Abstract
Whole genome amplification by the multiple displacement amplification (MDA) method allows sequencing of genomes from single cells of bacteria that cannot be cultured. However, genome assembly is challenging because of highly non-uniform read coverage generated by MDA. We describe an improved assembly approach tailored for single cell Illumina sequences that incorporates a progressively increasing coverage cutoff. This allows variable coverage datasets to be utilized effectively with assembly of E. coli and S. aureus single cell reads capturing >91% of genes within contigs, approaching the 95% captured from a multi-cell E. coli assembly. We apply this method to assemble a single cell genome of the uncultivated SAR324 clade of Deltaproteobacteria, a cosmopolitan bacterial lineage in the global ocean. Metabolic reconstruction suggests that SAR324 is aerobic, motile and chemotaxic. These new methods enable acquisition of genome assemblies for individual uncultivated bacteria, providing cell-specific genetic information absent from metagenomic studies.
A myriad of uncultivated bacteria are found in environments ranging from surface ocean 1 to the human body 2. Advances in DNA amplification technology have enabled genome sequencing directly from individual cells without requiring growth in culture. These genome-centric culture-independent studies are a powerful complement to gene-centric metagenomics studies.
Genome sequencing requires that the femtograms of DNA present in a single cell be amplified into the micrograms of DNA necessary for existing sequencing technologies. Genomic sequencing from single bacterial genomes was first demonstrated 3 with cells isolated by flow cytometry, using multiple displacement amplification (MDA) 4–6 to prepare the template. MDA is now the preferred method for whole genome amplification from single cells 7, 8. The first attempt to assemble a complete bacterial genome from one cell 9 further explored the challenges of assembly from MDA DNA, including amplification bias and chimeric DNA rearrangements. Amplification bias results in orders of magnitude difference in coverage 3, and absence of coverage in some regions. Chimera formation occurs during the DNA branching process by which the phi29 DNA polymerase generates DNA amplification in MDA 10, but increased sequencing coverage helps to alleviate this problem.
Single cell sequencing methods have enabled investigation of novel uncultured microbes 11–13. However, while recent studies have continued to improve assemblies 14–18, the full potential of single cell sequencing has not yet been realized. The challenges facing single cell genomics are increasingly computational rather than experimental 17. All previous single cell studies used standard fragment assembly tools 19, 20, developed for data models characteristic of standard (rather than single cell) sequencing. These algorithms are not ideal for use with non-uniform read coverage. Most existing fragment assembly tools implicitly assume nearly uniform coverage, and most produce erroneous contigs (linking non-contiguous genomic fragments) when the rate of chimeric reads (or chimeric read pairs) exceeds a certain threshold. Thus, there is a need to adapt existing fragment assembly tools for single cell sequencing.
We developed a specialized software tool for assembling sequencing reads from single cell MDAs. Applying it to assemble single cell datasets from two known genomes and an unknown marine genome yielded valuable assemblies that identified the majority of genes, with no efforts to close gaps and resolve repeats.
Results
Velvet-SC: improved assembly of single cell short reads with highly non-uniform coverage
There are basically two key algorithmic paradigms in fragment assembly: the overlap-layout consensus approach, which dominated assembly projects with Sanger reads, and the de Bruijn graph approach, which dominates next generation sequencing (NGS) assembly projects. Most existing NGS assemblers follow a two-stage procedure 21: error correction (correcting errors in reads prior to assembly) and assembly of the error-corrected reads using a “de Bruijn graph.” A de Bruijn graph (see Fig. 1c) represents each read as a series of k-mers (k consecutive bases) by rolling a k-base window along the length of the read. The k-mers are represented as “vertices” (shown as small circular points), and consecutively overlapping k-mers are represented by “edges” (shown as short lines). Reads will share vertices and edges when they have at least k consecutive bases in common. This construction reveals reads with shared k-mers and builds contigs in an efficient manner.
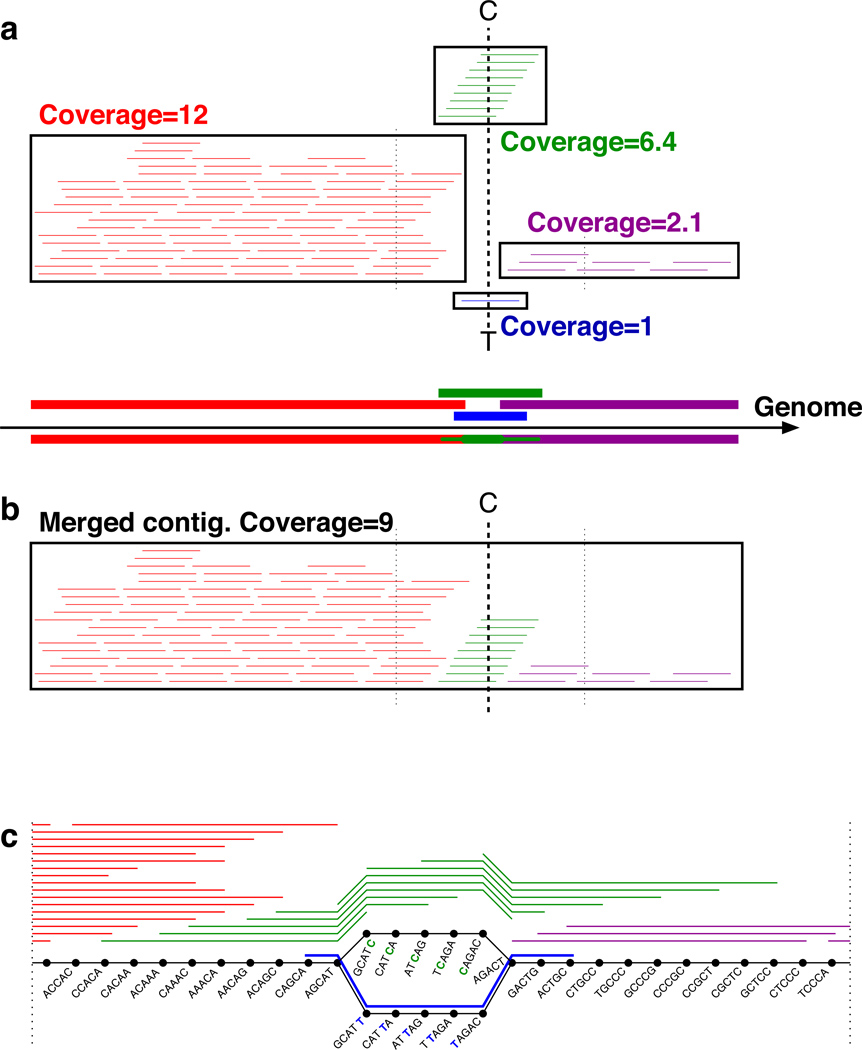
Figure 1.
