Abstract
Background
Recently, we have shown that peroxisomal protein expression was induced upon retinoic acid treatment in mouse embryonic stem cells during the process of neurogenesis. Thus, characterization of the respective promoter could elucidate the molecular aspects of transcriptional regulation of this gene.
Methods
Using the conventional software programs for promoter prediction, a putative promoter region was identified approximately 561 bp upstream of the peroxisomal protein coding sequence. In order to clone this region with a GC-content of 71.01%, a cocktail of ammonium sulfate buffer supplied with two additive components, betaine and dimethyl sulfoxide, and a high concentration of MgCl2 was used.
Results
The modulated polymerase chain reaction composition significantly improved the amplification of GC-rich DNA target sequences. Improved amplification of this region was due to reduction in the formation of secondary structures by the GC-rich region.
Conclusion
Therefore, this polymerase chain reaction composition could be generally used to facilitate the amplification of other GC-rich DNA sequences as verified by amplification of different GC rich regions.
Keywords: Amplification, GC-rich region, Peroxsiomal protein, Promoter
Introduction
Polymerase Chain Reaction (PCR) is a conventional method for amplification of DNA regions while templates with high percentages of GC bases including eukaryotic promoter regions are not easily amplified (1–4). In this state, PCR results in production of undesired truncated fragments instead of bona fide product (1, 5).
Several approaches have been proposed to overcome this problem including additives usage in different PCR procedures (2, 6). Different additives were utilized to amplify GC-rich regions including betaine (N,N,N- trimethylglycin), dimethyl sulfoxide (DMSO), formamide, glycerin, glycerol, dithiothreitol (DTT), Bovine Serum Albumin (BSA), non-ionic detergents such as Triton X-100, nucleotide analogs 7-deaza dGTP, ethylene glycol and 1, 2-propanediol, and reducing agents such as β-mercapto-ethanol (1–9). These additives not only prevent secondary structure formation of both templates and primers, but also increase the annealing chance of primers to plate strands at temperatures lower than the melting temperature of primers (1, 3, 5, 7, 9).
In the present study, we have utilized a combination of betaine and DMSO to amplify the putative promoter region of mouse peroxisomal protein (PeP). Mouse PeP which has been identified as a fibronectin type III domain containing protein (FNDC5; NCBI accession no: NC_000070) is supposed to undergo proteolytic cleavage and releasing to the extracellular space (10, 11). Our data have elucidated that its expression increased upon Retinoic Acid (RA) treatment during the process of neurogenesis (12).
Very recently Bostrom et al have indicated that the cleaved form of PeP named irisin is a secreting myokine which its production could be induced after exercise, may improve the glucose homeostasis by regulating the obesity (11). To find out molecular mechanisms involved in the regulation of PeP expression, a putative promoter region at position -561/+101 relative to the Transcription Start Site (TSS), upstream of the PeP gene was chosen for amplification and further cloning.
