Abstract
A20 is an immediate-early NF-κB target gene. Prior to NF-κB stimulation, the A20 promoter is bound by the polymerase II machinery to allow rapid transcription activation. Here we show that the basal A20 transcription is repressed at the level of elongation in a promoter-specific fashion. Immunodepletion in vitro and RNA interference in cultured cells suggest that the basal elongation inhibition is conferred by DRB sensitivity-inducing factor (DSIF). We have identified a negative upstream promoter element called ELIE that controls DSIF activity. Remarkably, following NF-κB stimulation, inhibition of the A20 promoter by DSIF persists, but it is now regulated by NF-κB rather than ELIE. Similar regulation by DSIF is shown for another NF-κB-responsive gene, the IκBα gene. These findings reveal an intimate and dynamic relationship between DSIF inhibition of elongation and promoter-bound transcription factors. The potential significance of the differential regulation of DSIF activity by cis-acting elements is discussed.
Transcription of mRNA-encoding genes is a multistep process divided into initiation, elongation, and termination. Although regulation of initiation plays a major role in gene expression, it has become apparent that the transcription elongation stage is also critical for regulation of gene expression (for reviews see references 5 and 16). Studies of transcription elongation have revealed the existence of several families of elongation factors governing the activity of polymerase II (Pol II) either positively or negatively. P-TEFb is the best-characterized positive elongation factor. It acts to induce Pol II processivity by facilitating the transition from the early to the late and productive elongation phase (for a review, see reference 9). Two negative transcription elongation factors, DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF), have recently been identified and characterized. DSIF is composed of two subunits, p160 and p14, which are homologs of the Saccharomyces cerevisiae transcription factors Spt5 and Spt4, respectively (12). NELF is composed of five polypeptides. DSIF and NELF function cooperatively to inhibit RNA Pol II elongation (15).
The family of NF-κB transcription factors is a central component of the cellular response to a broad range of extracellular signals including inflammatory cytokines, tumor promoters, and chemotherapeutic agents. A major pathway regulating NF-κB activity involves its nuclear transport. In unstimulated cells NF-κB is retained in the cytoplasm in an inactive form by IκB inhibitory proteins. Signals that activate NF-κB trigger ubiquitination and degradation of IκB, resulting in transport of NF-κB into the nucleus (for a recent review see reference 6).
The mechanism of rapid transcription activation of NF-κB-responsive genes in vivo and in vitro was recently analyzed (1). Chromatin immunoprecipitation (ChIP) assays showed that induction of NF-κB by extracellular signals does not enhance preinitiation complex formation, since general transcription factors, Pol II, and various coactivators were found already associated with these genes prior to NF-κB induction. Using the immediate-early NF-κB-responsive gene A20 as a prototype promoter, we found that preinitiation complex formation and the initiation step are mediated by the constitutive transcription factor Sp1, which is crucial for rapid transcriptional induction by NF-κB. NF-κB acts at the postinitiation stage, enhancing the transcription reinitiation rate. These findings suggest that the consecutive effects of Sp1 and NF-κB on the transcription process underlie the mechanism of their synergy, which allows rapid transcriptional induction in response to cytokines.
In the present study we investigated more deeply the transcriptional regulation of the NF-κB response gene A20 and found that the basal activity of the A20 gene is repressed at the level of elongation by DSIF. This elongation repression is controlled by the promoter through an upstream repressor element called ELIE. Unexpectedly, even after NF-κB stimulation of A20 transcription, DSIF retains its inhibitory effect. Under these conditions NF-κB is responsible for the negative elongation effect of DSIF. Our results reveal a regulatory link between upstream promoter-bound factors and the downstream elongation step. The significance of the dynamic regulation of DSIF by promoter-bound transcription factors is discussed.
MATERIALS AND METHOD
Plasmids
A20Luc and pBC-SK−A20 have been described previously (1). A20 promoter mutants m1, m2, m3, mNF-κBs, and m1+mNF-κBs were generated by replacing the PmlI-XmaI fragment in the A20 promoter corresponding to positions −74 to −46 with the synthetic oligonucleotides containing mutations as follows: m1, 5′-ACGACTTTGGAAAGTCCCGTGGAAATCC-3′; m2, 5′-GTGCACTTGGAAAGTCCCGTGGAAATCC-3′; m3, 5′-ACGAGGACGGAAAGTCCCGTGGAAATCC-3′; mNF-κBs, 5′-GTGACTTTGGAAAGATCTGTGGAAAAGA-3′; and m1+mNF-κBs, 5′-ACGACTTTGGAAAGATCTGTGGAAAAGA-3′.
To construct the m4 mutant, a similar strategy was applied, but the replaced fragment was located between the EagI and XmaI sites of the A20 promoter (−114 to −46), and the sequence of the oligonucleotide was 5′-GGCCGGCTGGACGCACTTCGCAGCCCGACCCAGAGAGTCACGTGACTTTGGAAAGTCCCGTGGAAATCC-3′. pSuper plasmid was a generous gift from R. Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). The DSIF RNAi vector was constructed according to the method of Brummelkamp et al. (4), using pSuper plasmid and a synthetic oligonucleotide targeting 5′-GTCATCTGTGGGAGAGACG-3′, corresponding to positions 594 to 612 of DSIF p160 mRNA. Rous sarcoma virus (RSV)-Renilla luciferase plasmid was a generous gift from M. Walker (Weizmann Institute of Science, Rehovot, Israel). ELIE-α-actin and mELIE-α-actin were generated by cloning synthetic double-stranded oligonucleotides composed of the sequence from −74 to −34 and −71 to −34, respectively, upstream from the α-actin core promoter (−38 to +80).
Antibodies
Anti-DSIF p160 and anti-NELF-A antibodies have been previously described (15). Anti-Pol II is the SWGI6 monoclonal antibody (Babco). Anti-Oct-2 antibody is C-20 rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.).
