Abstract
Over the years, the use of plastics has complicated the problem of disposal of solid wastes. One strategy to reduce plastic waste is the use of biodegradable plastics. A group of these plastics are polyhydroxyalkanoates (PHAs). To date more than 250 different microorganisms are known to synthesize and accumulate PHA. Most Pseudomonas strains are able to accumulate mcl-PHA. In previous studies, the phaC1 and phaC2 genes were identified in Pseudomonas aeruginosa (P.aeruginosa) PTCC 1310 and were cloned. The aim of this study was to express these genes and optimize the conditions for their expression. The inserts obtained from vectors pTZPHAC1 and pTZPHAC2 were subcloned into pET15b expression vector. After transformation of competent Escherichia coli (E.coli) BL21 (DE3) cells with recombinant plasmids, expression was induced using IPTG. By changing expression conditions such as IPTG concentration, time and temperature of incubation with IPTG, the expression conditions for these enzymes were optimized, and the obtained results were compared using proper statistical analysis. The PHA synthase genes were induced with IPTG and the expressed 62 kDa protein was observed and purified. By changing expression conditions, 1 mM IPTG, 37 °C and a 2 hr incubation provided the highest level of protein production in E.coli cells. These results suggest that induction condition of PhaC genes can influence expression of PHA synthase enzymes.
Keywords: PhaC1, PhaC2, Polyhydroxyalkanoate, Protein expression
Introduction
Poly (hydroxyalkanoates) (PHAs) are natural biopolymers produced by many microorganisms. These polymers are accumulated by cells as storage materials under stress conditions such as nitrogen limitation and excess supply of carbon sources (1–3).
PHAs have a wide range of applications because of their thermo-plasticity and degradability properties. Controlled drug release systems, scaffold materials in tissue engineering, absorbable surgical sutures, prosthesis and bioimplants are some of their uses in medicine (4). Due to the growing needs for these biopolymers in industrial applications, there is great interest to produce them in large amounts through recombinant DNA technology.
Although many wild-type microorganisms are known to produce PHAs, natural PHA producers have a long generation time, are often hard to lyse, and contain pathways for PHA degradation. There are several advantages of using recombinant E.coli for the production of PHAs such as fast growth, ease of lysis, very good understanding of their genetics and biochemical properties, and broad substrate utilizing abilities (5–8).
PHA synthases are the key enzymes of PHA biosynthesis. In our previous study, the PHA biosynthesis genes of P.aeruginosa PTCC 1310 (pha C1 and phaC2 genes) were cloned and sequenced (9). The aim of this study was to express these genes and optimize the conditions for their expression in E.coli.The optimization of conditions for phaC genes expression can serve as an important stage for establishing PHA production in E.coli.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains used were E.coli BL(DE3) and Pseudomonas citranolleise (P.citranolleise) (DSMZ 11735). E.coli was grown at 37°C in Luria-Bertani medium (LB). P.citranolleise was grown in Minimal Salt Medium (MSM) containing 1.5% glucose. When needed, ampicillin and Isopropyl-b-D thiogalactopyranoside (IPTG) were added to the medium.
Construction of expression plasmids
Two plasmids, pTZPHAC1 and pTZPHAC2, which contained the PHA synthase genes (phaC1 and phaC2) from P. aeruginosa PTCC 1310, were constructed from genomic DNA as described previously. (9) These plasmids were extracted and digested with restriction enzymes. The NdeI and BamHI enzymes were used to digest pTZPHAC1 and for digestion of pTZPHAC2 NdeI was used (BamHI was not used because it digests the phaC2 sequence). The obtained phaC1 and phaC2 inserts were ligated into pET15b (Novagen, USA) expression vector digested with NdeI, BamHI and NdeI, respectively. These recombinant vectors were transformed into competent E.coli BL21 (DE3) cells using the CaCl2 method (10).
Expression of phaCgenes and purification of His-tagged enzymes
For the expression of PHA synthase, a positive cell clone was inoculated into 5 ml of LB medium (100 µg/ml Amp). An overnight culture was used to inoculate (1% v/v) 50 ml of fresh LB medium in 250 ml flasks and incubated at 37°C to an OD600nm of 0.4-0.6. Then the expression of phaC genes was induced by the addition of IPTG to a final concentration of 1 mM. The expressed enzymes were purified with Ni-NTA His Bind Resin (Novagen, USA) as described by the manufac-turer.