Assembling single cell reads using Velvet-SC. (a) Coverage varies widely along the genome, between 1 and 12 in this cartoon example. Reads (short lines) and potential contigs (thick lines; boxes around the supporting reads) are positioned along the genome, with a box around the reads supporting each contig. There are two potential contigs to choose from in the middle, differing by a single nucleotide (C vs. T): a green contig with coverage 6.4, and a blue contig with coverage 1. With a fixed coverage threshold of 4, Velvet would delete the low coverage blue and purple contigs, and then merge the high coverage red and green contigs into a contig much shorter than the full genome. Velvet-SC instead starts by eliminating sequences of average coverage 1, which only removes the blue contig. The other contigs are combined into a single contig (b) of average coverage 9. The purple region is salvaged by Velvet-SC because it was absorbed into a higher coverage region coverage threshold increased. Velvet-SC repeats this process with a gradually increasing low coverage threshold. (c) A portion of the de Bruijn graph for the contigs described in (a). The black circles are the “vertices” and represent 5-mer strings derived from the reads, which are indicated by colored lines alongside the chains of vertices, including a blue read with an erroneous T. The lines between the vertices are termed “edges” and represent the overlaps between the 5-mers. The edges are directed from left to right in this example. The read with the C/T mismatch results in two alternative paths for assembly, both with 5 intermediate vertices. The lower of the two paths arises from the erroneous blue read and has coverage 1; it is the only part of the graph eliminated by Velvet-SC, leaving a single chain of vertices that gives a single contig for the entire genome. See Supplementary Figure S3 for an example of the condensing of contigs. An example of Velvet-SC handling of a chimeric read is presented in Supplementary Figure S4.
Non-uniform coverage poses a serious problem for existing de novo assembly algorithms. Of de Bruijn based assemblers21, for example, Velvet and ABySS 22 use an average coverage cutoff threshold for contigs to prune out low coverage regions, which tend to include more errors, while EULER-SR 23 uses a k-mer coverage cutoff. This pruning step significantly reduces the complexity of the underlying de Bruijn graph and makes the algorithms practical. Single cell read datasets (detailed below) have highly variable coverage, and a single coverage cutoff prevents assembly of a significant portion of the data (see assembly statistics in Table 1, Fig. 2). Supplementary Figure S2 plots the percentage of positions with given coverage. In the multicell E. coli dataset, most positions in the genome have coverage between 450–800x, and pruning by a coverage threshold helps eliminate erroneous reads; only 0.1% of positions have coverage below 450x. In contrast, in the single cell E. coli dataset (lane 1), 5% of positions have < 10x coverage and 11% of positions have < 30x coverage. Assembly of reads from a standard multicell sample by current NGS assemblers usually requires at least 30x coverage of a region for a successful assembly without gaps.
Table 1.
Comparison of assemblies of known genomes (for contigs > 110 bp): number of contigs, genome N50, the length of the largest contig, total nucleotides in the assembly, substitution error rate in the assembled contigs (per 100 kbp), number of genes completely or partially present in the assembly, and number of operons completely or partially present in the assembly. Partial means that a gene and a contig (or an operon and a contig) have an overlap of at least 100 nucleotides. Best by each criteria is indicated in bold. EULER-SR 2.0.1, Velvet 0.7.60, Velvet-SC, and EULER + Velvet-SC were run with k-mer size equal to 55. Edena 2.1.138 was run with a minimum overlap of 55. SOAPdenovo 1.0.4 39 was run with k=27–31. E+V-SC stands for EULER + Velvet-SC. Gene annotations were from http://www.ecogene.org/ (E. coli) and http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=ntsa10 (S. aureus). Operon annotations (E. coli) were from http://csbl1.bmb.uga.edu/OperonDB/displayNC.php?id=215 40. Some of the contigs in the single cell assemblies represent contaminants.