In vitro transcription assays
Jurkat cell nuclear extract used for in vitro transcription assays was prepared as described previously (1). Assembly of in vitro transcription was conducted in a total volume of 32 μl containing 18 μl of nuclear extract in HEMG buffer (100 mM KCl, 25 mM HEPES [pH 7.9], 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, and 1 mM dithiothreitol)-400 ng of DNA template-10 μM ZnSO4. The mixture was incubated at 30°C for 30 min to allow preinitiation complex formation. Transcription was initiated by the addition of 4 μl of nucleotide solution, containing 3.6 mM ATP, GTP, and CTP; 0.04 mM UTP; and 50 μCi (0.025 mM) of [α-32P]UTP for a runoff assay with the A20 gene or unlabeled 5 mM nucleoside triphosphates for the primer extension protocol. The reaction was terminated after 10 min by the addition of 100 μl of stop solution (0.2 M NaCl, 20 mM EDTA, 1% sodium dodecyl sulfate, 0.25 mg of yeast tRNA per ml, 50 μg of proteinase K per ml) and incubation at 42°C for 30 min. To release elongation inhibition, 0.2% Sarkosyl was added to the reaction mixture 1.5 min after the initiation of transcription, and incubation was continued for an additional 8.5 min. The RNA was extracted with phenol-chloroform (1:1) and then with chloroform and finally precipitated with ethanol. The RNA products were detected either by immediate separation on an 8% urea-polyacrylamide gel (runoff protocol) or by extension of an end-labeled primer complementary to specific regions of the respective mRNAs (primer extension protocol). The primers for primer extension assays were as follows: for the A20 promoter fused to the lacZ gene (pBC-SK−A20), primers complementary to positions +30 (5′-GGATCCTCTAGAGTCGACCT-3′), +60 (5′-CACCGC GGTGGCGGCCGCTC-3′), +90 (5′-CCTCACTAAAGGGAACAAAA-3′), and +120 (5′-GACCATGATTACGCCAAGCG-3′); for the A20 promoter fused to the Luc gene (A20pLuc), primer corresponding to position +109 (5′-GCCTTTCTTTATGTTTTTGGCG-3′). The RNA samples were resuspended in 10 μl of reaction mixture containing 25 fmol of labeled primer (an excess over the RNA template), 2 mM Tris HCl (pH 7.8), 0.2 mM EDTA, and 0.25 M KCl; denatured at 85°C for 5 min; and annealed at 50°C for 45 to 60 min. A 25-μl volume of extension mixture composed of 2.5 U of avian myeloblastosis virus (AMV) reverse transcriptase (RT; Promega), 20 mM Tris HCl (pH 8.7), 10 mM MgCl2, 5 mM dithiothreitol, and 0.33 mM deoxynucleoside triphosphates was added to the annealed primer-RNA duplexes, and an extension reaction was performed at 37°C for 15 min. The cDNA products were ethanol precipitated and separated on an 8 or 10% urea-polyacrylamide gel. Dried gels were visualized with a Phosphoimager (Fuji; BAS 2500).
The DNA for the transcription assays on immobilized template was generated by PCR with pBC-SK−A20 as template, a biotinylated T7 primer as forward primer, and the +120 primer described above as the reverse primer. The biotinylated A20 promoter fragment (400 ng for each transcription reaction) was bound to streptavidin-conjugated magnetic Dynabeads. Transcription was analyzed by run-on assay as described above.
The template for the runoff assay with the A20 gene was generated by genomic PCR. The primers used to produce this fragment were 5′-GCTTCCGAAATGCCCAGGTG-3′ and 5′-CTCCGGGCCCCGCGATCC-3′. The in vitro transcription using the 119-nucleotide U-less reporter was done as described previously (14)
Immunodepletion
Purified monoclonal antibodies to DSIF p160 and rabbit polyclonal anti-Oct-2 antibody (5 μl each) were incubated with 20 μl of protein A/G-Sepharose (Amersham Pharmacia) and 100 μl of HEMG buffer for 1 h at 4°C to immobilize the antibodies on the beads. After being washed three times with HEMG buffer, the immobilized antibodies were added to 200 μl of Jurkat cell nuclear extract containing 0.01% NP-40 and incubated for 2 h at 4°C. This depletion process was repeated three more times, each time with the addition of fresh antibody-immobilized beads.
The input, depleted extracts (2 μl each), and immunoprecipitated proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane, and the depletion of DSIF p160 was monitored by immunoblotting with anti-DSIF p160 antibody.
Transfection
293T cells (human embryonic kidney fibroblasts) were maintained in F-12-Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transfections were performed using the standard CaPO4 method. To avoid basal NF-κB activity, cells were kept from reaching confluence and replated not more than nine times. For reporter assays, subconfluent cells were transfected in a 24-well plate with 1.1 μg of pSuper or DSIF RNAi plasmid and 15 ng of the A20Luc reporter derivatives, 3 ng of RSV-Renilla luciferase, 10 ng of cytomegalovirus-green fluorescent protein, and 1 ng of p65-RelA (as indicated). Forty-eight hours after transfection cells were either left untreated or treated with human recombinant tumor necrosis factor (TNF; 5 ng/ml; Peprotech Inc.) for 4.5 h and harvested, and their luciferase and Renilla luciferase activities were measured. For the ChIP assay subconfluent cells in 100-mm-diameter dishes were transfected with 12 μg of DSIF RNAi or pSuper plasmid. Forty-eight hours later chromatin extract was prepared from the cells.
ChIP
The ChIP assay was performed as described previously (1). The primers used for the A20 promoter were 5′-CAGCCCGACCCAGAGAGTCAC-3′ (forward) and 5′-CTCCGGGCCCCGCGATCC-3′ (reverse). The IκB promoter primers were the same as described previously (1).
RNA preparation, RT-PCR, and real-time quantitative RT-PCR analysis
Human 293T cells in 35-mm-diameter plates were transfected with 3 μg of either pSuper or DSIF RNAi plasmid. Forty-eight hours after transfection cells were left untreated or treated with TNF alpha (TNF-α) for 1 h. Total RNA was prepared using the Trizol reagent (Gibco BRL), according to the manufacturer's instructions. RNA preparations were treated with RQ1 DNase I (Promega) to avoid contamination of genomic DNA.