Briefly, 1 ml of each sample was collected and centrifuged after an appropriate growth period. The pellets were resuspended in 200 µl of buffer A and clarified by centrifugation at 15000 g for 10 min and the supernatant was taken for the next steps. Fifty µl of the 50% 1X Ni-NTA His Bind Resin was added to each sample and mixed gently for 30 min at room temperature. After centrifugation of samples, the supernatant was removed and the pellet was washed two times with 250 µl of buffer C. Finally, the His-tagged enzyme was eluted with buffer E and 20 µl of each sample was electrophoresed on a 12% polyacrylamide gel using SDS-PAGE.
Optimization of conditions for phaCgenes expression
IPTG concentration, time and temperature of incubation with IPTG were optimized for phaC genes expression in E.coli BL21 (DE3) cells. For optimization of enzyme expression, IPTG concentration at four levels (0.5, 1, 1.5 and 2 mM), three different times of incubation with IPTG (1, 2 and 3 hrs) and the temperature of incubation with IPTG at three levels (25, 37 and 39°C) were examined. Each assay was repeated three times for pET-phaC1 and pET-phaC2 and one time for pET15b as the negative control (a total of 252 experiments). Expressed protein samples were analyzed on a 12% SDS-PAGE gel and the intensity of bands were quantified and compared using SPSS 10 version and differences at p<0.05 were considered as significant.
Detection of PHA accumulation
To confirm the expression of functional PHA synthase enzymes by E.coli BL21 (DE3) cells car-rying the phaC genes, the accumulation of PHA in bacteria was detected with Nile red (11). The E.coli BL21 (DE3) cells carrying the phaC gene, E.coli BL21 (DE3) without the phaC gene (as the nega-tive control) and P.citranolleise (as the positive control) were investigated for PHA production. E.coli BL21 (DE3) cells were streaked on an LB plate supplemented with sodium-gluconate (0.5% w/v), Nile red (0.05 µg/ml), IPTG (0.1 mM) and ampicillin (100 µg/ml). P.citranolleise was grown on MSM supplemented with sodium-gluconate (0.5% w/v), glucose (1.5% w/v) and Nile red (0.05 µg/ml). The agar plates were incubated at 37°C for 24 hr.
Results
Expression and purification of phaC synthase
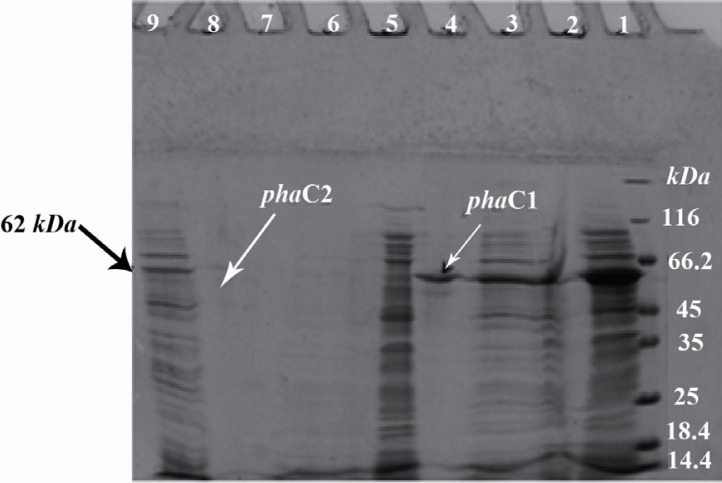
The PHA synthase enzymes were expressed by the addition of IPTG, then the cell suspension was centrifuged. The pellets were added to Ni-NTA His Bind resins. After purification, samples were analyzed by SDS-PAGE, which revealed that the target protein of 62 kD was expressed (Figure 1).
Figure 1.