| Dataset | Assembler | # contigs |
N50 (bp) |
Largest (bp) |
Total (bp) |
Subs. Error (per 100 kbp) |
Known genes |
Complete genes |
Partial genes |
Predicted operons |
Complete operons |
Partial operons |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
E. coli lane 1 |
EULER-SR Edena SOAPdenovo Velvet Velvet-SC E + V-SC |
1344 1592 1240 428 872 501 |
26662 3919 18468 22648 19791 32051 |
140518 44031 87533 132865 121367 132865 |
4369634 3996911 4237595 3533351 4589603 4570583 |
16.1 2.6 98.3 3.0 4.4 2.7 |
4324 | 3178 2425 3021 3055 3617 3753 |
627 1112 612 170 325 185 |
884 | 553 317 520 584 643 713 |
248 444 248 106 184 109 |
|
E. coli lane 6 |
EULER-SR Edena SOAPdenovo Velvet Velvet-SC E + V-SC |
1820 1536 1397 522 945 481 |
29551 4899 20319 18410 27113 36581 |
170385 42342 204730 168533 144462 173901 |
4469152 4147566 4576388 3753818 4688759 4668135 |
16.6 3.2 48.3 4.1 3.8 1.7 |
3339 2705 3353 3131 3779 3943 |
734 1075 646 253 409 158 |
561 368 586 566 694 749 |
283 428 244 145 161 101 |
||
|
E. coli lane normal |
EULER-SR Edena SOAPdenovo Velvet Velvet-SC E + V-SC |
295 1673 192 408 350 339 |
110153 3814 62512 31503 52522 54856 |
221409 20470 172567 129378 166115 166115 |
4598020 4611645 4529677 4569225 4571760 4571406 |
3.5 3.8 26.8 1.6 1.1 1.5 |
4119 3019 4128 4061 4121 4124 |
115 1189 81 139 157 66 |
788 317 802 760 804 808 |
80 538 53 108 58 57 |
||
| S. aureus | EULER-SR Edena SOAPdenovo Velvet Velvet-SC E + V-SC |
4398 1288 2470 625 1084 355 |
7247 1881 5385 15800 20163 32296 |
66549 37770 37397 67677 76884 107657 |
3376776 2358911 3273188 2807042 3001635 2962136 |
53.1 3.0 42.9 6.2 4.2 4.7 |
2622 | 1958 1222 482 2244 2100 2408 |
640 925 1740 268 458 173 |
- | nd | nd |
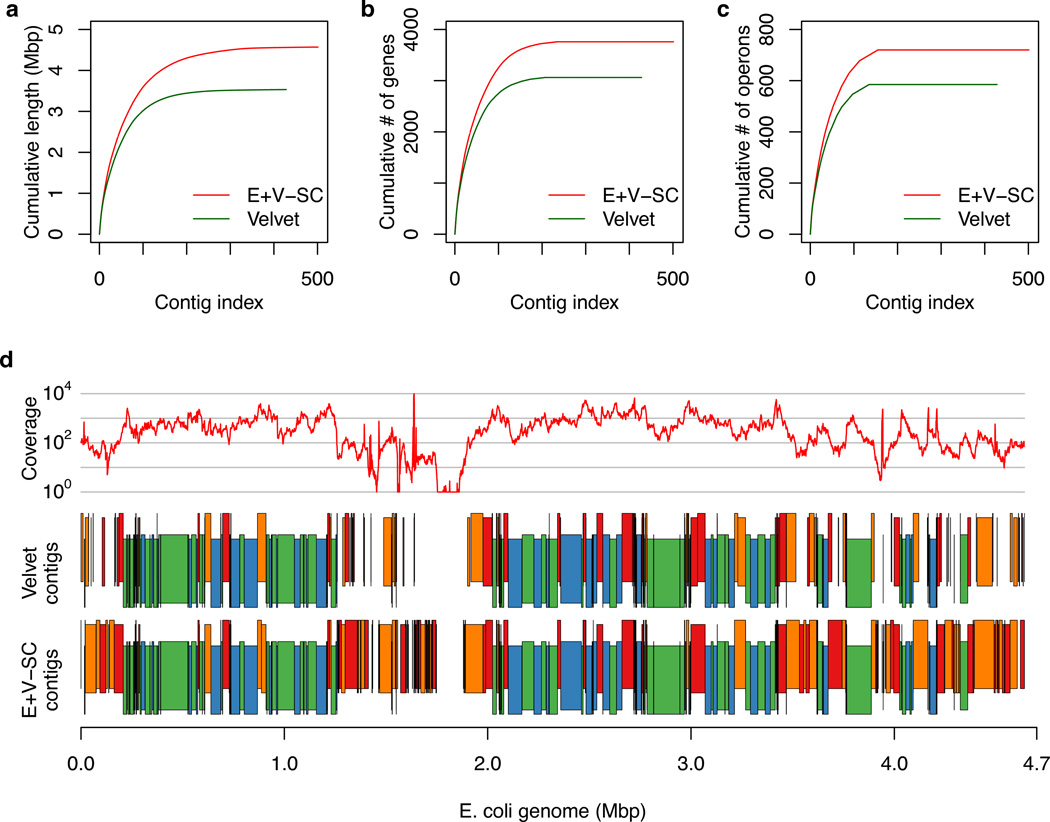
Figure 2.
Comparison of contigs generated by Velvet vs. EULER+Velvet-SC for single cell E. coli lane 1. (a,b,c) Contigs are those presented in Table 1 and are ordered from largest to smallest number of bases. The y-axis shows (a) the cumulative length, (b) the cumulative number of genes, and (c) the cumulative number of operons in the contigs. EULER+Velvet-SC improves upon Velvet in all three plots. (d) Average read coverage over a 1000 bp window (top, log scale), Velvet contigs (middle) and EULER+Velvet-SC contigs (bottom), mapped along the E. coli reference genome, with vertical staggering to help visualize small contigs. Contigs in blue or green match between the assemblies. Contigs in red or orange differ between the assemblies: they either have substantially different lengths, are broken into a different number of contigs, or are present in one assembly but missing in the other.
We developed the EULER+Velvet-SC algorithm specifically for single cell assembly. Velvet-SC (“Velvet Single Cell”; source code available in Supplementary Data 1 and at http://bix.ucsd.edu/singlecell/) is a modification of the popular open source assembly program Velvet that incorporates lower coverage sequences that most existing assemblers discard. Briefly, instead of pruning out low average coverage contigs of the de Bruijn graph based on a fixed cutoff, Velvet-SC uses a variable cutoff that starts at 1 and gradually increases. After the lowest coverage contigs are removed based on the current cutoff, some contigs may merge into a larger contig, whose average coverage is recomputed. This tends to incorporate low coverage contigs into higher coverage regions, sparing them from deletion. This process is iterated with a gradually increasing cutoff. See Methods, Figure 1 legend, Supplementary Methods, and Supplementary Figures S3-S5 for further explanation of Velvet and Velvet-SC. EULER+Velvet-SC combines the error correction from EULER-SR (source code available in Supplementary Data 2 and at http://bix.ucsd.edu/singlecell/) with Velvet-SC to further improve the assembly.