First-strand cDNA and PCR amplifications were performed in a single tube with the Access RT-PCR* Introductory System (Promega). Specific analysis of A20 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels was performed with 0.4 or 0.05 μg of total RNA, respectively.
The oligonucleotides used for RT-PCR were as follows: A20 forward, 5′-CACACAAGGCACTTGGATCC-3′, and reverse, 5′-TCCCCAGGAGTCCGTGCAGC-3′; GAPDH forward, 5′-TTGTCATCAATGGAAATCCC-3′, and reverse, 5′-TGTCGCTGTTGAAGTCAG-3′; and IκB forward, 5′-CCTTCCTCAACTTCCAGAACAACC-3′, and reverse, 5′-GGCTAAGTGTAGGCAGGTGTGGC-3′.
For real-time PCR, cDNA was synthesized from 4 μg of total RNA with oligo(dT)15 primer, AMV RT, and AMV RT buffer, all from the Reverse Transcription System (Promega). The PCR was performed in 20-μl glass capillary tubes with a LightCycler System (Roche Molecular Biochemicals), equipped with a thermal cycler and real-time detector of fluorescence. An 0.25-μl amount of total cDNA was amplified with A20 primers with the LightCycler-FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals), according to the manufacturer's instructions. The oligonucleotides used for real-time PCR were the same as those for RT-PCR.
RESULT
Specific inhibition of A20 promoter activity in vitro at the level of elongation
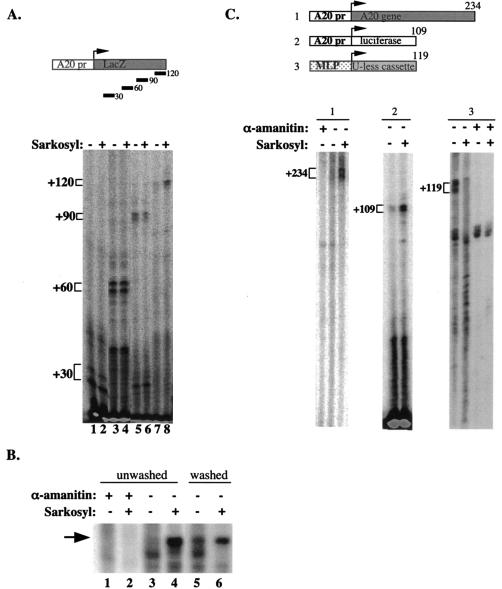
The promoter of the rapidly induced NF-κB target gene A20 is bound by the general transcription machinery and coactivators prior to NF-κB stimulation. This transcription complex is capable of initiation but not reinitiation (1). To investigate the regulation of the basal A20 transcription further, we studied the elongation step of transcription. We used the A20 promoter in in vitro transcription assays with nuclear extracts from unstimulated Jurkat cells and monitored the levels of transcripts of different lengths using primers complementary to positions +30, +60, +90, and +120 in primer extension assays. As shown in Fig. 1A, transcripts up to +90 are readily detected while the +120 transcript is barely visible (lanes 1, 3, and 5 compared to lane 7). These results suggest that Pol II is halted when it reaches a position around +120. To check the possibility that an inhibitor is blocking elongation, we used the detergent Sarkosyl, known to interfere with some protein-protein interactions but without effect on the elongating Pol II. The 120-nucleotide transcript was significantly induced by Sarkosyl while it had no effect on the 30-, 60-, and 90-nucleotide transcripts (compare lanes 2, 4, and 6 to lane 8). This finding is consistent with the idea that an inhibitor bound to the elongation complex is released by Sarkosyl, allowing extension transcription. The addition of Sarkosyl shortly after initiation inhibits not only the negative elongation activity but also reinitiation; thus, the resistance of the 30-, 60-, and 90-nucleotide transcripts to Sarkosyl confirms our previous finding (using the +90 primer) that, in the absence of NF-κB, the basal A20 transcription is limited to a single round of transcription regardless of the elongation inhibition (1).
FIG. 1.
(A) Transcription directed by the A20 promoter is inhibited at the level of elongation. In vitro transcription reactions were performed with nuclear extract prepared from nonstimulated Jurkat cells and analyzed by primer extension assay with primers corresponding to the +30, +60, +90, and +120 positions relative to the A20 transcription start site (as depicted in the upper panel). In lanes 2, 4, 6, and 8 Sarkosyl (0.2%) was added exactly 1.5 min after the addition of nucleotides (initiation). (B) In vitro transcription with immobilized template. A DNA fragment composed of the A20 promoter and 120 nucleotides of downstream sequence was immobilized on magnetic beads and incubated with nuclear extract. After addition of nucleotides (including [32P]UTP) Sarkosyl was added as indicated and either washed with transcription buffer 20 s later (lanes 5 and 6) or left untreated (lanes 1 to 4). Transcription from the washed template was then resumed with the addition of transcription buffer and unlabeled nucleotides. α-Amanitin (5 μM) was added to the reaction mixtures in the indicated lanes to verify that the synthesized transcript was Pol II specific. (C) A20 elongation inhibition is promoter driven and promoter specific. Shown are in vitro transcription reactions analyzed either by run-on assay (subpanels 1 and 3) or by primer extension (subpanel 2) with A20 promoter with its native downstream fragment (subpanel 1) or A20 promoter fused to the luciferase gene (subpanel 2) as the template. MLP U-less reporter was used as a control (subpanel 3). (The structure of the templates is shown schematically in the upper panel.) α-Amanitin (5 μM) was added where indicated to verify Pol II transcription. Sarkosyl (0.2%) was added exactly 1.5 min after the addition of nucleotides to the indicated reaction mixtures.
To confirm that the observed effect of Sarkosyl is associated with a removal of an elongation inhibitor, we performed transcription reactions on an immobilized DNA fragment consisting of the A20 promoter plus 120 nucleotides of downstream sequence (Fig. 1B). Sarkosyl was added shortly after initiation as before, and the beads were either left untreated (lanes 1 to 4) or immediately washed and reequilibrated with transcription buffer containing unlabeled nucleotides, and so transcription proceeded in the absence of Sarkosyl (lanes 5 and 6). Under both conditions transcription elongation was stimulated by Sarkosyl, confirming that the target of Sarkosyl is not the elongation complex itself. (The reduced intensity of the transcript derived from the washed template is most likely due to the absence of a labeled nucleotide during elongation after the washing step.) Moreover, washing of the beads without Sarkosyl also stimulated transcription elongation (compare lane 3 to lane 5), suggesting that this inhibitor is loosely associated with the elongation complex and can be partially washed off even without Sarkosyl.