Electrophoresis of the expressed phaC1 and phaC2 proteins on 12% SDS-PAGE. 1) Protein marker; 2) Total protein from induced E.coli BL21(DE3) containing pET-phaC1; 3) Total protein from induced E.coli BL21 (DE3) containing pET-phaC1 after incubation with Ni-NTA His Bind resin; 4) Purified his-tagged phaC1 protein formNi-NTA His Bind resin; 5) Total protein from induced E.coli BL21(DE3) containing pET15b without insert; 6) Total protein from induced E.coli BL21(DE3) containing pET15b without insert after incubation with Ni-NTA His Bind resin; 7) Purified sample from induced E. coli BL21 (DE3) containing pET15b without insert; 8: Purified his-tagged phaC2 protein form Ni-NTA His Bind resin; 9) Total protein from induced E.coli BL21(DE3) containing pET-phaC2 after incubation with Ni-NTA His Bind resin
Optimization of conditions for phaC gene expression
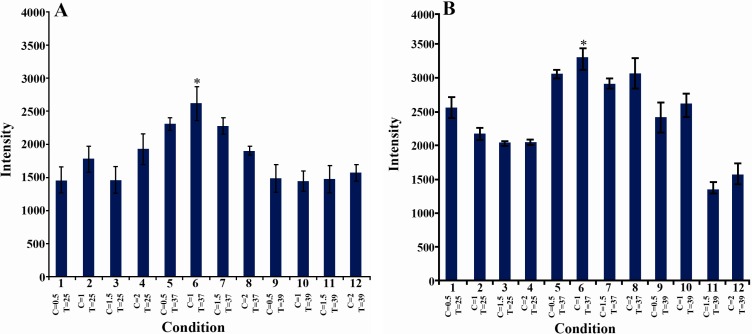
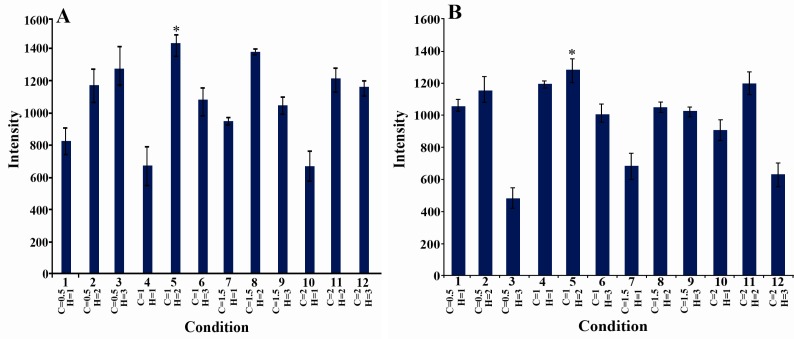
The effect of incubation temperature and IPTG concentration on SDS-PAGE band intensity of phaC1 and phaC2 were investigated and the results are shown in Figure 2. The maximum production of the PHA synthase enzymes occurred when 1 mM IPTG was used at 37°C. The effects of incubation time and IPTG concentration on SDS-PAGE band intensity of phaC1and phaC2 were evaluated (Figure 3). Maximum production of PHA synthase enzyme was observed when 1 mM IPTG was used for 2 hr.
Figure 2.
The effect of temperature of incubation with IPTG (°C) and IPTG concentration (mM) on SDS-PAGE band intensity of target protein: Error bars show standard errors of mean. C: IPTG concentration, T: temperature of incubation with IPTG. A) phaC1, the intensity of the band observed in lane 6 was significantly different from lanes 1, 3, 9,10, 11 and 12. B) phaC2, the intensity of the band observed in lane 6 was significantly different from lanes 1, 2, 3, 4, 9, 10, 11 and 12. (ANOVA, post hoc, n=3 and p<0.05)
Figure 3.
The effect of the time of incubation with IPTG (h) and IPTG concentration (mM) on SDS-PAGE band intensity of target protein: H: time of incubation with IPTG, C: IPTG concentration. A) PhaC1, the intensity of the band observed in lane 5 was significantly different from lanes 1, 4, 7 and10. B) PhaC2, the intensity of the band observed in lane 5 was significantly different from lanes 3, 7, 10 and 12. (ANOVA, post hoc, n=3 and p<0.05)
Detection of PHA accumulation
Nile red plate assay revealed functional expression of PHA synthase by recombinant E.coli BL21 (DE3) cells (Figure 4).