Materials and Methods
To predict a potential promoter region of mouse PeP (Accession No: NC_000070), approximately 2x104 bp upstream of the PeP gene was submitted to the Geno-matix (http://www.Geno-matix.de/en/index.html) (13) and Proscan (http://www.proscan.co.za/) (Version 1.7) software programs. DNA extraction was performed on i) one Caucasian individual after signing a consent form and ii) a heart tissue sample of NMRI mouse, which was obtained from Royan Institute (Tehran, Iran) using DNeasy Blood and Tissue Kit (Qiagen, Germany). The ethics approval to use specimen was obtained from Royan Institute Bioethics Committee (Approval ID. No: EC-90-1077). Then 857 bp upstream of the PeP gene including a part of the transcriptional region was chosen for amplification using specific primers in conventional PCR (Table 1). Then, a set of PCRs was performed to identify the optimal amplification conditions using betaine (Sigma), DMSO (Merck), glycerol (Merck), formamide (Scharlau), DTT (Sigma) (1–9). Moreover, different approaches of PCR including touch-down PCR (2, 6), slow-down PCR (2) and hot-start PCR were applied. Finally, the successful amplification condition for amplicon (875 bp), included 10x PCR buffer AMS [composed of 750 mM of tris-HCl (pH = 8.8), 200 mM of (NH4)2SO4, 0.1% Tween 20] (Cinna-Gen, Iran) and 4 mM Mg2+ (Cinnagen, Iran). To improve PCR efficiency, different concentrations of betaine (0.5 to 1 M) and DMSO (5-10% v/v) were applied. Simultaneously, similar amplification condition was repeated in Pfu buffer [200 mM of Tris-HCl (pH = 8.8), 100 mM of KCl, 100 mM of (NH4)2SO4, 1% Triton X-100 and 1 mg/ml of nuclease-free BSA] (Fermentas, Lithuania) instead of 10 x PCR buffer AMS. Touch-down PCR was carried out in an Eppendorf thermocycler (Eppendorf, Germany) with one step of initial denaturation at 95°C for 5 min followed by twenty repetitive cycles, including 94°C for 10 s, a variable annealing step and an elongation step at 72°C for 4 min. The variable annealing step was started at a temperature of 66°C which decreased by 0.5°C/ cycle to reach a temperature of 56 °C for 30 s. Subsequently, additional amplification was continued for 20 cycles with a constant annealing temperature of 56°C for 30 s. PCR was terminated after final elongation step at 72°C for 10 min.
Table 1.
Primers used for amplification of nucleotides
| Amplified DNA (species) | Primers sequence | Length of product | Product GC% | Accession No. |
|---|---|---|---|---|
| PeP Promoter (Mouse) | F: 5' GCT ATT AAT GAA AGG ACT GGT CTG AGG TCT CAG 3' R: 5' GTT GCT AGC CTA GGG TCA GGT TAG GAG AGT AGG 3' |
875 bp | 70.01% | NC_000070 |
| Androgen receptor (Exon I) (Human) | F: 5' ACC CTC AGC CGC CGC TTC CTC ATC 3' R: 5' CTG GGA TAG GGC ACT CTG CTC ACC 3' |
270 bp | 71.80% | NC_000023 |
| Eukaryotic releasing factor 3a (Exon I) (Human) | F: 5' CAT TTC TCG CTC TCT GTC CAC 3' R: 5' CTG GTC CCA GCA GTC AGG 3' |
142 bp | 75.94% | NC_000016 |
| Elongation factor 1a promoter (Human) | F: 5' GTT ATT AAT CGT GAG GCT CCG GT3' R: 5' GCC GCT AGC TCA CGA CAC CTG AA3' |
1186 bp | 60.09% | NC_000020 |
F and R, are referred as forward and reverse primers respectively. AseI (ATTAAT) and NheI (GCTAGC) restriction sites are underlined
To demonstrate whether optimized PCR condition was applicable for other GC enriched regions, similar procedure was repeated for amplification of different loci on human genome (Table 1). Secondary structures predictions of the putative promoter of PeP and other amplified loci were assessed by Mfold web server (http://mfold.Bio-info.rpi.edu/) (14).