Characteristics of single cell sequences
Single cell amplified DNAs display a range of genome representation due to amplification bias, template quality, and presence of contaminating DNAs as discussed previously 3, 17. For this study, sets of E. coli and S. aureus single cell amplified DNAs prepared in parallel from clonal populations were evaluated for genome representation using qPCR for 10 loci (Methods), as described previously 3. In some amplified DNAs a few loci were detected and in some all were detected (data not shown). We chose two E. coli and one S. aureus amplified DNAs with all loci detected for this study. An average of 93% of reads from the two single E. coli cells and one S. aureus cell mapped to the respective reference genomes (Supplementary Table S1) vs. 99% of the reads in the E. coli standard dataset. Nonmapping reads in MDA datasets can often be attributed to minor contaminating sequences 17. Analysis of read filtering, read mapping, and read pairs is presented in the Supplementary Tables S2 and S3 (also see Supplementary Data 3). Chimeric fragments (where the ends map to different regions of the genome) were 2% of the E. coli read pairs and 0.5% of the S. aureus read pairs (Supplementary Table S3; Supplementary Data 3). These data are consistent with previous data regarding chimeras in MDA sequence datasets 9, 10.
The single cell datasets (reads available at http://bix.ucsd.edu/singlecell/) display highly non-uniform coverage typical of single cell amplification 3 (Supplementary Figs. S1 and S2; Supplementary Table S4), including “blackout regions,” which are contiguous regions of the genome to which no reads aligned (coverage 0). Single cell sequencing at ~600x depth results in 94 and 50 blackout regions for E. coli lane 1 and lane 6, respectively, while sequencing of unamplified DNA results in no blackout. Genome regions with coverage 0 or 1 comprise ~116 kbp in E. coli lane 1 and ~13 kbp in lane 6. There are only two small blackout regions in the S. aureus ~2300x coverage dataset, comprising just 143 bases. These observations illustrate the substantial variability in coverage even for MDAs generated from single cells processed in parallel from the same culture (lane 1 vs. 6). As evident with the two E. coli datasets, blackout regions can potentially be eliminated by combining reads from multiple single cells when available 3,13, 16.
De novo single cell assembly of E. coli and S. aureus
De novo assemblies generated by Velvet, Velvet-SC (Velvet Single Cell), and EULER+Velvet-SC (EULER-SR’s error correction followed by Velvet-SC), were compared with those generated by several other assemblers (Table 1, Fig. 2, and Supplementary Fig. S6). The metrics compared were the percentage of the genome present in the final assembly (in terms of bases, genes, and operons); N50 (the contig length at which all longer contigs represent half of the total genome length); and substitution error rate per 100 kbp. We note that the single cell assemblies include some contigs that do not map to the E. coli or S. aureus genomes. As both nonmapping and mapping reads are informative with regard to assembler functionality, and for simplicity in presenting the data, we have not removed them from the analysis.
We find that EULER+Velvet-SC outperforms Velvet-SC, while Velvet-SC outperforms Velvet. For example, from the E. coli single cell lane 1 data set EULER+Velvet-SC assembled 4.57 Mb of contigs while Velvet assembled only 3.53 Mb. EULER+Velvet-SC assembled with an N50 of 32.1 kbp compared to an N50 of 22.6 kbp with Velvet. EULER+Velvet-SC achieved an error rate of 2.7 mismatches per 100 kbp compared to an error rate of 3.0 mismatches per 100 kbp with Velvet. For single cell S. aureus reads, Velvet assembled 2.81 Mb of contigs with an N50 of 15.8 kbp (6.2 mismatches per 100 kb) whereas EULER+Velvet-SC assembled 2.96 Mb of contigs with an N50 of 32.3 kbp (4.7 mismatches per 100 kb).
Single cell assembly of E. coli (lane 6) with EULER+Velvet-SC captured 91.2% of E. coli genes and 84.7% of E. coli operons in single contigs, slightly less than 95.4% and 91.4% respectively captured in a multicell E. coli assembly. Single cell assembly of S. aureus captured 91.8% of S. aureus genes in single contigs. EULER+Velvet-SC captured sequences from 2 of the 3 plasmids in this S. aureus strain 21 (pUSA02, 4439 bp in one contig; pUSA03, 37136 bp in 30 contigs) while Velvet only captured sequences from one plasmid. The EULER+Velvet-SC assembly of E. coli had no misassembled contigs as determined by BLAST analysis (also visualized in Supplementary Fig. S7), while the assembly of S. aureus had one misassembled contig. The ability of the Velvet-SC algorithm to extend contigs into regions of low coverage is illustrated in Figure 2 and Supplementary Figure S8. In Figure 2, we highlight the improvement of EULER+Velvet-SC over Velvet on single cell E. coli lane 1, in capturing more bases, genes, and operons, and in assembling regions with low read coverage.
By these tests, EULER+Velvet-SC outperformed the other assemblers, generating higher quality single cell assemblies.
To test the effect of sequencing with lower coverage, we randomly selected a fraction of the input reads ranging from 0.1 to 0.9 of the total and assembled them with both EULER+Velvet-SC and Velvet (Supplementary Fig. S9). As expected, for single cell E. coli datasets (lanes 1 and 6) increased coverage gives better results, and EULER+Velvet-SC outperforms Velvet for total bp assembled at all coverage depths. Assembly of half the reads allowed capture of 3579 of the 3753 complete genes captured in lane 1 using all the reads, with a similar result for lane 6 (Supplementary Fig. S9 and data not shown), suggesting sequencing effort could be diminished by half.
