent study (85) provides the first evidence that miRNAs control vascular inflammation.
In this study, miR-126 was demonstrated to inhibit the expression of vascular cell adhesion molecule 1 (VCAM-1), which is required to mediate leukocyte adherence to endothelial cells. In macrophages, miR-155 has been shown to be induced by cytokines such as TNFα and IFN-β (86, 87) and contribute to physiological granulocyte/ monocyte expansion during inflammation (88). Furthermore, miR-155 is reported to be required for B and T lymphocyte and dendritic cell function (89, 90). At the gene level, the transcription factor Pu.1 has been identified as a direct target of miR-155 in B cells (62, 90).
Another study (91) has demonstrated that miR-181 is involved in regulation of haematopoietic lineage differentiation and several miRNAs have been reported to regulate B-cell differentiation, including the miR-17-92 cluster and miR-150 (92–94). During monocyte differentiation, the transcription of miR-424 has been shown to increase and regulate the translation of the transcription factor NFI-A involved in monocyte/ macrophage differentiation (95). Moreover, miR-146 is found to be induced in macrophages by several microbial components and proinflammatory cytokines (96). Finally, the myeloid-specific miR-223 is reported to be involved in regulation of granulocyte differentiation and activation during inflammation (62, 97). The role of miRNAs in regulation of every major cell type important in modulation of the immune system indicates that they can be potentially used in immune therapies.
MicroRNAs in Neurodevelopmental Disease
MicroRNAs are highly expressed in human and other mammalian brains relative to other organs (98–100). The results of high-throughput sequencing experiments suggest that the number of miRNAs expressed in human brain should be over 1000, although currently this number stands at about 550 in all humans (101). The expression of miRNAs in brain changes during brain development. Therefore, some miRNAs are expressed more abundantly during early development in the mammalian brain, and some are expressed less during later development (102, 103). The changes in miRNA expression levels in brain during development may represent biochemical signals for cell fate determination, apoptosis and/or cell division programming (104). Studies (105) have shown that some miRNAs are differentially expressed in neuronal nuclei, and/or different cell populations in brain (106).
Because miRNAs are known to be dynamically regulated in neurogenesis and brain development (107, 108), it is believed that miRNAs are also involved in neural development and play an important role in mediating neuronal plasticity. One of the major common traits linking many of the neurodevelopmental disorders [e.g. intellectual disability, autism, Attention Deficit Hyperactivity Disorder (ADHD) and epilepsy] is that disease onset occurs during periods of maturation and development (109). Therefore, it is very likely that miRNAs contribute significantly to the pathogenesis of neurodevelopmental disorders at the molecular level (110).
As for the role of non-coding RNAs (especially mirRNAs) in human brain, it is speculated that these molecules may help the generation of complexity required for brain functions from approximately 25, 000 human protein-coding genes (106, 111, 112).
A. Fragile X syndrome
Fragile X Syndrome (FXS) is the first genetic disorder linked to the miRNA pathway (113–115). The clinical manifestations of FXS include: learning disabilities, severe cognitive or intellectual disabilities and delays in speech and language development (116). The fragile X mental retardation-1 (FMR1) gene is associated with a massive unstable CGG trinucleotide repeat expansion within the gene's 50 untranslated region (50-UTR) (117–119).
The functional FMR1 gene product, Fragile X Mental Retardation Protein (FMRP), belongs to a small and highly conserved RNA-binding protein family (120–124) and functions as a suppressor of target mRNA translation via binding of non-coding RNA structures within the UTRs of target mRNAs (110, 125–128).
Recombinant human FMRP has been shown to be able to act as an acceptor for Dicer-derived miRNAs, and significantly, endogenous miRNAs are found associated with FMRP in both flies and mammals (113–115). This interaction is presumed to regulate translation of target mRNAs and demonstrates the involvement of miRNA machinery in inducing the genetic Fragile X syndrome.
B. Rett syndrome
The X-linked dominant Rett syndrome (RTT) (129) is a progressive neurodevelopmental disorder and one of the most common causes of mental retardation and involves de novo mutations in MECP2 gene known to cause aberrations in the DNA methyl-CpG-binding protein, MeCP2 (110, 130). Interestingly, it has been demonstrated that in postnatally cultured rat neurons, miR-132 directly repress the expression of MeCP2. This observation may indicate that the role of miRNA is important in regulation of MeCP2 as a mechanism by which normal neuronal development and synaptic maturation in the postnatal brain is maintained (131). The role of MeCP2 in direct regulation of the expression of miRNA genes and the role of miRNA(s) in the pathogenesis of RTT is still being determined.