Results
Putative promoter prediction and amplification
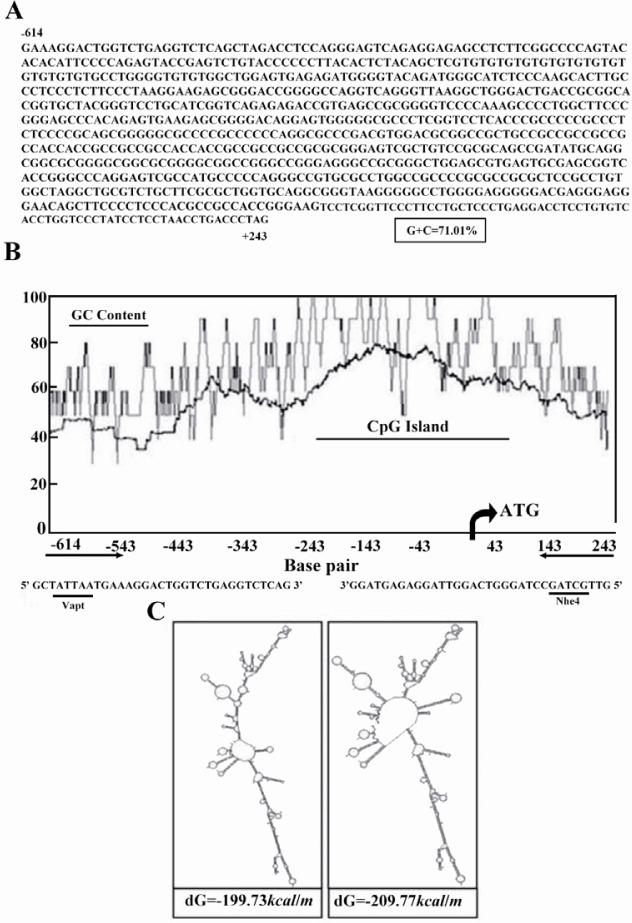
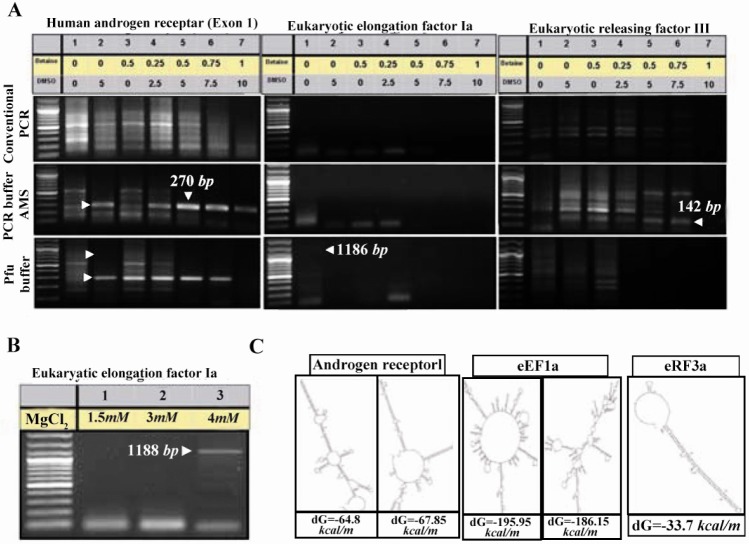
Computational analyses predicted a putative promoter region in location of -561 to +101 relative to the TSS of PeP gene (Figure 1A) with high GC content (approximately 71.01%) (Figure 1B). Secondary structure prediction of this region indicated formation of nine independent structures owing to its high internal energy in the range of -199.73 to -209.77 kcal/mol (data not shown). Two extreme structures with the highest and the lowest internal energy are shown in Figure 1C. Cloning of this region was carried out using different PCRs to obtain proper amplification. However, there were no product yields regardless of different additives (Figure 2A). Data indicated utilization of a mixture containing 10x PCR buffer AMS with different amounts of betaine (final: 0.5, 0.75 and 1 M), and 5-10% (v/v) of DMSO improved PCR yield (Figure 2B, lanes 5-7). Meanwhile, substitution of Pfu buffer with 5% (v/v) DMSO in the PCR reaction resulted in amplification of desired fragment (Figure 2C, lane 2).
Figure 1.
DNA sequence, GC and CpG nucleotide composition graph of the putative promoter of PeP gene. A) Sequence of DNA fragment consisting of PeP gene (Accession no: NC_000070) putative promoter. The initiation codon is underlined. B) Genetyx software was used to plot the GC-graph and CpG plot programs (http://www.ebi.ac.uk/Tools/es/cgi-bin/jobresults.cgi/cpgplot/cpgplot-20100823-1343338151.html). Translation start site (TSS) is located at position +21 relative to the transcription start site. Primer sequences are shown below the graph. C) Two different secondary structures predicted graphs of target DNA based on the Mfold web server (http://mfold.bioinfo.rpi.edu/) for nucleic acid folding and hybridization prediction as noted in the Materials and Methods section. The internal energy required for formation of these constructs is indicated as kcal/mole
Figure 2.