Single cell assembly of an uncultured Deltaproteobacterium
To demonstrate the performance of EULER+Velvet-SC with an uncultivated organism, a genome of a marine bacterium was sequenced from a single cell isolated from a marine sample collected at La Jolla, CA (see Methods). Single cell MDA reactions were screened by 16S PCR and an uncultured SAR324 Deltaproteobacterium was chosen for testing the de novo assembly methods. Like the reference cells, the SAR324_MDA reads (reads available at http://bix.ucsd.edu/singlecell/) were from a 100 bp paired end run of the Illumina GA pipeline, and 57,816,790 of 67,995,232 reads passed the Illumina purity filter.
Assembly statistics
As expected EULER+Velvet-SC outperforms Velvet and Velvet-SC in single-cell assembly of the uncultured Deltaproteobacterium (Table 2), with increased N50 and decreased total number of contigs. The ability of the assemblies to support ORF prediction was tested using MetaGene 22, a program designed for annotation of metagenomic sequences that uses less stringent criteria than traditional annotation tools. The decreased number of ORFs from Velvet to Velvet-SC to EULER+Velvet-SC suggests that both the increased bp incorporation and the EULER-SR error correction reduce spurious ORF calls. The EULER+Velvet-SC ORFs were of higher quality as evidenced by increased numbers of ORFs with taxonomic affiliations identified using BLAST and phylogenetic analysis via the Automated Phylogenetic Inference System (APIS, see Methods), by increased numbers of ORFs corresponding to orthologous genes in the COG database 23, and by increased numbers of single copy conserved genes detected. By all these criteria, EULER+Velvet-SC yields the most robust assembly for annotation.
Table 2.
Comparison of Velvet-based assembler results (k=55) on SAR324_MDA assembly: total number of contigs; assembly N50 (for contigs > 110 bp); length of the largest contig (for contigs > 110 bp); total nucleotides in the assembly (for contigs > 110 bp); number of ORFs >20 bp predicted by MetaGene22; number of ORFs with phylogenetic assignments by APIS (see Methods); number of ORFs with COGs identified via BLAST (see Methods); and number of 111 conserved single copy genes present 30. N50 is defined as the contig length such that using the same length or longer contigs produces half of the total assembly length.
| Assembler | # of contigs |
N50 (bp) | Largest (bp) |
Total (bp) |
# ORFs (MetaGene) |
# ORFs (APIS) |
# COGs | # Conserved single copy genes |
|---|---|---|---|---|---|---|---|---|
| Velvet | 1856 | 11531 | 100589 | 3921396 | 4575 | 2462 | 2160 | 55/111 (46%) |
| Velvet-SC | 933 | 23230 | 113282 | 4284882 | 4234 | 2627 | 2307 | 75/111 (67%) |
| E +V-SC | 823 | 30293 | 113282 | 4282110 | 4154 | 2604 | 2281 | 75/111 (67%) |
Assembly purity
Single cell MDAs may sometimes contain DNA from other organisms originating from the MDA reagents or the biological source material, as discussed in 17. Contaminants can potentially be identified from reads or contigs by analysis of GC content, nucleotide frequencies, and BLAST analysis vs. reference bacterial genomes. The purity of the SAR324_MDA assembly was assessed as follows (see Methods for details). BLAST analysis of contigs revealed that all top BLAST hits for contigs >500 bp were to uncultured marine organisms (data not shown), supporting the novelty of the single cell genome, its marine origin, and an absence of known DNA contaminants. Principal component analysis of nucleotide frequencies of the contigs examined in three-dimensional space is consistent with a single genome being present (Supplementary Fig. S10). A plot of the GC content of the reads forms a unimodal distribution, also consistent with the presence of a single genome (data not shown).
APIS was used to assess the purity of the genome. APIS attempts to generate a phylogenetic tree for each ORF based on BLAST analysis against reference genomes (see Supplementary Methods). Contaminating contigs might be revealed as having ORFS with phylogeny distinct from the rest of the assembly. Interestingly, the contigs tend to consist of ORFs with a variety of phylogenies, although putative operons with shared phylogeny are present, as expected. While clustering of ORFs with phylogenies distinct from the rest of a genome can be indicative of horizontal gene transfer, in this case, we suggest the variety of phylogenies may be due to divergence of the SAR324 genome from available reference genomes. Alternatively, it could be a characteristic of Deltaproteobacterial genomes, as similarly varied ORF phylogenies have been observed for other Deltaproteobacteria 24 (and J.H. Badger, unpublished).
SAR324_MDA Deltaproteobacterium genome: insights from single cell sequencing
Phylogenetic analysis of 16S sequences (Fig. 3) revealed that this organism is a member of the deeply branched and divergent clade of uncultured deltaproteobacteria designated SAR324 25. Spatial and temporal studies of oceanic bacterial diversity show that SAR324 is cosmopolitan, appearing in both surface and deep ocean 26, 27. Although sequences of several diverse 30–40 Kb fosmids for SAR324 are available (representative 16S fosmid sequences from 27 included in Fig. 2), the lack of even a draft reference genome for SAR324 prevents elucidation of its ecophysiological role. Pleocystis pacifica, the closest cultured relative for which a complete genome is available, as noted by 28, is an obligate aerobe with a chemoheterotrophic lifestyle 29, and phenotypic characteristics of myxobacteria. However, the significant phylogenetic distance between the SAR324 clade and P. pacifica suggests that the latter may have limited relevance to SAR324. Genome assembly attained using a culture-independent approach represents the best possibility for elucidating the ecological role of SAR324.
Figure 3.