Inhibition of A20 transcription elongation is regulated by the promoter
We next examined whether the sequences downstream of the initiation region have a role in elongation inhibition, as most of the transcribed sequence analyzed in the previous experiment is derived from the lacZ gene. To address this, we tested the effect of Sarkosyl on transcription using the A20 promoter with its native downstream sequence (−240 to +234), the A20 promoter (−240 to +12) fused to the luciferase gene, and the adenovirus major late promoter (MLP) fused to a U-less cassette. Figure 1C shows that in each of the templates driven by the A20 promoter the basal activity is low and transcription is induced by Sarkosyl, indicating that there is inhibitory elongation activity whenever the A20 promoter is present regardless of the transcribed sequence. By contrast, the basal activity directed by the adenovirus MLP is high and is not induced but rather inhibited by Sarkosyl, suggesting that the elongation-inhibitory activity is specific to the A20 promoter.
A20 elongation inhibition in vitro is exerted by DSIF
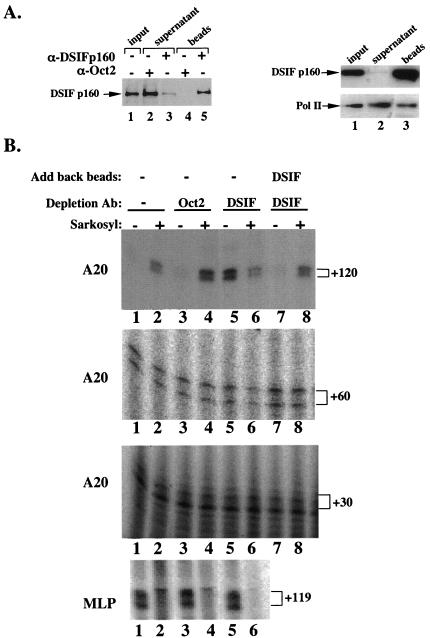
DSIF, a heterodimeric complex composed of the human homologs of the yeast Spt5 and Spt4 gene products, is a known inhibitor of Pol II elongation (15). Its inhibitory activity was observed in vitro on a model promoter system only when the kinase inhibitor DRB was added to the transcription reaction. In our system the negative elongation activity is integral to the A20 promoter and does not require the presence of DRB. To examine the possible involvement of DSIF in A20 transcription, we depleted DSIF from uninduced Jurkat cell nuclear extract with an antibody to the human p160 (Spt5) subunit of DSIF and analyzed the effect of immunodepletion on the level of the 30-, 60-, and 120-nucleotide transcripts directed by the A20 promoter and also on the 119-nucleotide transcript driven by MLP. As a control, a similar procedure was carried out with a nonrelevant antibody (Oct-2). Figure 2A shows immunoblot analyses of the immunodepleted extract confirming specific depletion of DSIF (Fig. 2A, left panel). DSIF depletion only slightly reduced the level of Pol II in the extract (Fig. 2A, right panel), consistent with a previous report showing that only a small fraction of Pol II is associated with DSIF (12). The in vitro transcription analysis (Fig. 2B) shows that immunodepletion of DSIF significantly increased basal activity of the 120-nucleotide transcript but had no effect on either 30- or 60-nucleotide transcripts or the MLP-directed transcript (compare lanes 1 to 5 in all panels). In the absence of DSIF Sarkosyl did not stimulate the 120-nucleotide transcript but rather inhibited it like MLP (lane 6). Adding back the beads containing the immunoprecipitated and washed DSIF reconstituted the elongation-inhibitory activity and the Sarkosyl inducibility only of the 120-nucleotide transcript with no effect on the 30- or 60-nucleotide transcripts (lanes 7 and 8 in all panels). These results strongly suggest that the elongation-inhibitory activity directed by the A20 promoter is being conferred by DSIF.
FIG. 2.
Inhibition of transcription elongation directed by the A20 promoter is mediated by DSIF. (A) Left panel: immunoblot analysis of Jurkat cell nuclear extract immunodepleted of DSIF p160 by using anti-DSIF p160 antibody. Anti-Oct-2 antibodies were used as the specificity control for the immunodepletion. Ten percent of precipitated proteins was loaded in lanes 4 and 5. Right panel: analysis of Pol II in DSIF-depleted extract. Shown are the results of immunoblot analysis of nuclear extract that was either untreated (lane 1) or immunodepleted of DSIF (lane 2) by using anti-DSIF p160 antibody and anti-Pol II antibody (large subunit). The beads (lane 3) represent the immunoprecipitated complex. (B) Transcription activity of the DSIF p160-depleted extract. Shown are the results of in vitro transcription reactions with either A20-lacZ or MLP U-less templates and Jurkat cell nuclear extract that was untreated (lanes 1 and 2) or immunodepleted with the indicated antibodies. The A20 transcripts were analyzed by primer extension assay with primers corresponding to +30, +60, and +120 relative to the transcription start site. The MLP transcript was analyzed by the run-on assay. In lanes 2, 4, 6, and 8 Sarkosyl (0.2%) was added 1.5 min after the addition of nucleotides. In lanes 7 and 8 the beads containing immunoprecipitated and washed DSIF were added back to the DSIF-depleted extract.