Figure 4.
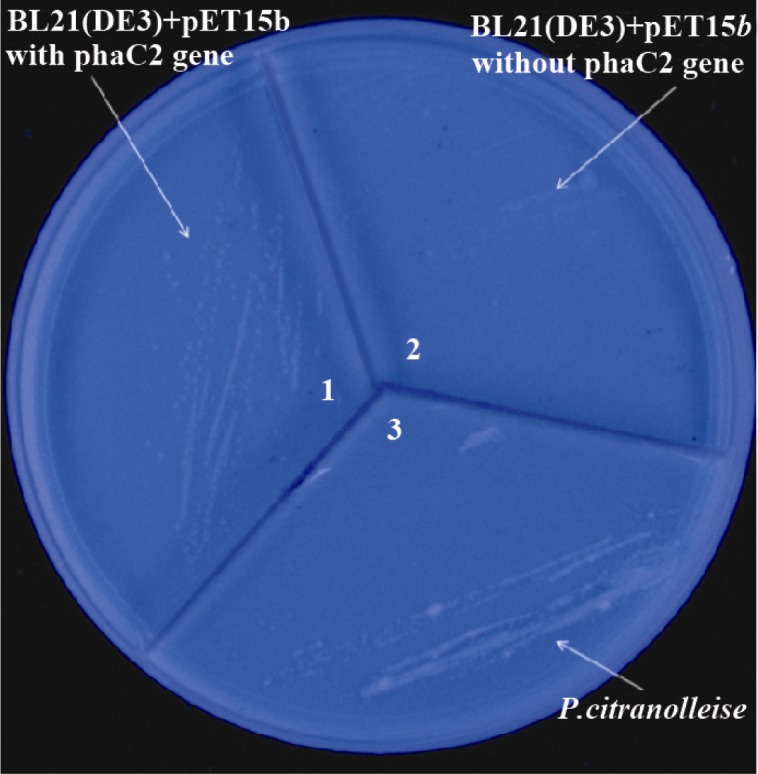
Expression of phaC gene as visualized by Nile Red plate assay.1) E.coliBL21(DE3) carrying the phaC2 gene, 2) E.coli BL21(DE3) cells without the phaC2 gene (as a negative control) and 3) P.citranolleise (as a positive control)
Discussion
Optimization of conditions for the expression of recombinant proteins has been performed in many studies. Bai et al evaluated the effect of concentration of IPTG and cell density on the level of expression of catalase (12). In another study performed by Wen and his coworkers, the effect of temperature and concentration of IPTG on the expression of PET-ILC-HIL-2-MGM was investigated (13). They found that maximum protein expression occurred when 0.3 mM IPTG was used at 42°C. Yang et al optimized four factors, including the initial cell density, concentration of IPTG, incubation temperature and induction time during the induction of the target proteins (14). According to their results, the expression of superoxide dismutase was optimal when E.coli cell density was OD600nm=0.6, IPTG concentration was 0.125 mM and incubation at 37°C was for four hours. In another research performed by Qun et al, two phaC genes from P.putida were expressed in E.coli and the effect of IPTG concentration on PHA content and PHA monomer composition was evaluated (15).
In the present study we investigated three factors: the concentration of IPTG, temperature, and time of induction of the target proteins. Our results showed that the expression of phaC1 and phaC2 with 1 mM of IPTG for 2 hr at 37°C was the optimum condition for the expression of these genes. In comparison with other studies, optimum incubation time for PHA synthase enzyme expression was shorter and IPTG concentration was greater than those studies.
Using a Nile red plate assay, the accumulation of PHA in bacteria was detected which confirmed the expression of functional PHA synthase enzymes in E.coli BL21(DE3) cells. To increase the yield of PHA production, using mutant strains of E.coli that could block the oxidation of fatty acids and accumulate greater level of PHA is also recommended (16).
Conclusion
In this study, PhaC genes from an Iranian isolate, P.aeruginosa, were expressed in E.coli and their expression was optimized. According to our findings, the concentration of IPTG, incubation temperature and incubation time during the induction with IPTG can affect the expression of these proteins. Therefore, it is now possible to continue our studies in producing these enzymes in larger quantities for industrial production of biodegradeable polymers.