A 16S maximum likelihood tree of Deltaproteobacterial 16S sequences including SAR324_MDA (red). Sequences with species identification are from representative Deltaproteobacterial reference genomes in GenBank. The environmental 16S sequences (designated uncultured SAR324 or uncultured deltaproteobacteria) were retrieved from GenBank based on their accession numbers (see Fig. S3 of 27). The sequences were aligned using MOTHUR 36. The tree was inferred using the nucleotide maximum likelihood feature of PAUP* 4.0b10 37. Branches drawn in thick lines are clades with bootstrap support of 75% or greater. Sequences present on fosmids with extensive nucleotide similarity to the SAR324_MDA assembly are indicated (red star), as is a SAR324 fosmid (yellow star) encoding CoxL homologs also present in the SAR324_MDA assembly (see Supplementary Fig. S13).
The SAR324_MDA assembly (assembly available at http://bix.ucsd.edu/singlecell/) includes about 4.3 Mb of non-redundant contigs yielding 3811 ORFs (Table 3). We searched the ORFs for two sets of conserved genes typically found in single copy within bacterial genomes, which can be used to estimate genome size 30–32. Seventy-five of a set of 111 (67%) conserved single copy genes 30 were represented, and 58 out of 66 (87%) single copy gene clusters 31, 32 were represented. A criterion of 90% of the 66 gene clusters is a suggested “passing” metric for draft genomes of cultured strains 32. Extrapolating from these results suggests a complete genome size of 4.95–6.42 Mb, and that the assembly contains a majority of the gene complement and should provide significant insight into SAR324. That the genome is not complete is typical of the majority of single cell genomes published to date 17, 33, and most likely due to the absence of sequences in the amplified DNA (as in the E. coli and S. aureus datasets (Supplementary Fig. S1) as described previously 3, 17.
Table 3.
Features of the SAR324_MDA single cell assembly (EULER + Velvet-SC). 3811 genes are those > 180 bp in length.
| Genome size (bp in assembly) | 4.3 Mb |
| Estimated genome size | 4.9-6.4 Mb |
| % GC | 43% |
| # tRNA genes | 20 types |
| # tRNA synthetases | 17 of 21 types |
| # rRNAs | 1 each of 5S, 16S, 23S |
| # genes | 3811 |
| # conserved single copy genes | 75/111 (67%) |
| # conserved single copy gene clusters | 58/66 (87%) |
The SAR324_MDA assembly has sequences in common (84–99% identity) with two SAR324 fosmids, HF0010_10I05 and HF0070_07E19 27, and some synteny is evident in alignments (Supplementary Fig. S11). The 16S sequences from these two fosmids and SAR324_MDA cluster tightly (Fig. 3).
The assembly appears to contain a majority of the genome by other criteria as well: all 20 tRNA types, 17 of the 21 types of tRNA synthetases (including selenocysteine), and full biosynthetic pathways for all amino-acids and most vitamins are present (see Table 3 for partial data). Complete glycolytic/gluconeogenesis, tricarboxylic acid, and pentose pathways are present, supporting a chemoheterotrophic lifestyle. Many of the components of chemotaxis and flagella synthesis and operation are encoded (Supplementary Fig. S12), as are the components of aerobic metabolism (e.g., cytochrome c oxidase). Putative formate dehydrogenase and carbon monoxide (CO) dehydrogenases within the SAR324 contigs that indicate the potential for anaerobic metabolism were examined in more detail. Phylogenetic analysis shows that the putative formate dehydrogenases and orthologs found in other marine aerobes, including P. pacifica, form a clade quite divergent from the biochemically-characterized anaerobic formate dehydrogenases (Supplementary Fig. S13a). This suggests that they act in an unknown aerobic pathway, expanding the metabolic diversity of this ancient protein family. Similarly, phylogenetic analysis (Supplementary Fig. S13b) of SAR324_MDA putative CO dehydrogenases, and similar proteins encoded by two recently sequenced fosmid clones of SAR324 27 and the P. pacifica genome shows they are divergent from functionally characterized versions. Protein alignment shows these deltaproteobacterial oxidoreductases contain the Mo-cofactor (MoCo) binding site but lack CO dehydrogenase consensus sequences 34. All the deltaproteobacterial putative CO dehydrogenase genomic clusters encode both the MoCo-binding large subunit and the Fe-S binding small subunit, but not the flavoprotein-binding medium subunit, providing more support that they are not CO dehydrogenases. In summary, the more detailed sequence analysis of these proteins is inconsistent with roles in anaerobic metabolism. One of the most striking features of the SAR324 assembly is the presence of eighteen putative phytanoyl dioxygenases, which catalyze the degradation of the lipid chain on chlorophyll a. The metabolic features of SAR324, and its dominance in the upper mesopelagic, suggest they track and degrade sinking photosynthetic biomass as it leaves the sunlit surface ocean.
Discussion
A main challenge for single cell genome assembly is the non-uniformity of coverage, particularly when combined with increased error rates and chimeras. EULER-SR’s error correction algorithm 35 was employed to correct read errors prior to Velvet assembly with Velvet-SC, a modified Velvet assembler tailored for single cell data. Validation of de novo assembly by EULER+Velvet-SC with single cells from reference genomes shows that EULER+Velvet-SC successfully copes with the non-uniformity of coverage, incorporating significantly more bases in the assembly than Velvet, and increasing the quality of the assembly. Although using a lower cutoff initially creates a noisier graph with more contig fragmentation, the iterative cutoff approach used by Velvet-SC overcomes the fragmentation and results in longer contigs. The addition of EULER-SR error correction upstream of assembly further enhances contig size and assembly quality.
