Abstract
MicroRNAs (miRNAs) are a class of small non coding regulatory RNAs that have key functions in multiple cell processes. Deregulation of these tiny miRNAs is involved in various human diseases. MiR-155 is one of the multifunctional miRNA that its over-expression has been found to be associated with different kinds of cancer such as leukemia, breast and colon cancers. It is thought that deregulation and over-expression of this microRNA may be associated with PC12 cell proliferation. So, the aim of this study was to investigate the role of miR-155 expression on PC12 cell growth. For this reason, PC12 cells were cultured and transfected by 3 different concentration (25, 50 and 75 nmol) of either LNA anti-miR-155 or scramble antisense in 24-well plate. Then, total RNA was extracted from transfected cells. miRNA cDNAs were synthesized from isolated total RNA. In the second step, miR-155 expression level was analyzed using the quantitative real-time polymerase chain reaction (QRT-PCR). MTT test was performed to evaluate cell viability. In the next step, apoptosis assay was assessed to investigate anti miR-155 effect on PC12 cells death. Obtained results were analyzed with t-test. MTT test revealed that cell viability of transfected cells with 75 nM of anti-miR- 155 to be reduced by half of the control and scramble groups (0.5 vs. 0.97 and 0.94). Our data suggest that miR-155 over-expression is associated with PC12 cell growth. So, miR-155 down regulation by anti-miR-155 could open up new ways to restrain brain tumor growth, as anti-miR-155 causes PC12 cells to repress.
Keywords: Apoptosis, miR-155, MTT test, PC12 cells
Introduction
microRNAs are tiny regulatory molecules that have important role in various biological processes such as proliferation, cell death, differentiation and metabolism (1–5). Misregulation of these molecules is correlated with different pathogenic situations including tumorogenesis (6). In cancer, miRNAs act as oncogenes or tumor suppressors. Over expression of oncogenic miRNAs (oncomiRs) can down regulate tumor suppressors and/or other genes involved in cell differentiation, thus leading to tumor formation by stimulating proliferation, angiogenesis, and invasion. Similarly, tumor suppressor miRNAs can down regulate different proteins with oncogenic activity (7–11). Recent studies point out that miR-155 frequently up regulates in various cancers, so it is considered as oncomiR (12).
Brain tumors are created by abnormal proliferation of cells that normally exist in the brain such as neurons or spread from cancers primarily located in other organs. However, any brain tumor is inherently dangerous and life-threatening. So, there is considerable interest in identifying more effective therapeutic strategies for treatment of brain tumors (10).
Anti-miRs are chemically modified, single stranded nucleic acids designed to specifically bind to and inhibit endogenous microRNA (miRNA) molecules. Thus, they are miRNA Inhibitors. In general, anti-miRs have many benefits in cancer treatment including efficiency, precision, versatility, speed and cost-effectiveness (13–19).
Therefore in this study, we investigated the anti miR-155 effect on repression of PC12 tumor cell growth and proliferation. The PC12 is a cell line that is established from rat pheochromocytoma. PC12 cells divide and resemble sympathetic neurons, when grown in Nerve Growth Factor (NGF) containing media. Thus, the PC12 cell line is used as a model system for neuronal cells.
Materials and Methods
Cell cultures
Pc12 cell line was maintained in RPMI 1640 supplemented with 10% Fetal Bovine Serum (FBS) and 50 mg/ml penicillin/ strep-tomycin in a humidified incubator containing 5% CO2 at 37 °C. Cells were fed three times a week and passaged every 7 days.
Oligonucleotide sequences
5' fluorescent antisenses: anti-155 (CCTA TCACGATTAGCATT, EXIQON, product no:410078-04);5'fluorescent scramble siRNA (GTGTAACACGTCTATACGCCCA,EXIQON product no:199004-08). QRT-PCR primers: miR-155 forward primer (5'-TTAATGCTA ATCGTGATAGG-3'); U6 forward primer (5'- CTCGCTTCGGCAGCACACATATAC-3'),U6 reverse primer (5'-ACGCTTCACGAATT TGC GTGTC-3').
