Abstract
Metazoan replication-dependent histone mRNAs end in a stem-loop sequence. The one known exception is the histone mRNA in amphibian oocytes, which has a short oligo(A) tail attached to the stem-loop sequence. Amphibian oocytes also contain two proteins that bind the 3′ end of histone mRNA: xSLBP1, the homologue of the mammalian SLBP, and xSLBP2, which is present only in oocytes. xSLBP2 is an inhibitor of histone mRNA translation, while xSLBP1 activates translation. The short A tail on histone mRNAs appears at stage II to III of oogenesis and is present on histone mRNAs throughout the rest of oogenesis. At oocyte maturation, the oligo(A) tail is removed and the xSLBP2 is degraded, resulting in the activation of translation of histone mRNA. Both SLBPs bind to the stem-loop with the oligo(A) tail with similar affinities. Reporter mRNAs ending in the stem-loop with or without the oligo(A) tail are translated equally well in a reticulocyte lysate, and their translation is stimulated by the presence of xSLBP1. In contrast, translation of the reporter mRNA with an oligo(A) tail is not activated in frog oocytes in response to the presence of xSLBP1. These results suggest that the oligo(A) tail is an active part of the translation repression mechanism that silences histone mRNA during oogenesis and that its removal is part of the mechanism that activates translation.
During Xenopus oogenesis and early embryogenesis, much of the control of gene activity is a result of programmed translational regulation (4, 12, 36). Since there is no transcription until the midblastula transition (21), the oocyte and early embryo rely on regulation of the stored maternal mRNAs to modulate the pattern of protein synthesis. One set of proteins whose synthesis is regulated during oogenesis and early embryogenesis is that of the histone proteins (1, 39). The mRNAs encoding all five classes of histone proteins are unique among metazoan mRNAs in that they are the only known mRNAs that lack poly(A) tails and end instead in a conserved stem-loop sequence (16, 18). This stem-loop is a critical cis-acting element involved in all aspects of histone mRNA biosynthesis across different species (17). Over 25 years ago it was reported that histone mRNAs from Xenopus oocytes are polyadenylated (15). These amphibian transcripts have an A tail added directly to the stem-loop (2).
Histone mRNAs are synthesized early in oogenesis, and synthesis is complete by the end of stage II of oogenesis. The histone mRNAs are also translated in stage II to provide a maternal store of histone proteins (39). In later stages of oogenesis there is little or no translation of the histone mRNA (39). When full-grown stage VI oocytes are treated with progesterone, they undergo maturation, initiating progression through meiosis, and also activate a translational regulation program necessary for the initial stages of embryogenesis. One of the mRNAs activated at oocyte maturation is histone mRNA (1).
The 3′ end of histone mRNA is essential for efficient translation (9, 25). In Xenopus oocytes there are two stem-loop binding proteins (SLBPs) that can bind the 3′ end of histone mRNAs: xSLBP1 and xSLBP2 (33). xSLBP1, which is the orthologue of the mammalian SLBP, is required for histone pre-mRNA processing (13), and it is also necessary for efficient translation of histone mRNA (25). In contrast, xSLBP2 is found only in oocytes and does not support translation of histone mRNA. xSLBP2 is associated with the maternally stored histone mRNA and is destroyed during oocyte maturation (33). The destruction of xSLBP2 may allow xSLBP1 to bind to the mRNA, activating its translation.
Histone mRNAs in Xenopus oocytes contain a short A tail, which is attached to the stem-loop (2). We report here that the oligo(A) tail plays an active role in translation repression. Histone mRNAs ending with an oligo(A) tail are not translated in the presence of xSLBP1 in Xenopus oocytes, although they are translated in reticulocyte lysates. In addition to xSLBP2, the oligo(A) tail also plays a role in repression of histone mRNA translation.
MATERIALS AND METHOD
Construction of SLBP, histone, and luciferase clones
The construction of vectors encoding xSLBP1, xSLBP2, human SLBP (hSLBP), MS2, and MS2-hSLBP and the luciferase mRNAs ending in the stem-loop, tetraloop (TL), and MS2 sequences have been described previously (25). To generate the Luc-SLA8/SLC8/SLN8 genes, two complementary oligonucleotides encoding the histone stem-loop sequence followed by eight adenosines, eight cytosines, or the sequence GATTACAG were annealed and ligated into the Luc-SL vector digested with BamHI and AflII. A similar strategy was used to construct the Luc-MS2A8 gene.