In nonstimulated cells regulation of A20 transcription by DSIF is mediated by an upstream regulatory element
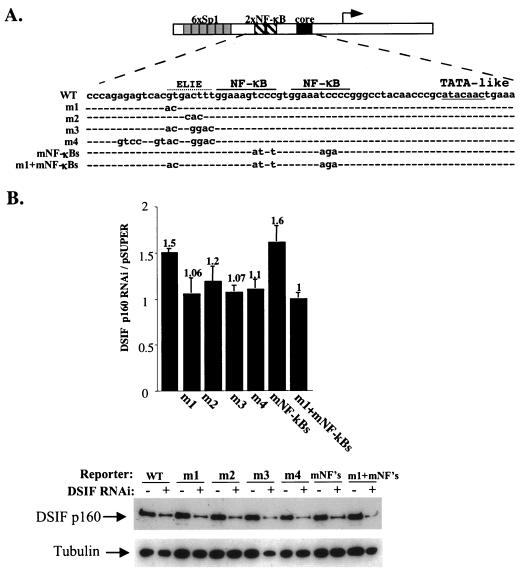
To investigate the role of DSIF in A20 promoter activity in the cell, we used an RNA interference strategy (RNAi) to reduce the endogenous DSIF levels. We transfected into 293T cells a plasmid directing expression of a 21-nucleotide double-stranded RNA specific for the p160 (Spt5) subunit of DSIF. Analysis of DSIF p160 by immunoblotting (Fig. 3B) shows its specific depletion from the cells transfected with DSIF p160 RNAi plasmid but not from cells transfected with the parental plasmid (Fig. 3B, bottom panel). Interestingly, this level of depletion did not affect the survival or the proliferation of the transfected cells during the course of the experiment. Next we examined the effect of DSIF p160 RNAi on the basal A20 promoter activity (Fig. 3B, lanes WT). The results show that reduced levels of DSIF in the cell enhanced A20 promoter activity by 1.5-fold (a statistically significant effect; P < 0.0001) but had no effect on transcription directed by the RSV promoter that was used as a reference, confirming regulation of A20 transcription by DSIF in the cell.
FIG. 3.
In unstimulated cells DSIF inhibits the A20 transcription and is regulated by a specific upstream negative element. (A) A20 promoter mutants. The schematic of the A20 promoter is shown at the top. The DNA sequence of the region in the A20 promoter that was subjected to mutations is expanded in the top (WT) row, and only the substituted nucleotides are denoted. The two NF-κB binding sites, the TATA-like sequence, and the putative ELIE are indicated. (B) The effect of DSIF p160 knockdown on the transcription activity of the A20 promoter. 293T cells were transfected with a plasmid expressing DSIF p160 RNAi or the empty expression plasmid pSuper together with a luciferase reporter gene controlled by A20 promoter variants shown in panel A. Promoter activity is presented as the ratio between the luciferase reporter activity in the presence of DSIF RNAi and its activity in the presence of the RNAi parental vector (pSuper). Luciferase activities were normalized to the activity of cotransfected RSV promoter-driven Renilla reporter luciferase that was used to correct for differences in transfection efficiencies. The data represent the means and standard deviations of seven transfection experiments, each done independently in duplicate. The bottom panel shows results of an immunoblot analysis of a representative transfection experiment using anti-DSIF p160 antibody and antitubulin, which served as a control for protein loading. (C) The effect of the mutations on the basal activity of the A20 promoter is presented as relative luciferase units (RLU). (D) Effect of DSIF RNAi on the endogenous A20 promoter occupancy by DSIF and Pol II. 293T cells were transfected with a plasmid expressing DSIF p160 RNAi or the empty expression plasmid pSuper. Forty-eight hours later ChIP assays were performed on these cells with DSIF, Pol II, and control antibodies. The bound A20 promoter region was analyzed by PCR (right panel). The left panel shows results of immunoblot analysis of DSIF and tubulin of these cells. WT, wild type; Ab, antibody.
To investigate the mechanism by which the A20 promoter mediates the DSIF effect, we set to identify the responsible promoter element. We introduced mutations in the region between Sp1 and NF-κB (Fig. 3A), since initial experiments excluded the involvement of the region between −240 and −74 containing the six Sp1 sites or the NF-κB binding sites (see below) in elongation regulation of A20 in unstimulated cells (data not shown). Mutated A20 promoter constructs were cotransfected with either DSIF RNAi or empty expression vector, and 48 h later luciferase activity was measured (Fig. 3B and C). Promoter constructs containing mutations in the two NF-κB binding sites (mNF-κBs column) responded similarly to the wild-type promoter to DSIF RNAi. On the other hand mutation in sequences adjacent to the 5′ NF-κB binding site either reduced (m2) or abolished (m1, m3, and m4) the stimulatory effect of DSIF RNAi. These results suggest that a DNA element composed of the minimal sequence GTGACTTT mediates the inhibitory effect of DSIF under basal conditions. We have called this element ELIE for elongation-inhibitory element. Consistent with ELIE being a negative regulatory element, the m2 and m3 mutations of ELIE enhanced basal A20 promoter activity of A20 (Fig. 3D). The m1 mutation decreased A20 promoter activity but also abolished DSIF inhibition; thus, it is likely that this mutation affects an overlapping positive regulatory element. This possibility is consistent with the small net effect of the large m4 mutation, which probably destroys both positive and negative regulatory sites. In the absence of NF-κB stimulation, the mutations in NF-κB sites had no effect on the basal promoter activity. In a computer search for transcription factor binding sites, we found that ELIE's sequence does not resemble that of any known transcription factor, raising the possibility that it binds a novel and as yet unidentified repressor. The sequence just upstream from ELIE (the overlapping positive element) contains an E-box motif.
To confirm direct regulation of A20 gene transcription by DSIF, we tested whether it physically associates with the A20 promoter-proximal region. We performed ChIP using 293T cells transfected with either pSuper or DSIF RNAi. The results of ChIP (Fig. 3D) show that DSIF, like Pol II, is associated with the promoter-proximal region of the A20 gene in nonstimulated cells, and the amount of DSIF is decreased by DSIF RNAi. These findings are consistent with DSIF regulating the A20 gene transcription and confirm the specificity of DSIF RNAi. We also observed some reduction of Pol II on the A20 promoter-proximal region. Given the enhanced transcriptional activity in the absence of DSIF, it is unlikely that less Pol II is recruited to the promoter. A possible explanation is that Pol II that normally pauses at a promoter-proximal region moves downstream to synthesize the mRNA, causing reduced occupancy at a promoter-proximal region.