C. Down syndrome
Down Syndrome (DS) results from triplication of all or part of human chromosome 21 and affects 1 in 700 newborns and is manifested with variable phenotypes such as congenital heart defects, craniofacial abnormalities and cognitive impairment (132). Recently, bioinformatic analyses of chromosome 21 have revealed that five miRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802) are encoded on this chromosome. Interestingly, all five miRNAs have been shown to be overexpressed in fetal brain and heart tissues from DS individuals, suggesting a role in the cognitive and cardiac defects observed in DS (Table 4) (110, 133).
In general, individuals with DS exhibit lower blood pressure and lower angiotensin II type 1 receptor (AGTR1) protein levels than those without DS. Recent study (134) has reported that miR-155 downregulates AGTR1, a human gene associated with hypertension; thus providing potential evidence that miRNAs contribute to the DS phenotypes.
D. Alzheimer's disease
Dysregulated miRNA expression has been reported in the brain of Alzheimer patients (135). Specific miRNAs have been demonstrated to be linked to the pathogenesis of Alzheimer's Disease (AD). For example, loss of miR-29 cluster has been shown to be associated with increased (beta-amyloid precursor protein-converting enzyme) BACE1/ beta-secretase expression in sporadic AD patients (136). Furthermore, miR-298 and miR-328 have been identified to directly interact with the 30-UTR of the BACE1 transcript that is involved in b-amyloid production (137). Recently, the miR-20a family (i.e. miR-20a, miR-17-5p and miR-106b) has been demonstrated to regulate the expression of Amyloid Precursor Protein (APP) raising the possibility of a role for these micro RNAs during AD development (138, 139).
DNA arrays were employed to analyze and evaluate the expression of a subset of 12 miRNAs in the AD hippocampus in comparison with non-demented controls and fetal brain (140, 141). The results of the expression profiling showed that miR-9, miR-128a and miR-125b are elevated in AD hippocampus (Table 4) (140). In another study (141), cultured human fetal brain-derived primary neural (HN) cells were shown to be induced for production of Reactive Oxygen Species (ROS) in presence of metal salts such as aluminum and iron sulfates. Simultaneously, these cells were demonstrated to have increased expression of miR-9, miR-128 and to a lesser extent miR-125b, suggesting that ROSs influence AD brain through pathways specifically mediated by miRNAs. Further clarifications of the role of miRNAs in development of AD remain to be elucidated in the future.
E. Huntington's Disease
Huntington's Disease (HD) is a neurodegenerative disease resulted from CAG expansion in the gene encoding the protein huntingtin (Htt). The manifestations of HD include cognitive defects and motor control impairment which lead to neuronal dysfunction characterized by progressive loss of cortical and striatal neurons (142). How this process is regulated is not precisely known; however many potential miRNA targets have been predicted in the brains of HD sufferers. In the cortex of the mouse model of HD (R6/2), out of the seven target miRNAs which were found to be expressed after 40 cycles of qPCR, four microRNAs (mir-29a, mir-124a, mir-132 and mir-135b) displayed significant reduction in expression (143).
Interestingly, among the dysregulated miRNAs found in the cortex of the mouse model of HD, mir-124a and mir-132 are known for their important neuronal-specific activity. These results indicate that widespread and significant dysregulation of target miRNAs takes place in the brains of R6/2 animals. In tissue samples from human unaffected individuals (‘WT’) and HD-sufferers (‘HD’), target miRNAs, mir-29a, mir-124a, mir-132 and mir-330 have been detected but not shown to be significantly over or under expressed at any specific pathologic state in HD (143).
Some investigators have suggested that the results from mouse and human samples cannot be compared because they are taken from different regions and are from different species. Although no experimentally validated target mRNAs have been reported for the miRNAs in the brain of HD patients, transcriptional dysregulation of miRNAs in human HD cortex has been shown and further studies are needed to establish the association between miRNAs and HD in the clinic.
F. Schizophrenia
Schizophrenia is a neuropsychiatric disorder that approximately affects 1 in 100 individuals in a general population (144). Schizophrenia is a heritable disorder (145, 146) and Brain-Derived Neurotrophic Factor (BDNF) has been shown to play an important role in the regulation of the development and synaptic maintenance of a variety of neurons in the CNS including GABA and glutamatergic neurons (147, 148). Furthermore, BDNF may regulate type 3 dopamine receptor (DRD3) expression during brain development (149–151).