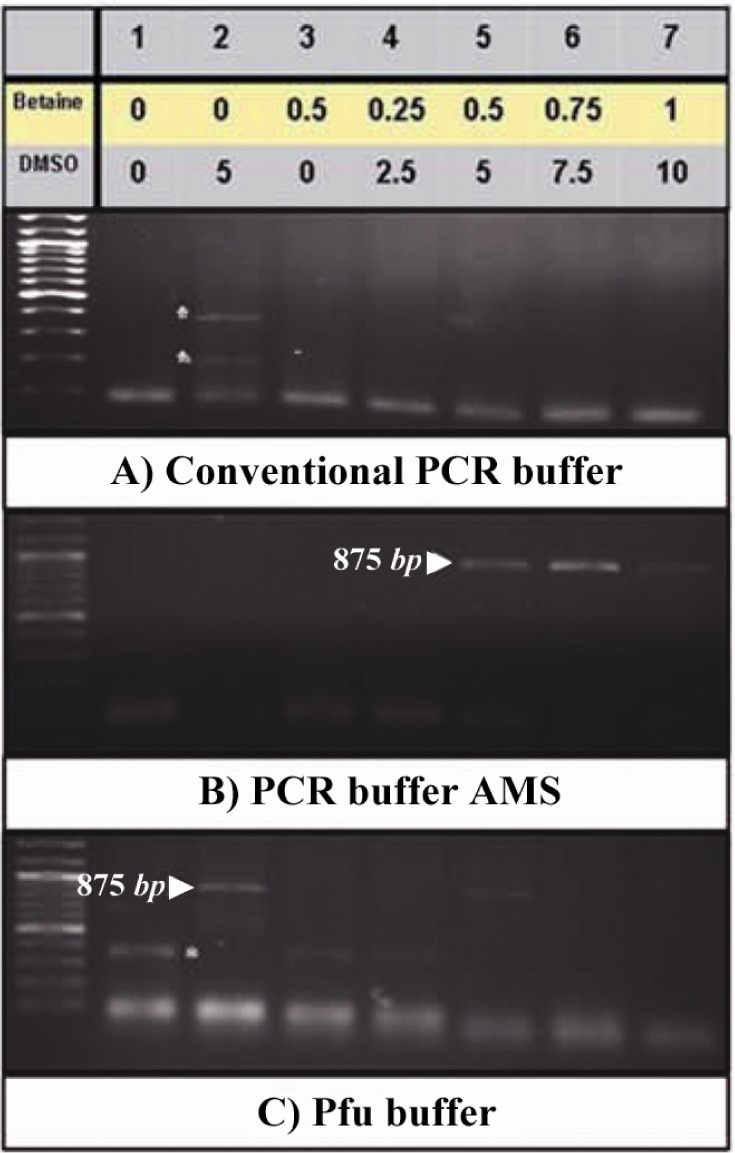
Effects of additives and PCR buffers on amplification of the putative promoter of PeP gene. Betaine (0-1 M) and DMSO (0-10% v/v) (lanes 2-8) were added to the different PCR reaction buffers to obtain amplification of the putative promoter region of the PeP gene. The desired PCR products (875 bp) are indicated by arrow heads while the star represents the nonspecifc band. Lane one represents the DNA ladder (100 bp)
Improvement of the PCR yield
Interestingly increased amounts of MgCl2 (3 and 4 mM) improved PCR yields. Particularly, PCR buffer AMS supplied with DMSO (10% v/v), betaine (final concentration: 1 M) and 4 mM of MgCl2 improved PCR efficiency (Figure 3). While, utilization of touch down PCR did not enhance the PCR product amounts compared to the conventional PCR approach (Figure 4).
Figure 3.