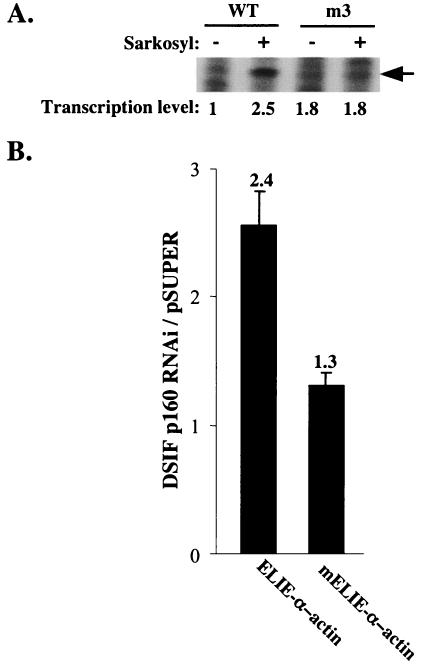
To test further the regulation of elongation by the A20 promoter via ELIE, we examined the effect of Sarkosyl on the m3 mutation of ELIE in in vitro transcription assays (Fig. 4A). Consistent with the idea that ELIE is directly involved in inhibition of elongation, mutation in ELIE increased the basal activity (1.8-fold) of the full-length transcript and eliminated the Sarkosyl stimulatory effect on elongation. The overall level of transcription directed by this mutant is lower than wild-type template, presumably because the m3 mutation also affects an overlapping positive regulatory element (Fig. 3B).
FIG. 4.
Analysis of ELIE. (A) In vitro transcription reactions analyzed by run-on assay using wild-type or m3 A20 promoter variants (Fig. 3). The correctly initiated transcript (shown by arrow) was quantified and is presented as a ratio to the activity of the wild-type (WT) A20 promoter in the absence of Sarkosyl. (B) Small DNA fragments derived from the A20 promoter containing either full-length ELIE or ELIE with 3 nucleotides deleted (ELIE-α-actin and mELIE-α-actin, respectively) were cloned in front of a minimal α-actin core promoter driving a luciferase reporter gene. These reporter plasmids we cotransfected into 293T cells along with either the plasmid expressing DSIF p160 RNAi or the empty expression plasmid pSuper, and 48 h later luciferase activity was measured and normalized to the activity of cotransfected RSV-Renilla luciferase and is presented as relative luciferase units. The data represent the means and standard deviations of five transfection experiments done independently in duplicate.
Next we examined ELIE's activity in a heterologous context. We cloned a small region of the A20 promoter containing either the complete ELIE (−74 to −34) or ELIE with 3 nucleotides deleted (−71 to −34) upstream of an α-actin core promoter (−38 to +80) that drives a luciferase reporter gene. These reporter plasmids we cotransfected into 293T cells along with either the plasmid expressing DSIF p160 RNAi or the empty expression plasmid pSuper (Fig. 4B). Depletion of DSIF from the cells significantly stimulated ELIE-α-actin reporter but was significantly less effective in stimulating the mutated ELIE reporter plasmid (2.4- versus 1.3-fold, respectively). These results further support the idea that ELIE is a promoter element that negatively regulates transcription elongation via DSIF.
Transcription of endogenous A20 and IκBα genes is negatively regulated by DSIF before and during TNF-α stimulation
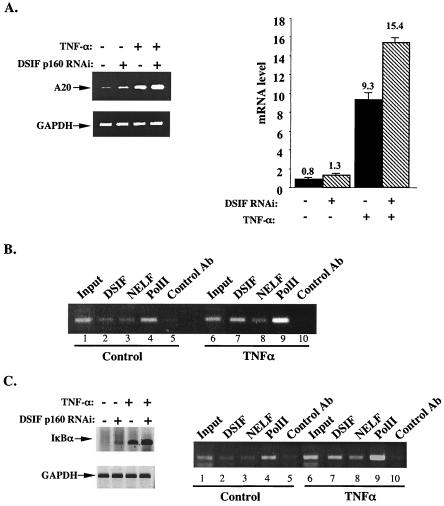
To examine the effect of DSIF on the endogenous A20 gene transcription, we analyzed the levels of A20 mRNA in the presence of DSIF RNAi, using either standard RT-PCR (Fig. 5A, left panel) or quantitative real-time PCR (Fig. 5A, right panel). Consistent with the effect of DSIF on the A20 reporter gene, reduction in DSIF levels elevated the basal A20 mRNA levels by 1.6-fold.
FIG. 5.
DSIF controls the transcription of the endogenous A20 gene in unstimulated and TNF-α-stimulated cells. (A) 293T cells transfected with either pSuper or DSIF p160 RNAi plasmids were left untreated or treated with TNF-α for 1 h. Total RNA prepared from these cells was used either for standard RT-PCR (left panel) or quantitative real-time PCR (right panel) analysis of A20 mRNA level. For a control, analysis of GAPDH mRNA level was performed as well. (B) ChIP assay using soluble chromatin extract from control or TNF-α-treated Jurkat cells and antibodies directed against DSIF p160, NELF-A, and Pol II. The precipitated DNAs were used for PCR amplifications with primers spanning the core and promoter-proximal regions of the A20 gene. (C) The endogenous IκBα gene was analyzed similarly to the A2O gene by RT-PCR (left panel) and by chromatin immunoprecipitation (right panel) as described for panels A and B. Ab, antibody.
The A20 gene is activated by NF-κB following TNF-α induction. NF-κB interacts with and recruits the positive elongation factor P-TEFb (2), a factor capable of antagonizing the negative effect of DSIF in vitro (10, 11, 13). It was therefore expected that, after TNF-α stimulation, DSIF would not inhibit transcription of NF-κB-induced genes. However, after 1 h of TNF-α treatment, DSIF RNAi augmented TNF-α-induced mRNA levels. This finding suggests that DSIF retains its inhibitory activity for the A20 gene even after NF-κB induction by TNF-α.