A test with a novel uncultured organism confirms that a useful genomic draft can be obtained from a single lane of Illumina paired end sequencing reads. The SAR324 single cell genome provides an excellent example of an assembly obtained with minimal effort and reasonable cost (1 or ½ of an Illumina lane, with no closure efforts), that vastly exceeds the information about genomes of uncultivated bacteria that can be extracted via traditional metagenomic approaches. This approach will enable draft assemblies of large numbers of single cell bacterial genomes at affordable cost. The ability to generate high-quality draft assemblies that support annotation of the majority of the gene complement from Illumina reads will drive advances in characterizing uncultured organisms from the human microbiome (including pathogens), and from the environment (including bacteria producing antibiotics and bacteria with potential for biofuel production). Where the organism is of sufficient interest to warrant additional effort, several publications have investigated strategies to approach completion of assemblies 9, 17, 18. Mate pair sequencing can also assist in assembly; however, the presence of chimeric rearrangements occurring at about one per 10–30 kb 9, 10 of amplified DNA may limit the useful length of inserts. The optimal use of mate pairs for single cell sequencing remains to be investigated. The rapid improvement of sequencing technologies and reduction of cost also promises to accelerate progress.
A major goal of single cell genomics is to complement the large volume of gene level metagenomic data with genome level assemblies and to apply this emerging technology to study uncultured organisms from various environments including marine, soil, and the human microbiome. The cost effective approach demonstrated here should contribute to exploration of microbial taxonomy and evolution, and facilitate the mining of environmental organisms for genes and pathways of interest to biotechnology and biomedicine. We also envision applications and further development of EULER+Velvet-SC to applications in metagenomics and transcriptome sequencing projects, which also are characterized by highly non-uniform coverage.
Methods
Velvet-SC: Modifications to Velvet assembly algorithm
While Velvet19, ABySS41, and EULER-SR42 generate many correct contigs, they also generate many erroneous regions (caused by errors in reads as well as assembly errors) during intermediate stages of assembly that must be removed in the final assembly. In normal multicell assembly, coverage throughout the genome is fairly uniform, so all these tools use a fixed coverage cutoff to eliminate erroneous contigs. This strategy, however, fails in single-cell assembly since coverage is highly non-uniform (see Supplementary Figs. S1 and S2; Supplementary Table S4), and low coverage regions can represent correct contigs.
The Velvet-SC (http://bix.ucsd.edu/singlecell/) algorithm is designed to salvage low coverage regions. We give an informal explanation of Velvet-SC here (for detailed pseudocode, see Supplementary Fig. S5). Velvet combines reads using a “de Bruijn graph” 35. Vertices of the de Bruijn graph correspond to all k-mers present in reads and edges correspond to all (k +1)-mers present in reads. An edge corresponding a (k +1)-mer a1a2 … akak+1connects a vertex a1a2 … ak (prefix k-mer) with a vertex a2 … ak-1ak+1 (suffx k-mer). The coverage of an edge ((k +1)-mer) in the de Bruijn graph is the number of times this (k +1)-mer appears in all reads.
The resulting de Bruijn graph is very complex (even for small bacterial genomes), necessitating a step of removing low-coverage edges. Velvet removes such edges (and entire low-coverage regions) using a fixed threshold; this is a critical assembly step that attempts to remove errors, but it assumes that coverage is uniform across the genome. However, this approach leads to removal of many correct edges with low coverage since such edges are prevalent in many low-coverage genomic regions in single cell assembly projects. The Velvet-SC algorithm instead uses a variable threshold that starts at 1 and gradually increases. Some contigs may potentially be linked by two possible intermediate linker sequences, one with high coverage and one with low coverage; see Figure 1. Velvet-SC removes the low coverage linker sequence, allowing the neighboring sequences to be merged into a longer contig. This procedure is iterated with a gradually increasing low coverage cutoff. Since single-cell sequencing results in a mosaic of short low coverage regions and (typically longer) higher coverage regions, Velvet-SC typically merges low coverage regions with high coverage regions (resulting in a region with high coverage), thus rescuing low coverage regions from elimination.
EULER+Velvet-SC is EULER-SR’s error correction 43 combined with Velvet-SC. To test EULER-SR+Velvet-SC, sequencing reads were generated from MDAs performed on single cultured cells of E. coli K-12 and S. aureus USA300. The E. coli (lane 1 and lane 6) and S. aureus datasets are 600x-coverage and 2300x-coverage 100 bp paired-end runs of the Illumina Genome Analyzer IIx pipeline, respectively (~270 bp average insert length for E. coli and ~220 bp average insert length for S. aureus). A standard unamplified genomic DNA-derived E. coli K-12 dataset was used as a control (EMBL-EBI Sequence Read Archive, ERA000206, average insert length ~215 bp).
Single cell isolation
Single cells of Escherichia coli (ATCC 700926) and Staphylococcus aureus MRSA USA300 strain FPR3757 (21 ATCC 25923) were isolated by micromanipulation as described in Supplementary methods. Marine cells were sorted by flow cytometry. A marine water sample from the Scripps Research Pier (Scripps Institute for Oceanography, La Jolla, CA, 6 m depth, collected on October 8, 2008 at 9:00 am) was filtered (0.8 µm pore size), flash frozen and stored at −80°C in 30% glycerol. Prior to sorting, the thawed sample was stained with 10x SYBR Green I nucleic acid stain (Invitrogen). Single cells were sorted using a FACS Aria II flow cytometer (BD Biosciences) equipped with a custom forward scatter (FSC)-PMT using detection by the FSC-PMT and green fluorescence, and at the highest purity setting and a low flow rate to avoid sorting of coincident events. Cells were sorted into 384-well plates containing 4 µl of TE buffer per well, and stored at −80°C.
Multiple Displacement Amplification (MDA) and selection of candidate marine amplified DNAs
MDA of single cell genomes was performed using GenomiPhi HY reagents (GE Healthcare) as detailed in Supplementary methods. 16S rRNA gene was amplifed and sequenced (see Supplementary Methods) and marine MDAs of interest were selected by BLAST analysis of their 16S sequences against a curated marine 16S rRNA database derived from the Global Ocean Sampling (GOS) 16S data 28. MDAs with 16S rRNA sequences with > 97% identity to operational taxonomic units in the dataset were selected for sequencing, including the SAR_324 MDA described here.