Transfection and MTT experiments
Twenty four hr before transfection 5×105 PC12 cells were seeded per well in 24 well plate and allowed to grow overnight. The cells were then transfected with three different concentration of anti-155 (25 nM, 50 nM and 75 nM, respectively) and highest concentration of scramble siRNA(75 nM) using lipo-fectamine 2000 (Invitrogene) according to the manufacture's protocol. Scramble siRNA dose not target any known miRNA in trans-criptome. These anti-miRs were purchased from Exiqon Company.
The day after transfection, medium was replaced with fresh RPMI containing 10% FBS. Cultures were incubated for the next day at 37 °C. Then, the cells were treated with MTT solution (5 mg/ml) for 4 hr. Thereafter, Dimethyl Sulfoxide (DMSO) were added to each well and absorption at wavelength of 540 nm measured by a spectrophotometer.
miRNA extraction and real-time
Total RNAs were extracted from transfected cells using biozol reagent (Bioneer Company). miRNAs cDNA were synthesized by Stratagene kit according to manufacturer's protocol. Thereafter QRT-PCR of miR-155 was performed with general reverse primer of Stratagene kit and specific forward primer for miR-155. The miR-155 expression was normalized to an endogenouse control U6. Relative expression was computed with relative standard curves for miR-155 and U6.
Apoptosis assay
Apoptosis assay was carried out in duplicate experiments using caspase-3 colorimetric activity assay kit (Millipore Company). Briefly, anti miR-155 transfections were performed as mentioned above. After 72 hr of transfection,0.5×106 cells were suspended in 100µl of chilled 1X cell lysis buffer and incubated on ice for 10 min and then centrifuged for 5 min at 10000 g. Assay mixtures were prepared in a 96-well plate. Twenty µl of 5X assay buffer and 10 µl of caspase-3 substrate (Ac-DEVD-pNA) were added to each test samples. Test samples were incubated for 2 hr at 37 °C. Then absorbance at 405 nm was measured with a microtiter plate reader. Fold increase in caspase-3 activity was calculated by comparing the OD reading from the induced apoptotic test samples with the level of the un-induced control.
Statistical analysis
The t-test was performed to investigate the differences in the obtained results of the three groups in the MTT and apoptosis analysis. Probability of 5% was assumed statistically significant.
Results
PC12 cells were divided in 3 groups: control group, scramble group that was transfected by 75 nM of scramble antisense and test group that was transfected by 25 nM, 50 nM and 75 nM of anti miR-155. Transfection efficiency was determined using inverted fluorescence microscope. Transfected cells with at least 70% efficiency were selected for further analysis. QRT-PCR analyses revealed that miR-155 expression was decreased in cells transected by different concentrations of anti-miR compared to control group (3 fold decrease for 25 nM, 5 fold decrease for 50 nM and 6.75 fold decrease for 75 nM concentrations of anti miR-155). No statically important differences were observed between miR-155 expression in control and scramble groups (Figure 1).
Figure 1.