Recent bioinformatic investigations have indicated that two newly described miRNAs, hsa-mir-1 and hsa-mir-206, may target the 30-UTR of BDNF (152). This suggestion provides credence to the hypothesis that miRNA could regulate BDNF protein synthesis by interfering with BDNF mRNA translation during brain development.
MicroRNAs in Autoimmune Disease
Results from both cell culture and animal studies indicate that miRNAs have an important role in regulation of normal immune functions and prevention of autoimmunity. What is not clear yet is the precise mechanism(s) by which miRNA dysregulation could lead to pathogenesis in an autoimmune disease. In the following sections some possible roles for miRNAs in regulation of two of the most significant autoimmune diseases [i.e. rheumatoid arthritis (RA)] and Systemic Lupus Erythematosus (SLE) will be reviewed (153).
A. Rheumatoid arthritis (RA)
In a recent study (154), an abnormal expression of miRNAs was reported in patients with RA. Specifically, two microRNAs (miR-155 and miR-146) were found to be expressed at a significantly higher level in synovial tissues and synovial fibroblasts isolated from patients with RA, relative to healthy controls (Table 4). Interestingly, the levels of both these miRNAs were found significantly upregulated in synovial fibroblasts from patients with RA following TNF/ interleukin (IL)-1β stimulation (155, 156).
Evidence from mouse 3’-UTR mutational studies has implicated an interaction between miRNAs and TNF transcripts (157, 158). In the near future, the identification of candidate miRNAs that target genes implicated in rheumatic disorders should increase our molecular understanding responsible for rheumatic disease. Furthermore, Inducible Costimulator (ICOS) was recently reported to be a target of miR-101 (159), and ICOS-deficient mice are shown resistant to collagen-induced arthritis without any signs of joint tissue inflammation (156).
Pathogenesis of RA has been associated with the viral infections such as Epstein–Barr Virus (EBV), chronic Hepatitis C Virus (HCV), HIV and Kaposi's-sarcoma associated herpes (160, 161). Variety of reports indicates that viruses have the ability to encode their own miRNAs and these viral-encoded miRNAs can control the expression of viral transcripts and suppress the host immune response during infection (162). For example, miR-UL112-1 expressed by the human cytomegalovirus has been demonstrated to target the major histocompatibility complex class 1-related chain B (162). Also, miR-K12- 11 encoded by the Kaposi's-sarcoma-associated herpes virus, has been reported to down-regulate the expression of numerous similar cellular target mRNAs (163). Therefore, it is possible that viral-encoded miRNAs by targeting different host proteins can induce inflammatory arthritis (156).
Three studies have demonstrated alteration of miRNA expression in RA patients compared to controls (154, 155, 164). Specifically, increased miR-155 and miR-146a expression in RA synovial fibroblasts have been in comparison to those patients with Osteoarthritis (OA) (155).
B. Systemic Lupus Erythematosus (SLE)
SLE is a systemic inflammatory autoimmune disease with diverse clinical manifestations including photosensitivity, arthritis, glomerulonephritis, and neurological disorders. In a recent microarray analysis of miRNA expressions in SLE patients, seven miRNAs (miR-196a, miR-17-5p, miR-409-3p, miR-141, miR-383, miR-112, and miR-184) were found downregulated and nine miRNAs (miR-189, miR-61, miR-78, miR-21, miR-142-3p, miR-342, miR-299-3p, miR- 198, and miR-298) upregulated compared to healthy controls (Table 4) (165). The miRNA profiling of kidney biopsies from lupus nephritis patients showed sixty six differentially expressed miRNAs (36 upregulated and 30 downregulated) when compared to healthy controls (166). Of course, further studies are required to determine if the differential expression of these miRNA in SLE patients are reproducible (153).
MicroRNAs in Liver Disease
The liver is composed of many cell types and each cell type may have a distinct miRNA expression profile. In recent years, an increasing understanding of miRNA functions in liver physiology and disease and identification and validation of miRNA targets have gained a lot of attention (167).