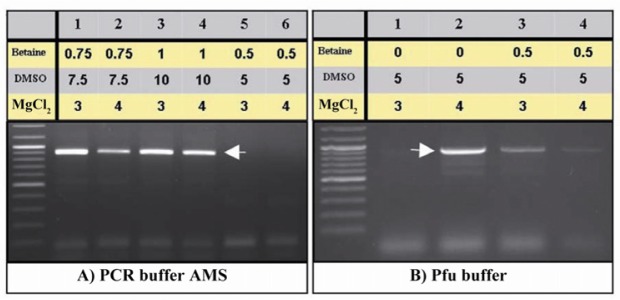
Optimization of amplification of the PeP gene putative promoter region by increasing MgCl2. The same PCR conditions of figures 2B (lanes 5-7) and 2C (lanes 2 and 5) were repeated using 3 and 4 mM MgCl2, respectively (A, B)
Figure 4.
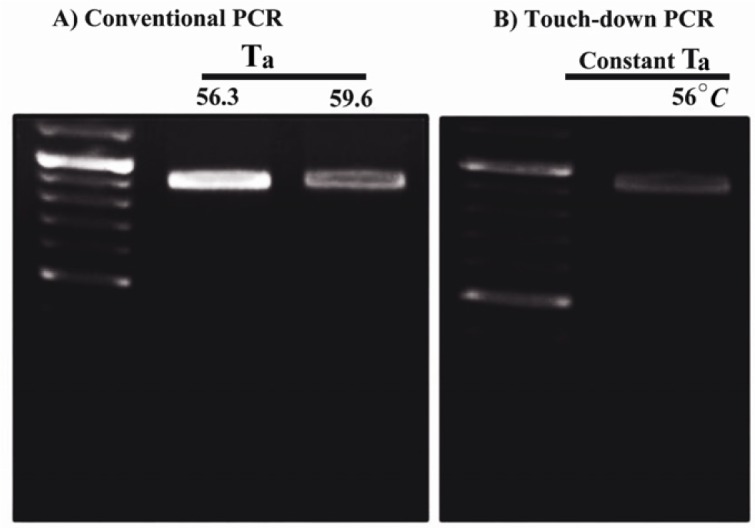
Optimized condition for amplification of the GC-rich putative promoter of the mouse PeP gene by different PCRs. The amplification of the 875 bp fragment was carried out by conventional (A) and touch-down PCR (B) using the mentioned annealing temperatures (Ta) and PCR conditions as described for figure 3a (lane 4)
To verify the efficacy of our approach, Androgen Receptor (Exon 1) and Eukaryotic Releasing Factor 3a (Exon 1) genes from a human source and a portion of human Elongation Factor 1a Promoter encompassing 71.8, 75.94 and 60.09% GC bases respectively were chosen for amplification (Table 1). The expected 270 bp PCR product band for exon 1 of human Androgen receptor was observed significantly when 5-10% DMSO, 0.5-0.75 M betaine was used in PCR buffer AMS or Pfu buffer (Figure 5A, left panel). Similarly, the amplified region of Eukaryotic Releasing Factor 3a (142 bp) was visualized when 5-7.5% DMSO, 0.5-0.75M betaine in PCR buffer AMS reaction was used (Figure 5A, right panel). Moreover, amplification for promoter region of Elongation Factor 1a was achieved when 4 mM of MgCl2 was added to the Pfu buffer (Figure 5A, middle panel and Figure 5B). Secondary structure prediction of aforementioned amplicons indicated formation of secondary structures due to presence of high internal energy (Figure 5C).
Figure 5.