Acknowledgement
This work was part of Dr Maryam Beheshti's PharmD thesis and was supported by the Isfahan University of Medical Sciences (grant no. 185085).
References
- 1.Verlinden RAJ, Hill DJ, Kenward M, Williams CD, Radecka I. Bacterial synthesis of biodegradable polyhydroxyalkanoates. J Appl Microbiol. 2007;102(6):1437–1449. doi: 10.1111/j.1365-2672.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- 2.Rehm BH, Steinbüchel A. Polyhydroxyalkanoate (PHA) synthases: the key enzymes of PHA synthesis. Biopolymers Online. 2002;3:173–215. [Google Scholar]
- 3.Rehm BH, Steinbuchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol. 1999;25(1-3):3–19. doi: 10.1016/s0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 4.Philip S, Keshavarz T, Roy I. Polyhydroxyalkanoates: biodegradable polymers with a range of applications. J Chem Technol Biotechnol. 2007;82(3):233–247. [Google Scholar]
- 5.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72(2):211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 6.Li R, Chen Q, Wang PG, Qi Q. A novel-designed Escherichia coli for the production of various polyhydroxyalkanoates from inexpensive substrate mixture. Appl Microbiol Biotechnol. 2007;75(5):1103–1109. doi: 10.1007/s00253-007-0903-2. [DOI] [PubMed] [Google Scholar]
- 7.Solaiman DKY. PCR cloning of Pseudomonas resinovorans polyhydroxyalkanoate biosynthesis genes and expression in Escherichia coli. Biotechnol Lett. 2000;22(9):789–794. [Google Scholar]
- 8.Andreessen B, Lange AB, Robenek H, Steinbuchel A. Conversion of glycerol to poly (3-hydroxypropionate) in recombinant Escherichia coli. Appl Environ Microbiol. 2010;76(2):622–626. doi: 10.1128/AEM.02097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abedi D, Najafabadi AJ, Sadeghi HM, Vallian S. Cloning and partial sequencing of phac1 and phac2 genes encoding poly(3-hydroxyalkanoate) synthases from Pseudomonas aeruginosa PTCC 1310. Biotechnology. 2007;6(4):497–504. [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrok J, Russell DW. Molecular Cloning a Laboratory Manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 11.Serafim LS, Lemos PC, Levantesi C, Tandoi V, Santos H, Reis MAM. Methods for detection and visualization of intracellular polymers stored by polyphosphate-accumulating microorganisms. J Microbiol Methods. 2002;51(1):1–18. doi: 10.1016/s0167-7012(02)00056-8. [DOI] [PubMed] [Google Scholar]
- 12.Bai Y, Zhang YL, Jin JF, Wang JD, Zhang ZS, Zhou DY. Recombinant Helicobacter pylori catalase. World J Gastroenterol. 2003;9(5):1119–1122. doi: 10.3748/wjg.v9.i5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen Q, Ma L, Wang X. Culture condition optimization of engineered E. coli BL21/pET-11c/hIL-2-mGM-CSF. Nan Fang Yi Ke Da Xue Xue Bao. 2006;26(4):418–420, 424. [PubMed] [Google Scholar]
- 14.Yang L, Yang Q, Liu PG, Li S, Li H. Optimization of the Trichoderma Harzianum Cu Zn superoxide dismutase gene expressicn in E. coli. KMITL Sci Tech J. 2007;7(2):179–184. [Google Scholar]
- 15.Ren Q, Beilen JB, Sierro N, Zinn M, Kessler B, Witholt B. Expression of PHA polymerase genes of Pseudomonas putida in Escherichia coli and its effect on PHA formation. Antonie Van Leeuwenhoek. 2005;87(2):91–100. doi: 10.1007/s10482-004-1360-x. [DOI] [PubMed] [Google Scholar]
- 16.Aldor IS, Keasling JD. Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates. Curr Opin Biotechnol. 2003;14(5):475–483. doi: 10.1016/j.copbio.2003.09.002. [DOI] [PubMed] [Google Scholar]