A. Viral hepatitis
Viral genes encode miRNAs and these miRNAs have a regulatory effect on the viral protein-coding genes (168). Using computational approaches, Hepatitis B Virus (HBV) has been found to encode a candidate pre-miRNA, suggesting that HBV has the capacity to use viral miRNAs to regulate its own gene expression (169). miRNAs from the host cells may also play a role in regulating viral genes (168, 170, 171). It has recently been reported that miRNA-122 facilitates the replication of Hepatitis C Virus (HCV) by targeting the viral 5’ non-coding region (172).
Expression of a total of 30 cellular miRNAs in hepatocytes has been shown to be influenced by IFN-α/β or IFN-γ. Interestingly, in this study, eight of the miRNAs (miR-1, miR-30, miR-128, miR-196, miR-296, miR-351, miR-431 and miR-448) were shown to be upregulated which also have an almost perfect complementarity with HCV RNA genomes. This suggests that these miRNAs are capable of inhibiting HCV replication and infection (167).
B. Polycystic liver diseases
The polycystic liver and kidney diseases have heterogeneous etiologies and are caused by mutations in two genes, PKD1 and PKD2 (173). Autosomal Recessive Polycystic Kidney Disease (ARPKD) can present in neonates with massive renal cysts and recent study by Lee et al, 2008, provides data to support a role for miRNA. In this study, it was demonstrated that levels of the miR15a was decreased in livers of patients with ARPKD. The findings in this study indicate that changes in miRNA expression contribute to the phenotypic changes found in cystic liver disease (173).
MicroRNAs in Skeletal Muscle Disease
The muscular dystrophies are a heterogeneous group of disorders involving degeneration of skeletal muscle. Recent studies provide evidence to support a role for miRNAs in the regulatortion of muscle development. The roles of miRNAs in myogenesis have been mainly from studies on muscle-specific miR-1, miR-133 and miR-206 (174). Recently unique miRNA signatures have been found in Duchenne muscular dystrophy (175, 176).
It has been demonstrated that diagnosis of Facioscapulohumeral muscular dystrophy (FSHD) could be distinguished from Duchenne muscular dystrophy based on the level of miRNAs-381 and miRNAs-382 expressions in FSHD patients (175). Other studies have also shown a significant up-regulation of miRNAs-100, 103 and 107 in certain myopathies (176).
MicroRNAs in Skin Disease
The involvement of miRNAs in hair follicle morphogenesis, autoimmune and chronic inflammatory diseases affecting skin has been proposed (177–181). Dermal fibroblasts are important cells involved in the wound healing process (182). Analysis of the potential involvement of miRNAs in regulating the transition to proliferation has been performed and a cluster of 33 miRNAs were reported to be involved in regulation of expression of target genes required for the entry of fibro-blasts into the cell cycle and proliferation (178). A recent review has outlined the potential importance of miRNAs’ involvement in wound angiogenesis and abnormal healing sequence in chronic wounds (179, 183).
Psoriasis
Psoriasis is a chronic inflammatory skin disease and genetic and environmental factors are thought to be involved in pathogenesis of the disease. It is widely accepted that psoriasis results from impaired communications between the immune system and the structural cells of the skin (180, 184, 185). The psoriasis-associated miRNAs have been identified in the skin and miR-203 was found to be expressed more than 100-fold higher in skin compared to other organs (Table 4). Expression analyses of psoriasis-associated miRNAs (miR-203, miR- 146a, miR-21, and miR-125b) in cells that present in healthy and psoriatic skin (keratinocytes, dermal fibro-blasts, melanocytes, and leukocyte/ immune cell subsets) have been shown to have a distinctive expression pattern (183). Such studies demonstrate that psoriasis-specific miRNA are involved in the the pathogenesis of psoriasis.
Perspective
One of the most exciting findings of human genome project has been the discovery of a major class of non-coding protein RNA molecules. The findings of human genome project have demonstrated that the DNA molecules previously known as “junk DNA”, is now actively transcribed and code for miRNAs. Studies now provide evidence that miRNAs play a pivotal role in a variety of developmental processes and disease as reviewed in the present article.miRNAs as a special class of non-coding RNAs can post-transcriptionally regulate gene expression in a negative manner; therefore, it is predicted to have a great potential in diagnosis and treatment of diseases in the future. For instance, in the last few years, several techniques such as miRNA silencing, antisense blocking and miRNA modification have been considered for potential therapeutic treatment of several types of cancers. As the role of miRNAs are further clarified and established in each disease, we will surely see new methods developed to address the diagnosis and treatment of major human diseases. This new knowledge appears to have a major role in the practice of personalized medicine in the near future.