To examine further the regulation of the endogenous A20 gene by DSIF in NF-κB-induced cells, we tested its association with the A20 promoter-proximal region in cells stimulated with TNF-α for 1 h by the ChIP assay. Figure 5B shows that DSIF is associated with the promoter-proximal region of the A20 gene not only before but also after TNF-α stimulation. Furthermore, the amount of DSIF is increased concomitantly with Pol II after stimulation. These findings confirm that the A20 gene is regulated by DSIF in NF-κB-induced cells.
The IκBα gene promoter is associated with Pol II and the basal transcription machinery before NF-κB stimulation like the A20 gene (1). To test whether regulation by DSIF is specific to the A20 gene or is common to other NF-κB response genes, we carried out a similar analysis on the IκBα gene. As shown in Fig. 5C (left panel), expression of DSIF RNAi enhanced both the basal and the TNF-α-induced mRNA levels of IκBα. Chromatin immunoprecipitation analysis of the IκBα proximal promoter region shows that it is bound by DSIF before and after TNF-α stimulation similar to the A20 gene (Fig. 5C, right panel). Looking at the sequence of the IκBα promoter, we found an element that is closely related to ELIE, GTGACCCT, located adjacent to one of the NF-κB binding sites (positions −173 to −166 relative to the transcription start site). It remains to be determined whether this element is involved in regulation of IκBα gene transcription by DSIF under basal conditions.
In stimulated cells, DSIF inhibitory activity is mediated by NF-κB
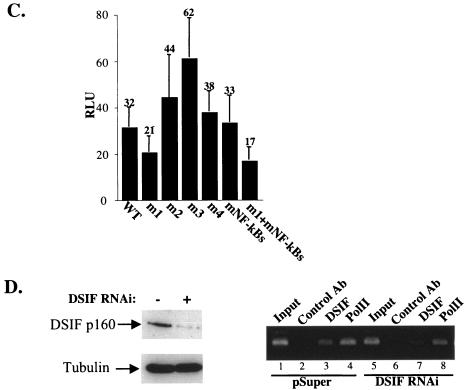
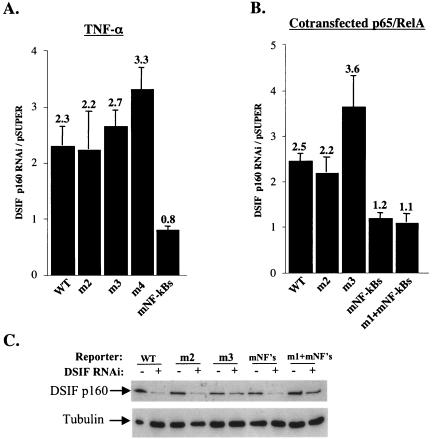
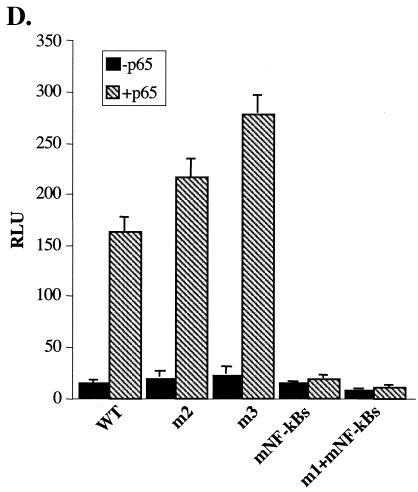
To investigate the mode of regulation of A20 gene transcription by DSIF in NF-κB-activated cells, we examined the effect of DSIF RNAi on the A20 reporter gene activity in cells stimulated with TNF-α for 4.5 h. DSIF RNAi increased TNF-α-induced activity by more than twofold (Fig. 6A, WT column), similar to its effect on the endogenous A20 mRNA. We next tested the effect of DSIF RNAi on the A20 promoter mutants after TNF-α stimulation. Remarkably, under these conditions, the mutations in the NF-κB sites, but not ELIE, eliminated the DSIF RNAi effect (Fig. 6A). These results suggest that, upon TNF-α induction, NF-κB is responsible for the inhibitory effect of DSIF. To confirm this possibility, A20 promoter derivatives were cotransfected with NF-κB protein p65-RelA in the presence or absence of DSIF RNAi (Fig. 6B and C). DSIF inhibition of A20 transcription persisted in the presence of p65-RelA and was eliminated only by mutation in NF-κB binding sites. The lack of DSIF RNAi effect on the mutated NF-κB promoter construct (mNF-κBs) is surprising given that this promoter has an intact ELIE. The reason for that is unknown. A possible explanation might be that NF-κB itself indirectly controls the activity of the factor that binds ELIE. Figure 6D shows the responsiveness of the different mutants to NF-κB, confirming that the mutations just upstream of the NF-κB site do not interfere with NF-κB activation. Together these findings suggest that the elongation-inhibitory activity of DSIF is regulated by the promoter via two specific elements, ELIE and NF-κB, which alternate according to the physiological conditions.
FIG. 6.
DSIF inhibitory activity in NF-κB-stimulated A20 promoter is mediated by NF-κB. (A) 293T cells were cotransfected with various A20 promoter reporters and either pSuper or DSIF p160 RNAi, and 48 h posttransfection cells were treated with TNF-α for 4.5 h. The effect of DSIF p160 depletion is presented as the ratio of the relative luciferase activity in the presence of DSIF RNAi to the activity in the presence of the RNAi parental vector (pSuper). (B) 293T cells were cotransfected with various A20 promoter reporters, p65-RelA expression vector, and either pSuper or DSIF p160 RNAi, and the effect of DSIF p160 depletion on the A20 promoter variants stimulated by NF-κB (p65-RelA) is presented as in panel A. (C) Immunoblot analysis of DSIF and tubulin from a representative transfection experiment. (D) Responsiveness of the various A20 promoter mutants to NF-κB (p65-RelA). Shown is the relative luciferase activity (luciferase units divided by the activity of cotransfected RSV promoter-driven Renilla reporter luciferase) of the transfection experiment described for panel B. The data represent the means and standard deviations of three to seven transfection experiments done independently in duplicate. WT, wild type.