Library generation and sequencing
Short insert paired end libraries were generated from amplified single E. coli cell DNA following the standard Illumina protocol 44. PCR-free paired end libraries were generated for S. aureus and Deltaproteobacteria (to avoid possible uneven representation of AT rich sequences) from 15 µg of amplified DNA using the adapters: 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ and 5’-GATCGGAAGAGCACACGTCTGAACTCCAGTCACATCACGATCTCGTATGCCGTCTTCTGCTTG-3’, and selecting an average insert size of ~250 bp. Sequencing was carried out on a Genome Analyzer IIx using standard reagents. PCR-free libraries were sequenced using the sequencing primer 5’-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’ in read 2.
Analysis and annotation of the single cell assembly
Contigs were analyzed by BLAST against a nucleotide sequence database with entries from GenBank and RefSeq (excluding whole genome shotgun assemblies). Contigs will be submitted to GenBank prior to publication. Annotation of ORFs, tRNAs, rRNA genes and tRNA synthetases was performed using the JCVI metagenomics annotation pipeline 45 without manual curation. Phylogenetic analysis of select proteins was conducted in Bosque 46, with substantial manual creation. Gene identifiers used in KEGG pathway analysis 47 at http://www.genome.jp/kegg/pathway.html were generated at the KEGG Automatic Annotation Server (KAAS, 48) using the bidirectional best hit settings. Of 3811 MetaGene ORFs submitted to KAAS, 1415 yielded a gene identifier.
Supplementary Material
Acknowledgements
This work was partially supported by grants to R.S.L. from the National Human Genome Research Institute (NIH-2 R01 HG003647) and the Alfred P. Sloan Foundation (Sloan Foundation-2007-10-19), and by a grant to P.A.P. and G.T. from the National Institutes of Health (NIH grant 3P41RR024851-02S1). We thank Maria Kim (JCVI) for bioinformatics support.
Footnotes
Author contributions
All authors analyzed data.
H.C. and G.T. wrote software.
M.N., J.Y.-G, M.-J.L., and L.J.F. performed wetlab experiments.
H.C., J.Y.-G., G.T., C.L.D., M.-J.L., L.J.F., N.A.G., P.A.P., and R.S.L. wrote the manuscript.
H.C., G.T., M.-J.L., C.L.D., J.H.B., D.B.R., and N.A.G. created figures and tables.
R.S.L. and M.-J.L. supervised the JCVI group. P.A.P. and G.T. supervised the UCSD group. N.A.G. and D.J.E. supervised the Illumina group. G.P.S. initiated the Illumina-JCVI collaboration.
Note: Supplementary information is available on the Nature Biotechnology website.
References
- 1.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;13(3):e77. doi: 10.1371/journal.pbio.0050077. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 2005;71:3342–3347. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasken RS. Single cell genomic sequencing using Multiple Displacement Amplification. Curr. Opin. Microbiol. 2007;10:1–7. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Ishoey T, Woyke T, Stepanauskas R, Novotny M, Lasken RS. Genomic sequencing of single microbial cells from environmental samples. Curr. Opin. Microbiol. 2008;11:198–204. doi: 10.1016/j.mib.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 10.Lasken RS, Stockwell TB. Mechanism of chimera formation during the Multiple Displacement Amplification reaction. BMC Biotechnol. 2007:19. doi: 10.1186/1472-6750-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasken RS, et al. In: in Whole Genome Amplification: Methods Express. Hughes S, Lasken R, editors. UK: Scion Publishing Ltd.; 2005. pp. 119–147. [Google Scholar]
- 12.Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl. Microbiol. Biotechnol. 2007;74(4):926–935. doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- 13.Mussmann M, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5:e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcy Y, et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podar M, et al. Targeted access to the genomes of low abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 2007;73(10):3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hongoh Y, et al. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc. Natl. Acad. Sci. USA. 2008;105:5555–5560. doi: 10.1073/pnas.0801389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldman BS, et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright TD, Vergin KL, Boyd PW, Giovannoni SJ. A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl. Environ. Microbiol. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 27.Rich VI, Pham VD, Eppley J, Shi Y, Delong EF. Time-series analyses of Monterey Bay coastal microbial picoplankton using a 'genome proxy' microarray. Environ. Microbiol. 2010 doi: 10.1111/j.1462-2920.2010.02314.x. [DOI] [PubMed] [Google Scholar]
- 28.Yooseph S, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 29.Iizuka T, et al. Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int J Syst Evol Microbiol. 2003;53:189–195. doi: 10.1099/ijs.0.02418-0. [DOI] [PubMed] [Google Scholar]
- 30.Callister SJ, et al. Comparative bacterial proteomics: analysis of the core genome concept. PLoS One. 2008;3:e1542. doi: 10.1371/journal.pone.0001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitreva M. Bacterial core gene set. 2008 http://www.hmpdacc.org/doc/sops/reference_genomes/metrics/Bacterial_CoreGenes_SOP.pdf.
- 32.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woyke T, et al. One bacterial cell, one complete genome. PLoS One. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King GM. Microbial carbon monoxide consumption in salt marsh sediments. FEMS Microbiol Ecol. 2007;59:2–9. doi: 10.1111/j.1574-6941.2006.00215.x. [DOI] [PubMed] [Google Scholar]
- 35.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Chapter 6, Unit 6 4. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez D, Francois P, Farinelli L, Ostera M, Schrenzel J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37:D459–463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaisson MJ, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanenbaum DM, et al. The JCVI standard operating procedure for annotating prokaryotic metagenomic shotgun sequencing data. S.I.G.S. 2010;2 doi: 10.4056/sigs.651139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez-Flandes S, Ulloa O. Bosque: integrated phylogenetic analysis software. Bioinformatics. 2008;24:2539–2541. doi: 10.1093/bioinformatics/btn466. [DOI] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.