References
- 1.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. The Lancet. 2002;359(9306):572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 2.Ardekani AM, Lance A, Liotta LA, Petricoin EF. Clinical potential of proteomics in the diagnosis of ovarian cancer. Expert Rev Mol Diagn. 2002;2(4):312–320. doi: 10.1586/14737159.2.4.312. [DOI] [PubMed] [Google Scholar]
- 3.Liotta LA, Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, et al. General Keynote: proteomic patterns in sera serve as biomarkers of ovarian cancer. Gynecol Oncol. 2003;88(1 Pt 2):25–28. doi: 10.1006/gyno.2002.6679. discussion S37-42. [DOI] [PubMed] [Google Scholar]
- 4.Petricoin EF, III, Ornstein DK, Paweletz CP, Ardekani AM, Hackett PS, Hitt BA, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst. 2002;94(20):1576–1578. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- 5.Leak LV, Petricoin EF, Jones M, Paweletz CP, Ardekani AM, Fusaro VA, et al. Proteomic technologies to study disease of the lymphatic vascular system. Ann NY Acad Sci. 2002;979:211–228. doi: 10.1111/j.1749-6632.2002.tb04881.x. [DOI] [PubMed] [Google Scholar]
- 6.Petricoin EF, Rajapaske V, Herman EH, Ardekani AM, Ross S, Johann D, et al. Toxicoproteomics: serum proteomic pattern diagnostics for early detection of drug induced cardiac toxicities and cardioprotection. Toxicol Pathol. 2004;32(Suppl 1):122–130. doi: 10.1080/01926230490426516. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Conklin L, Alex P. New serological biomarkers of inflammatory bowel disease. World J Gastroenterol. 2008;14(33):5115–5124. doi: 10.3748/wjg.14.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurian S, Grigoryev Y, Head S, Campbell D, Mondala T, Salomon DR. Applying genomics to organ transplantation medicine in both discovery and validation of biomarkers. Int Immunopharmacol. 2007;7(14):1948–1960. doi: 10.1016/j.intimp.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moslemi Naeini M, Ardekani AM. Noncoding RNAs and Cancer. Avicenna J Med Biotech. 2009;1(2):55–70. [PMC free article] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 12.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian micro RNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17(3):189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 14.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci. 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 15.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Suppl 1):R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 16.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22(1):1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Suppl 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 18.Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001;13(3):302–309. doi: 10.1016/s0955-0674(00)00212-x. [DOI] [PubMed] [Google Scholar]
- 19.Morey C, Avner P. Employment opportunities for non-coding RNAs. FEBS Lett. 2004;567(1):27–34. doi: 10.1016/j.febslet.2004.03.117. [DOI] [PubMed] [Google Scholar]
- 20.Bilen J, Nan L, Bonini NM. A new role for micro RNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5(24):2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- 21. microRNA.org- http://www.microrna.org/microrna/searchMirnas.do. Accessed on December 13, 2010.
- 22.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 23.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucl Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 27.Smalheiser NR, Torvik VI. Mammalian miRNAs derived from genomic repeats. Trends Genet. 2005;21(6):322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 29.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7(4):597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Devor EJ. Primate microRNAs miR-220 and miR- 492 lie within processed pseudogenes. J Hered. 2006;97(2):186–190. doi: 10.1093/jhered/esj022. [DOI] [PubMed] [Google Scholar]
- 31.Herbert A. The four Rs of RNA-directed evolutions. Nat Genet. 2004;36:19–25. doi: 10.1038/ng1275. [DOI] [PubMed] [Google Scholar]
- 32.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 33.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 34.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochimica et Biophys Acta. 2005;1756(1):65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Bartel D. MicroRNAs: genomics, biogenesis, mechanism, function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucl Acids Res. 2008;36(Suppl 1):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74(4):296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 38.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2 like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 40.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berezikov E, Plasterk RH. Camels and zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet. 2005;14(Suppl 2):R183–R190. doi: 10.1093/hmg/ddi271. [DOI] [PubMed] [Google Scholar]
- 42.Bartel B. MicroRNAs directing siRNA biogenesis. Nat Struct Mol Biol. 2005;12(7):569–571. doi: 10.1038/nsmb0705-569. [DOI] [PubMed] [Google Scholar]