A similar protocol comprising (NH4)2SO4, MgCl2, betaine and DMSO in PCR reaction was applied for efficient amplification of different loci in human genome. The same PCR reaction conditions of figure 2 was fulfilled to amplify several regions from human Androgen receptor, Eukaryotic Releasing Factor 3a, and Elongation Factor 1a (A). The same PCR conditions of figure 5A (lanes 1) was repeated using 1.5-4mM MgCl2 in Pfu buffer, to obtain a product band for Elongation Factor 1a with higher intensity (B). Two different predicted secondary structures graphs of target DNA as notified in figure 1 were obtained for nucleic acid folding and hybridization prediction. The internal energy required for formation of these constructs is indicated as kcal/mole. Please note that in case of Eukaryotic Releasing Factor 3a, only one type of secondary structure was suggested by software
Discussion
Amplification of GC-rich genomic regions requires time-consuming work to be optimized. Additives such as betaine (1, 3, 5–9), DMSO (1, 3, 5, 6, 8), form-amide (1, 3), glycerol (1, 6), glycerin (7), BSA (3), non-ionic detergents such as Triton X-100 (1), nucleotide analogs 7-deaza dGTP (5–7), DTT (3), ethylene glycol (9), 1,2-propanediol (9), reducing compounds such as β-mercapto-ethanol (3) and their combination have been utilized to optimize amplification. However, effects of these additives on amplification of different fragments are not similar (9). For instance, betaine not only acts to reduce the DNA thermal melting transition dependent on base pair composition, but also does not allow DNA polymerase to pause near hairpin-loop structures (7). Thus, a combination of betaine and DMSO make GC-rich fragments of DNA accessible for DNA polymerase (7). However, in this research no efficient amplification was seen using both betaine and DMSO. Previous reports have indicated that amplification of 71%> GC-rich DNA fragments was achieved by the addition of 16.6 mM (NH4)2SO4, 3.1 mM MgCl2 and 0.01% Tween 20 (3). In the present study, in order to amplify mouse PeP promoter with 71.01% GC, two different reaction buffers (10x PCR reaction buffer AMS and Pfu buffer) containing (NH4)2SO4 were utilized. The application of these reaction buffers resulted in efficient amplification due to reduction in the formation of secondary structure due to GC-rich regions. Computational analysis confirmed that secondary structure formation of a putative region of mouse PeP promoter was highly possible. Previous studies have shown that proper concentration of Mg2+ ion increased the productivity and fidelity of DNA polymerases due to the increased DNA/ DNA interactions and formation of complexes with dNTPs. However, higher amounts of MgCl2 may inhibit polymerase activity (15–17). In the present study, increased MgCl2 concentration (4 mM) was efficient. In some instances, modifications of the PCR procedure including a touch-down PCR is needed to gain satisfactory PCR products. However, utilization of this procedure did not yield more product than the conventional PCR. Our data demonstrated that addition of (NH4)2SO4 in PCR reaction with suitable concentrations of MgCl2, betaine and DMSO would yield the highest amount of several amplicons.
Acknowledgement
We thank Mr. Mohammad Hadi Sekhavati for his excellent technical assistance and helpful advice. This project was financed by a grant-in-aid of research from Royan Institute (Grant No. 148) to Kamran Ghaedi as the principal investigator and in support of Ms. Tahere Seifi for obtaining her MSc degree from the University of Isfahan.
References
- 1.Sahdev S, Saini S, Tiwari P, Sexena S, Saini KS. Amplification of GC-rich genes by following a combination strategy of primer design, enhancers and modified PCR cycle conditions. Mol Cell Probs. 2007;21(4):303–307. doi: 10.1016/j.mcp.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Hube F, Reverdiau P, Lochmann S, Gruel Y. Improved PCR method for amplification of GC-rich DNA sequences. Mol Biotech. 2005;31(1):81–84. doi: 10.1385/MB:31:1:081. [DOI] [PubMed] [Google Scholar]
- 3.Ralser M, Querfurth R, Warnatz HJ, Lehrach H, Yaspo ML, Krobitsch S. An efficient and economic enhancer mix for PCR. Biochem Biophys Res Commun. 2006;347(3):747–751. doi: 10.1016/j.bbrc.2006.06.151. [DOI] [PubMed] [Google Scholar]