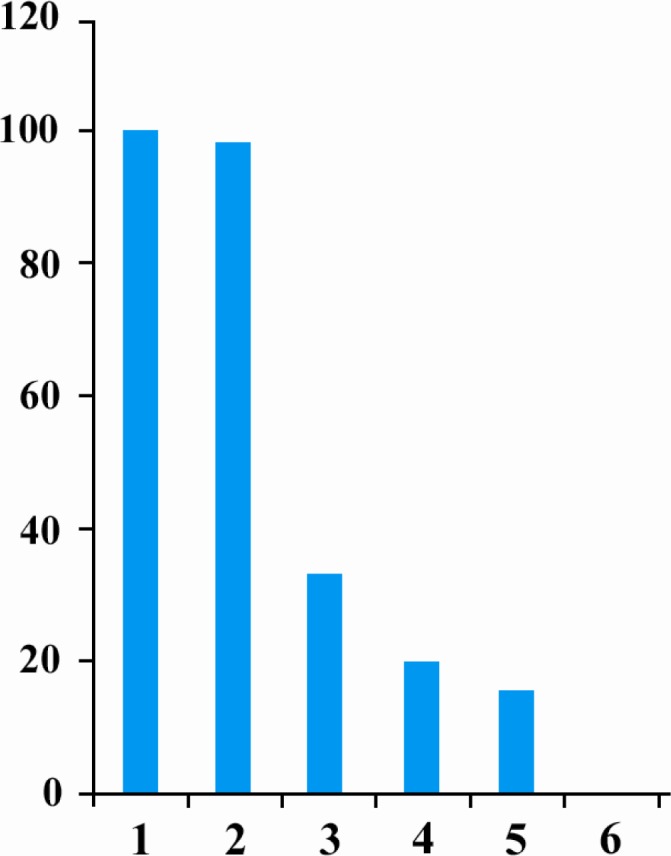
Relative Q-PCR of miR-155 level in PC12 cell line. (1) Control PC12 cells. (2) PC12 cells transfected by 75 nM of scramble. (3) PC12 cells transfected by 25 nM of anti miR-155(4) PC12 cells transfected by 50 nM of anti miR-155(5) PC12 cells transfected by 75 nM of anti miR-155
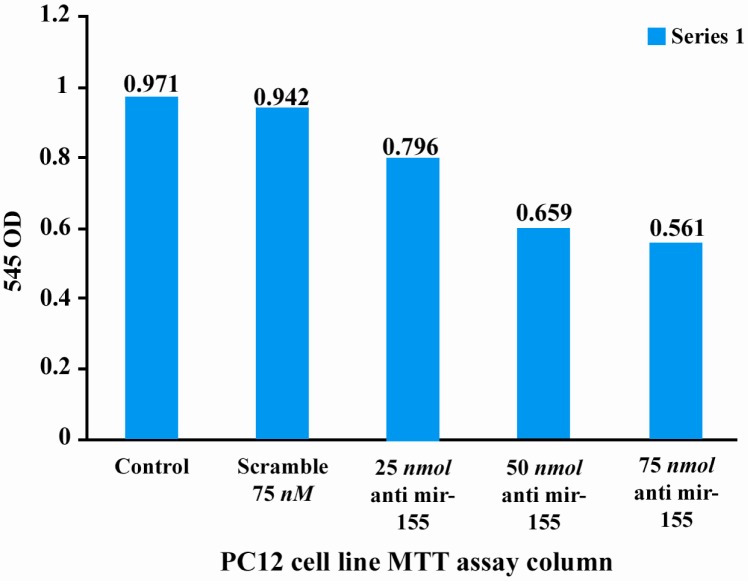
In the next step, cytotoxic effect of different concentration of anti miR-155 and scramble antisense were evaluated 48 hr after transfection by microscopy, trypan blue staining and MTT test. Optical densities at 540 nm were obtained for 3 groups: 0.971 for control group 0.962 for scramble group and 0.796, 0.659 and 0.561, respectively for 25 nM, 50 nM and 75 nM concentrations of anti miR-155 (Figure 2).
Figure 2.
MTT assay on control, scramble and test groups
These results demonstrated that optical density and therefore cell viability was similar in control and scramble groups (t = 3.60, p = 0.06). However, there were significant differences between optical density and cell viability in anti miR-155 group compared to control groups (t = 55.9723, p = 0.0003, t = 68.44, p = 0.0002, t = 182.01, p = 0.0001, respectively for 25 nM, 50 nM and 75 nM concentrations of anti miR-155).
Optical density and cell viability gradually decrease with increase in anti miR-155 concentration (0.796, 0.659 and 0.561 respectively). At 75 nM concentration of anti miR-155, optical density and cell viability were half of these parameters in control group. These data indicated that higher concentration of anti miR-155 had higher toxicity effect on PC12 cells and could decrease the cell viability and proliferation.