- 43.Zamore PD, Haley B. Ribo-genome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 44.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 45.Klein ME, Impey S, Goodman RH. Role reversal: the regulation of neuronal gene expression by microRNAs. Curr Opin Neurobiol. 2005;15(5):507–513. doi: 10.1016/j.conb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5732):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 47.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 48.Hatfield SD, Shcherbata HR, Fischer KA, Naka-hara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 49.Mattick JS, Igor V. Makunin. Non-coding RNA. Hum Mol Genet. 2006;15(Suppl 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 50.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 51.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 52.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochimica et Biophysica Acta. 2005;1756(1):65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Soifer HS, Rossi JJ, Sætrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Mulle CM, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009;33(4):667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang B, Pan X, Cobb GP, Anderson TA. Micro RNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 56.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumor suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 58.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of dicer and drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 59.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(3):1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine-kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation and angiogenesis. Cardiovasc Res. 2008;79(4):581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 63.Ziche M, Morbidelli L, Masini E, Amerini S, Granger HJ, Maggi CA, et al. Nitric oxide mediates angiogenesis in vivo and endothelial cell growth and migration in vitro promoted by substance P. J Clin Invest. 1994;94(5):2036–2044. doi: 10.1172/JCI117557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murohara T, Witzenbichler B, Spyridopoulos I, Asahara T, Ding B, Sullivan A, et al. Role of endothelial nitric oxide synthase in endothelial cell migration. Arterioscler Thromb Vasc Biol. 1999;19:1156–1161. doi: 10.1161/01.atv.19.5.1156. [DOI] [PubMed] [Google Scholar]
- 65.Rudic RD, Shesely EG, Maeda N, Smithies O, Segal SS, Sessa WC. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J Clin Invest. 1998;101(4):731–736. doi: 10.1172/JCI1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murohara T, Asahara T, Silver M, Bauters C, Masuda H, Kalka C, et al. Nitric oxide synthase modulates angiogenesis in response to tissue ischemia. J Clin Invest. 1998;101(11):2567–2578. doi: 10.1172/JCI1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeiher AM. Endothelial vasodilator dysfunction: pathogenetic link to myocardial ischaemia or epiphenomenon? Lancet. 1996;348(Suppl 1):S10–S12. doi: 10.1016/s0140-6736(96)98004-6. [DOI] [PubMed] [Google Scholar]
- 68.Aicher A, Heeschen C, Mildner-Rihm C, Urbich C, Ihling C, Technau-Ihling K, et al. Essential role of endothelial nitric oxide synthase for mobilization of stem and progenitor cells. Nat Med. 2003;9:1370–1376. doi: 10.1038/nm948. [DOI] [PubMed] [Google Scholar]
- 69.Iwakura A, Luedemann C, Shastry S, Hanley A, Kearney M, Aikawa R, et al. Estrogen-mediated, endothelial nitric oxide synthase-dependent mobilization of bone marrow-derived endothelial progenitor cells contributes to reendothelialization after arterial injury. Circulation. 2003;108:3115–3121. doi: 10.1161/01.CIR.0000106906.56972.83. [DOI] [PubMed] [Google Scholar]
- 70.Landmesser U, Engberding N, Bahlmann FH, Schaefer A, Wiencke A, Heineke A, et al. Statin-induced improvement of endothelial progenitor cell mobilization, myocardial neovascularization, left ventricular function, and survival after experimental myocardial infarction requires endothelial nitric oxide synthase. Circulation. 2004;110:1933–1939. doi: 10.1161/01.CIR.0000143232.67642.7A. [DOI] [PubMed] [Google Scholar]
- 71.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 73.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovas Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 75.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 76.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a micro RNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 79.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 80.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 81.Catalucci D, Latronico MV, Ellingsen O, Condo-relli G. Physiological myocardial hypertrophy: how and why? Front Biosci. 2008;13:312–324. doi: 10.2741/2681. [DOI] [PubMed] [Google Scholar]
- 82.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci. 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 83.Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 84.Silvestre JS, Mallat Z, Tedgui A, Levy BI. Post-ischaemic neovascularization and inflammation. Cardiovasc Res. 2008;78(2):242–249. doi: 10.1093/cvr/cvn027. [DOI] [PubMed] [Google Scholar]