- 4.Dutton CM, Paynton C, Sommer SS. General method for amplifying regions of high GC content. Nucleic Acids Res. 1993;21(12):2953–2954. doi: 10.1093/nar/21.12.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musso M, Bocciardi R, Parodi S, Ravazzolo R, Ceccherini I. Betaine, dimethyl sulfoxide, and 7-Deaza-dGTP, a powerful mixture for amplification of GC-rich DNA sequences. J Mol Diagn. 2006;8(5):544–550. doi: 10.2353/jmoldx.2006.060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei M, Deng J, Feng K, Yu B, Chen Y. Universal method facilitating the amplification of extremely GC-rich DNA fragments from genomic DNA. Anal Chem. 2010;82(14):6303–6307. doi: 10.1021/ac100797t. [DOI] [PubMed] [Google Scholar]
- 7.Henke W, Herdel K, Jung K, Schnorr D, Loenong SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic acid Res. 1997;25(19):3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen MA, Fukushima M, Davis RW. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PLoS One. 2010;5(6):e11024. doi: 10.1371/journal.pone.0011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Yang X, Meng L, Liu F, Shen C, Yang W. Enhanced amplification of GC-rich DNA with two organic reagents. Biotechniques. 2009;47(3):775–779. doi: 10.2144/000113203. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer-Martinez A, Ruiz-Luzanp P, Chien KR. Mouse PEP: A novel peroxisomal protein linked to myoblast differentiation and development. Dev Dyn. 2002;224(2):154–167. doi: 10.1002/dvdy.10099. [DOI] [PubMed] [Google Scholar]
- 11.Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC-α-dependent myokine that derives brown-fat-like development of white fat and thermogenesis. Nature. 2012;148(7382):463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostadsharif M, Ghaedi K, Nasr Esfahani MH, Mojbafan M, Tanhaie S, Karbalaie K, et al. Retinoic acid increased peroxisomal protein gene expression during neural differentiation. Differentiation. 2011;81(2):127–132. doi: 10.1016/j.diff.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Sobocki T, Jayman F, Sobocka MB, Marmur JD, Banerjee P. Isolation, Sequencing, and Functional Analysis of the TATA-Less murine ATPase II promoter and structural analysis of the ATPase II gene. Biochem Biophys Acta. 2007;1769(1):61–75. doi: 10.1016/j.bbaexp.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. http://www.med.yale.edu/genetics/ward/tavi/p08.html.
- 16. http://www.protocol-online.org/biology-forums/posts/4854.html.
- 17. http://wiki.answers.com/Q/why is a variable in PCR.
- 18.Brito M, Malta-Vacas J, Carmong B, Aires C, Costa P, Martins AP, et al. Polyglycine expansions in eRF3/GS PT1 are associated with gastric cancer susceptibility. Carcinogenesis. 2005;26(12):2046–2049. doi: 10.1093/carcin/bgi168. [DOI] [PubMed] [Google Scholar]
- 19.Malta-Vacas J, Chauvin C, Gonçalves L, Nazaré A, Carvalho C, Monteiro C, et al. eRF3a/GSPT1 12-GGC allele increases the susceptibility for breast cancer development. Oncol Rep. 2009;21(6):1551–1558. doi: 10.3892/or_00000387. [DOI] [PubMed] [Google Scholar]
- 20.Malta-Vacas J, Ferreira P, Monteiro C, Brito M. Differential expression of GSPT1 GGCn alleles in cancer. Cancer Genet Cytogenet. 2009;195(2):132–142. doi: 10.1016/j.cancergencyto.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Wijesekera LC, Leigh PN. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischbeck KH, Lieberman A, Bailey CK, Abel A, Merry DE. Androgen receptor mutation in Kennedy's disease. Philos Trans R Soc Lond B Biol Sci. 1999;354(1386):1075–1078. doi: 10.1098/rstb.1999.0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen CJ, Fischbeck KH. Clinical aspects and the genetic and molecular biology of Kennedy's disease. In: Wells RD, Warren ST, Sarmiento M, editors. Genetic instabilities and hereditary neurological diseases. Boston: Academic Press; 2006. pp. 211–222. [Google Scholar]
- 24.Kennedy WR, Alter M, Sung JH. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology. 1968;18(7):671–680. doi: 10.1212/wnl.18.7.671. [DOI] [PubMed] [Google Scholar]
- 25.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]