DISCUSSION
DSIF elongation factor has been implicated in controlling Pol II elongation both positively and negatively (12, 15). The elongation-inhibitory effect of DSIF was originally observed in vitro in the presence of the protein kinase inhibitor DRB. In this study we show that the NF-κB response gene A20 is directly targeted by DSIF for inhibition of elongation in vitro and in vivo, supporting the notion that DSIF functions as a negative elongation factor in vivo. The inhibitory effect of DSIF on A20 transcription in vitro was observed only after Pol II had transcribed more than 90 nucleotides. Nevertheless, the inhibitory effect of DSIF was dependent on the type of promoter used and not on the transcribed sequence, indicating that the negative effect of DSIF on elongation is promoter specific and not general.
Dissection of the A20 promoter activity, under basal conditions, led to the identification of ELIE, a specific negative element in the promoter that mediates DSIF inhibition. ELIE is likely to bind a novel type of repressor that negatively controls transcription elongation. The question arising is how does this promoter-bound elongation repressor regulate the activity of DSIF, which joins Pol II after initiation (3, 8) and inhibits elongation downstream? One possibility is that while Pol II is on the promoter it is modified by this repressor (directly or indirectly), rendering it much more sensitive to inhibition by DSIF. Alternatively, this repressor is involved in recruitment of either DSIF to the preinitiation complex or a specific form of Pol II that is already engaged with a factor that facilitates DSIF activity. The elongation inhibitor NELF could be such a factor, since it was previously shown to cooperate with DSIF in elongation inhibition and to interact with the initiating form of Pol II (15). We tested this possibility by immunodepletion of NELF, but since A20 basal activity was not enhanced (data not shown), NELF is unlikely to be involved in inhibition of A20 elongation. A third possibility is that this promoter-bound repressor functions in a manner analogous to the drug DRB to induce DSIF activity.
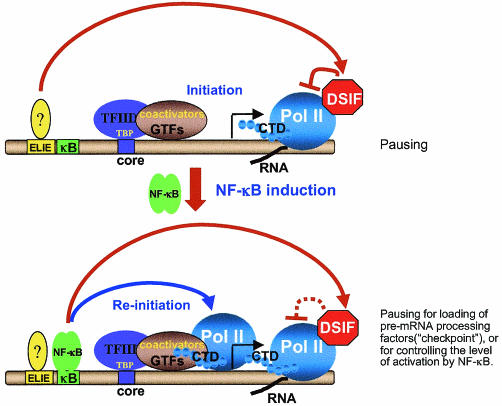
Before stimulation of NF-κB, the general transcription machinery and coactivators are associated with the A20 promoter. This transcription complex is capable of initiation whereas reinitiation is restricted (1) (Fig. 1). Given that ELIE negatively regulates the basal activity of A20 via DSIF, it is likely that, before NF-κB induction, DSIF plays a true inhibitory role, rendering the prebound transcription machinery less active until NF-κB is stimulated (Fig. 7).
FIG. 7.
Model explaining the differential role of DSIF inhibition of elongation in nonstimulated and stimulated cells. In unstimulated cells (upper panel), the promoters of rapidly induced NF-κB target genes, like A20, are bound by the entire Pol II transcription apparatus, but the basal activity is low since this complex is capable of initiation but not reinitiation (1). Under these conditions, a factor that binds ELIE on the A20 promoter inhibits the basal transcription at the level of elongation via DSIF. This effect is required to keep the basal activity of the prebound transcription complex much lower until NF-κB is stimulated. After NF-κB is induced, it enhances the reinitiation rate but, on the other hand, is responsible for inhibition of elongation by DSIF. This inhibition might serve to load the elongating polymerase with pre-mRNA processing factors in order to ensure that each transcript that has been initiated will be completely processed into mature and translatable mRNA. Alternatively, this inhibition may be used to control the level of activation of the A20 gene by NF-κB, which might be important under certain physiological conditions. CTD, C-terminal domain; GTFs, general transcription factors; TBP, TATA binding protein.
Although transcription of the A20 gene is induced by NF-κB in response to TNF-α, DSIF retains its inhibitory activity on the A20 promoter. Remarkably, after TNF-α induction, regulation of the inhibitory effect of DSIF on the A20 promoter is switched from ELIE to NF-κB. What is the function of the NF-κB-mediated inhibition of elongation by DSIF? Given the strong positive effect of NF-κB on A20 gene transcription, one possibility might be that this regulation serves an important purpose in transcription, rather than inhibition per se (Fig. 7). This role may be linked to the recently proposed “checkpoint” model for coordinating transcription with pre-mRNA processing (7). Slowing down the processivity of Pol II by DSIF may be used to guarantee that each initiated transcript will be completed properly, by providing enough time to load the pre-mRNA processing complexes onto the C-terminal domain of Pol II (Fig. 7). Alternatively, DSIF may be used to control the level of activation of the A20 gene by NF-κB. Whether these are indeed the purposes of the elongation control mechanism described here is yet to be determined. Nonetheless, we demonstrate here a dynamic and unique mechanism of regulation of Pol II elongation by DSIF, which takes place downstream from the initiation site, yet is tightly regulated by upstream promoter-bound transcription factors.
.
Acknowledgments
We thank Sandra Moshonov and Merav Revach for critical reading of the manuscript.
This work was supported by research grants from the Minerva Foundation, Germany; the Israel Science Foundation; and the Crown Endowment Fund for Immunological Research. Rivka Dikstein is an incumbent of the Martha S. Sagon Career Development Chair.
REFERENCE
- 1.Ainbinder, E., M. Revach, O. Wolstein, S. Moshonov, and R. Dikstein. 2002. The mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-κB. Mol. Cell. Biol. 22:6354-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-555. [DOI] [PubMed] [Google Scholar]
- 5.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 6.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 7.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 8.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 9.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 11.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 12.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolstein, O., A. Silkov, M. Revach, and R. Dikstein. 2000. Specific interaction of TAFII105 with OCA-B is involved in activation of octamer-dependent transcription. J. Biol. Chem. 275:16459-16465. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 16.Zorio, D. A., and D. L. Bentley. 2001. Transcription elongation: the ‘Foggy' is lifting… . Curr. Biol. 11:R144-R146. [DOI] [PubMed] [Google Scholar]