- 85.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion. Proc Natl Acad Sci USA. 2008;105(5):1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, et al. Modulation of miR-155 and miR-125b levels following lipopolysac-charide/ TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 88.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/ microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 92.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 94.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci USA. 2007;104(50):19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 98.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, et al. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucl Acids Res. 2007;35(7):52. doi: 10.1093/nar/gkl1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–1377. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 102.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nelson PT, Baldwin DA, Kloosterman WP, Kaup-pinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8(8):971–978. doi: 10.2217/14622416.8.8.971. [DOI] [PubMed] [Google Scholar]
- 105.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21(6):1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 106.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18(1):130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 109.Ehninger D, Li W, Fox K, Stryker MP, Silva AJ. Reversing neurodevelopmental disorders in adults. Neuron. 2008;60(6):950–960. doi: 10.1016/j.neuron.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang S, Wen S, Chen D, Jin P. Small regulatory RNAs in neurodevelopmental disorders. Hum Mol Genet. 2009;18:R18–R26. doi: 10.1093/hmg/ddp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fields RD, Nelson PG. Activity-dependent development of the vertebrate nervous system. Int Rev Neurobiol. 1992;34:133–214. doi: 10.1016/s0074-7742(08)60098-7. [DOI] [PubMed] [Google Scholar]
- 112.Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle”. J Neuropathol Exp Neurol. 2007;66(6):461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 113.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 116.Warren ST, Sherman SL. The fragile X syndrome. In: Scriver CR, Beaudet AL, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic & Molecular Bases of Inherited Disease. 1st ed. New York: McGraw-Hill Companies; 2001. pp. 1257–1290. [Google Scholar]
- 117.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 118.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 119.Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 120.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 121.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1(1):109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 122.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 123.Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucl Acids Res. 2001;29(11):2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17(5):1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, et al. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107(4):489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 128.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24(33):7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 130.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 131.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 132.Epstein CJ. Down syndrome (trisomy 21) In: Scriver CR, Beaudet AL, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic & Molecular Bases of Inherited Disease. 1st ed. New York: McGraw-Hill Companies; 2001. pp. 1223–1249. [Google Scholar]
- 133.Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370(3):473–477. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 134.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 30 untranslated region: a mechanism for functional single nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 136.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alz-heimer's disease correlates with increased BACE1/ beta-secretase expression. Proc Natl Acad Sci USA. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boissonneault V, Plante I, Rivest , Provost P. Micro RNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hebert SS, Horre K, Nicolai L, Bergmans B, Papa-dopoulou AS, Delacourte A, et al. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol Dis. 2009;33(3):422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 139.Weinberg MS, Wood MJA. Short non-coding RNA biology and neurodegenerative disorders: novel disease targets and therapeutics. Hum Mol Genet. 2009;18:R27–R39. doi: 10.1093/hmg/ddp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lukiw WJ. Micro RNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 141.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101(9):1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 143.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29(3):438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 144.Perkins DO, Jeffries CD, Jarskog LF, Thomson JM, Woods K, Newman MA, et al. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sullivan PF, Kendler KS, Neale MC. Schizophre-nia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 146.Harrison PJ, Owen MJ. Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet. 2003;361(9355):417–419. doi: 10.1016/S0140-6736(03)12379-3. [DOI] [PubMed] [Google Scholar]
- 147.Egan MF, Weinberger DR, Lu B. Schizophrenia, III: brain-derived neurotropic factor and genetic risk. Am J Psychiatry. 2003;160:1242. doi: 10.1176/appi.ajp.160.7.1242. [DOI] [PubMed] [Google Scholar]
- 148.Ashe PC, Berry MD, Boulton AA. Schizophrenia, a neurodegenerative disorder with neurodevelopmental antecedents. Prog Neuropsychopharmacol Biol Psychiatry. 2001;25(4):691–707. doi: 10.1016/s0278-5846(01)00159-2. [DOI] [PubMed] [Google Scholar]