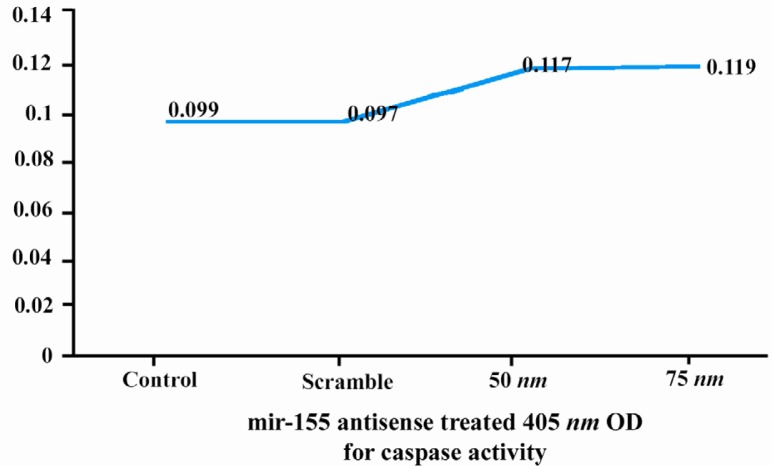
In the next step, we tried to identify the toxic effect of anti miR-155 (due to its effect on apoptosis pathway). For this reason, caspase-3 activity was calculated in 3 groups by caspase-3 colorimetric activity assay kit. Optical densities at 405 nm were 0.099 for control group, 0.097 for scramble group and 0.117 and 0.119 for anti-miR-155 group, that respectively was transfected by 50 and 75 nM of anti-miR (Figure 3). According to these data, there were no statically differences between optical density and caspase-3 activity between control and scramble groups (t = 0.44, p = 0.70) and control and anti miR-155 groups (t = 1.34, p = 0.31 for 50 nM concentration of anti miR-155 and t = 1.32, p = 0.31 for 75 nM concentration of anti miR-155).
Figure 3.
Effect of anti miR-155 on caspase-3 activity in control, scramble and test groups
Discussion
microRNAs are a class of small non-coding single-stranded RNA molecules that participate in control of gene expression by repression protein translation or by destabilizing target mRNA via cleavage or deadenylation. Any changes in the expression level of specific miRNAs are associated with multiple human diseases including diabetes, heart diseases, neurological diseases, hypertension and cancer (20–22).
Cancer is the multistep process caused by uncontrolled cell proliferation. Recent studies demonstrate that some miRNAs are considered as oncogenes or tumor suppressors and have important roles in tumor initiation, progression and metastasis (11, 22).
In 2002 for the first times, Calin and his partners indicated that down regulation of miR-15 and miR-16 was linked with chronic lymphocytic leukemia. So, these genes are thought to function as tumor suppressor (23–24).
On the other hand, some miRNAs act as oncogenes and are called oncomirs. Up regulation of these miRNAs could cause the tumor progression (25). miR-155 can be regarded as an oncomir whose expression is increased in different cancers including T cell lymphoma, leukemia, breast, thyroid and colon cancers (26–29). So, in this study, we utilized anti-miR-155 for repression of PC12 growth and proliferation. Transfection of PC12 cells with either anti miR-155 or scramble were done successfully by at least 70% efficiency. MTT test was performed and results indicated that although 25 nM and 50 nM of anti miR-155 have toxic effect, on PC12 cells, 75 nM of anti miR-155 could repress tumor cell proliferation strongly (half of the control group).
On the other hand, apoptosis analysis demonstrated that caspase-3 activity was similar in control group (0.099), scramble group (0.097) and anti miR group (0.117 for 50 nM and 0.119 for 75 nM of anti-155). Thus, anti miR-155 did not influence PC12 apoptosis. Consequently, anti miR-155 could decrease PC12 cells development, but could not induce apoptosis in these cells.
Previous studies have indicated that a number of genes that are regulated by miR-155 are involved in biological processes such as immune response (for instance Pu1 and SHIP1), inflammation (BACH1 and ZIC3) and cell growth/survival (TP53NP1, FOXO3 and GSK3β) (12, 26, 30–32)333435. Thus, miR-155 could promote cancer development and proliferation by targeting genes that control cell cycle arrest such as tumor protein 53-induced nuclear protein 1(TP53INP1), (25, 31). However, there is little information from direct targets or exact pathways through which miR-155 signals function to promote the PC12 tumor cell development and proliferation.
Conclusion
In summary, our data for the first time indicated that down regulation of miR-155 by anti-miRNA, that has sequence complementary to mature miR-155, may be a useful approach for inactivating this microRNA in brain tumors and cause slow tumor growth, development and proliferation. Meanwhile further investigation would be necessary for identification of the exact mechanism through which anti miR-155 influence the PC12 tumor cells growth and proliferation.
Acknowledgement
This study was accomplished at the University of Tarbiat Modares and supported by the Hematology department. The authors are grateful to the department for their support.
References
- 1.Abba M, Allgayer H. MicroRNAs as regulatory molecules in cancer: a focus on models defining miRNA functions. Drug Discov Today Dis Models. 2009;6(1):13–19. [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28(10):534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs-micro RNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Garofalo M, Condorelli GL, Croce C, Condorelli G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010;17:200–208. doi: 10.1038/cdd.2009.105. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13(4):253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 11.Shenouda K, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 12.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an oncomiR in breast cancer by targeting the suppressor of cyto-kine signaling 1 gene. Cancer Res. 2010;70:3119–3125. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discovery. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 14.Esau CC, Monia BP. Therapeutic potential for microRNAs. Adv Drug Deliv Rev. 2007;59(2-3):101–114. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Mei J, Gao Y, Zhang L, Cai X, Qian H, Huang H, et al. VEGF-siRNA silencing induces apoptosis, inhibits proliferation and suppresses vasculogenic mimicry in osteosarcoma in vitro. Exp Oncol. 2008;30(1):29–34. [PubMed] [Google Scholar]
- 16.Huang KH, Wu Y, Chen YT, Deng H, Lian GD, Shuai XT. PEI-PEG as a siRNA genetic vector demonstrating interference in the expression of CD44v6 protein in gastric cancer cells. Clin Oncol Cancer Res. 2010;7(3):187–192. [Google Scholar]
- 17.Cheng Q, Wang WL, Yan W, Li QL, Wang L, Wang WY. Knockdown of survivin expression by siRNA induces apoptosis of hepatocellular carcinoma cell LINE SMMC-7721. World J Gastro-enterol. 2005;11(5):756–759. doi: 10.3748/wjg.v11.i5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patutina O, Mironova N, Popova N, Kaledin V, Nikolin V, Zenkova M. The siRNA targeted to mdr1b and mdr1a mRNAs in vivo sensitizes murine lymphosarcoma to chemotherapy. BMC Cancer. 2010;10(1):204–206. doi: 10.1186/1471-2407-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Zhou Y, Xie C, Zhou F, Chen Y, Han G, et al. STAT6 specific shRNA inhibits proliferation and induces apoptosis in colon cancer HT-29 cells. Cancer Lett. 2006;243(1):38–46. doi: 10.1016/j.canlet.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh SK, Pal Bhadra M, Girschick HJ, Bhadra U. MicroRNAs-micro in size but macro in function. FEBS. 2008;275(20):4929–4944. doi: 10.1111/j.1742-4658.2008.06624.x. [DOI] [PubMed] [Google Scholar]
- 22.Xia PG. Great potential of microRNA in cancer stem cell. J Cancer Mol. 2008;4(3):79–89. [Google Scholar]
- 23.Calin GA, Dumitru CD, Shimizu M, Bichi R, Rai K. Frequent deletions and down-regulation of microRNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faraoni I, Romana F, Cardone J, Bonmassar E. miR-155 gene: A typical multifunctional micro RNA. Biochimica et Biophysica Acta (BBA) 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–17879. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor micro RNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39(2):167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 28.Rai D, Karanti S, Jung D, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet. 2008;181(1):8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56(3):248–253. doi: 10.1016/j.phrs.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]