- 149.Sokoloff P, Guillin O, Diaz J, Carroll P, Griffon N. Brain-derived neurotrophic factor controls dopamine D3 receptor expression: implications for neurodevelopmental psychiatric disorders. Neurotox Res. 2002;4(7-8):671–678. doi: 10.1080/1029842021000045499. [DOI] [PubMed] [Google Scholar]
- 150.Guillin O, Diaz J, Carroll P, Griffon N, Schwartz JC, Sokoloff P. BDNF controls dopamine D3 receptor expression and triggers behavioral sensitization. Nature. 2001;411:86–89. doi: 10.1038/35075076. [DOI] [PubMed] [Google Scholar]
- 151.Perkins DO, Jeffries C, Sullivan P. Expanding the ‘central dogma’: the regulatory role of nonprotein coding genes and implications for the genetic liability to schizophrenia. Mol Psychiatry. 2005;10:69–78. doi: 10.1038/sj.mp.4001577. [DOI] [PubMed] [Google Scholar]
- 152.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian micro RNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 153.Pauley KM, Cha S, Chan EKL. MicroRNA in auto-immunity and autoimmune diseases. J Autoimmun. 2009;32(3-4):189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nakasa T, Shigeru Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of micro RNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stanczyk J, Pedrioli DML, Brentano F, Pernaute OS, Kolling C, Gay RE. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 156.Tili E, Michaille JJ, Costinean S, Croce CM. Micro RNAs, the immune system and rheumatic disease. Nat Clin Pract Rheumatol. 2008;4:534–541. doi: 10.1038/ncprheum0885. [DOI] [PubMed] [Google Scholar]
- 157.Jacob CO, Lee SK, Strassmann G. Mutational analysis of TNF-alpha gene reveals a regulatory role for thec3’-untranslated region in the genetic predisposition to lupus-like autoimmune disease. J Immunol. 1996;156(8):3043–3050. [PubMed] [Google Scholar]
- 158.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 159.Yu D, Tan AHM, Hu X, Athanasopoulos V, Simpson N, Silva DG, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 160.Leirisalo-Repo M. Early arthritis and infection. Curr Opin Rheumatol. 2005;17(4):433–439. doi: 10.1097/01.bor.0000166388.47604.8b. [DOI] [PubMed] [Google Scholar]
- 161.Callan MF. Epstein–Barr virus, arthritis, and the development of lymphoma in arthritis patients. Curr Opin Rheumatol. 2004;16(4):399–405. doi: 10.1097/01.bor.0000126149.96627.82. [DOI] [PubMed] [Google Scholar]
- 162.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JTA, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EKL. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16(12):939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 166.Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y. Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int. 2009;29(7):749–54. doi: 10.1007/s00296-008-0758-6. [DOI] [PubMed] [Google Scholar]
- 167.Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15(14):1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Berkhout B, Jeang KT. RISCy business: Micro RNAs, pathogenesis, and viruses. J Biol Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 169.Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31(2):124–126. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 170.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucl Acids Res. 2009;37:1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308(5721):557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 172.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver specific microRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 173.Chu AS, Friedman JR. A role for microRNA in cystic liver and kidney diseases. J Clin Invest. 2008;118(11):3585–3587. doi: 10.1172/JCI36870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McCarthy JJ, Esser KA. MicroRNA-1 and micro RNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 175.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eisenberg I, Alexander MS, Kunkel LM. miRNAs in normal and diseased skeletal muscle. Cell Mol Med. 2009;13(1):2–11. doi: 10.1111/j.1582-4934.2008.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and micro RNA: mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185(1-3):157–161. doi: 10.1159/000101316. [DOI] [PubMed] [Google Scholar]
- 178.Gu J, Iyer VR. PI3K signaling and miRNA expression during the response of quiescent human fibro-blasts to distinct proliferative stimuli. Genome Biol. 2006;7:R42. doi: 10.1186/gb-2006-7-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shilo S, Roy S, Khanna S, Sen CK. MicroRNA in cutaneous wound healing: a new paradigm. DNA Cell Biol. 2007;26(4):227–37. doi: 10.1089/dna.2006.0568. [DOI] [PubMed] [Google Scholar]
- 180.Sonkoly E, Wei T, Janson PCJ, Saaf A, Lundeberg L, Linder MT, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS ONE. 2007;2(7):610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 182.Krasna M, Domanović D, Tomsic A, Svajger U, Jeras M. Platelet gel stimulates proliferation of human dermal fibroblasts in vitro. Acta Dermatoven Alp Panonica Adriat. 2007;16(3):105–110. [PubMed] [Google Scholar]
- 183.Bostjancic E, Glavac D. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatoven Alp Panonica Adriat. 2008;17(3):95–102. [PubMed] [Google Scholar]
- 184.Lebwohl M. Psoriasis. Lancet. 2003;361(9364):1197–1204. doi: 10.1016/S0140-6736(03)12954-6. [DOI] [PubMed] [Google Scholar]
- 185.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]