S1 nuclease mapping
To map the 3′ end of the histone mRNA, we used two different probes. The S1 probe used to map the 3′ end of the histone xH2a mRNA (Fig. 1B) was generated by PCR, using a previously cloned Xenopus H2a cDNA ending in 20 adenosines as the template (33). Standard RNAs ending in the histone stem-loop or in the stem-loop followed by 25 As were transcribed with T7 RNA polymerase from appropriate DNA templates. The 184-nucleotide (nt) PCR product contains 130 nt of histone mRNA and the 20-nt poly(A) tract followed by 34 nt that had no complementarity to the histone xH2a mRNA. This fragment was labeled at the 3′ end with the Klenow fragment of DNA polymerase I (New England Biolabs) and [α-32P]dCTP. For the S1 analysis whose results are shown in Fig. 1C and 2A, the plasmid encoding the xH2a gene was digested with XmaI and labeled with the Klenow fragment of DNA polymerase I and [α-32P]dCTP. Hybridization was done at 56°C, and S1 digestion was performed as previously described (10, 13) except that the digestion was carried out on ice with 4 U of S1 nuclease to preserve the oligo(A-T) hybrids (35). The protected fragments were resolved on a 10% polyacrylamide-7 M urea gel and detected by autoradiography.
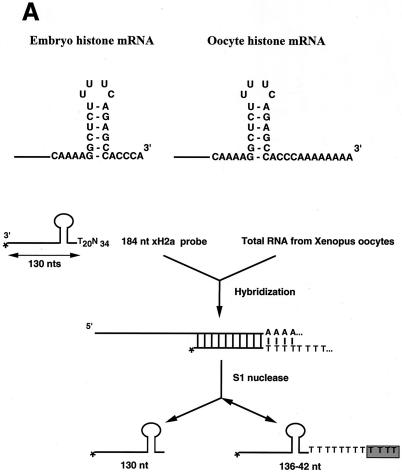
FIG. 1.
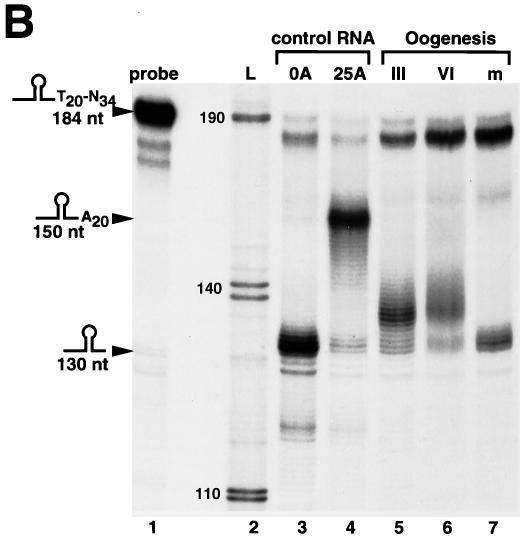
Oocyte histone mRNA contains an oligo(A) tail. (A) The structures of the 3′ ends of the egg (left panel) and oocyte (right panel) histone mRNAs are shown. The S1 nuclease protection assay used to determine the length of the poly(A) tail is diagrammed. (B) The S1 nuclease assay (as shown in panel A) was used to map the 3′ end of Xenopus histone H2a mRNA in stage III and stage VI oocytes and matured oocytes. Synthetic RNAs ending in the ACCCA sequence formed by histone pre-mRNA processing (lane 3) or ending in the ACCCA followed by 25 As (lane 4) were incubated with the probe containing 20 Ts after the stem-loop. The reaction mixtures were treated with S1 nuclease, resulting in a fragment of 130 nt resulting from protection of the ACCCA (lane 3) and a 150-nt fragment resulting from protection of the A20 tail (lane 4). For lanes 5 to 7, total cell RNA from one oocyte equivalent of RNA from stage III, stage VI, or matured oocytes (m) was incubated with the probe, the reaction mixtures were treated with S1 nuclease, and the protected fragments were analyzed on a 60-cm-long 8% polyacrylamide-7 M urea gel. Lane 1, a 184-nt probe; lane 2 (L), pUC18 digested with HpaII and labeled with [α-32P]dCTP. The sizes of these fragments are indicated. The fragment at about 175 nt in lanes 1 to 5 is derived from the probe. (C) RNA from one oocyte or egg was analyzed at the indicated stages by S1 nuclease mapping as described in Materials and Methods; the results are presented in lanes 1 to 6. The protected fragments were analyzed on a 10-cm-long 8% polyacrylamide-7 M urea gel. The results are representative of analyses of oocytes from four different frogs. The results of analysis of RNA from oocytes matured by treatment with progesterone are shown in lane 7, and lane 8 shows the results of analysis of RNA from Xenopus eggs. Lanes 9 and 10 show the results of analysis of synthetic RNAs ending in the histone stem-loop and in the stem-loop followed by 25 As, respectively. Lane 11 shows the results of analysis of 10 μg of yeast tRNA. (D) Stage VI oocytes were incubated with progesterone. At 1 h intervals, 20 oocytes were removed and pooled. The oocytes were homogenized in buffer, and RNA was prepared from 80% of the homogenate. The rest of the homogenate (four oocytes) was analyzed by sodium dodecyl sulfate (SDS)-gel electrophoresis, and xSLBP2 was detected by Western blotting. The S1 analysis was done using RNA from four oocytes. The percentages of histone mRNA containing oligo(A) tails and the amounts of xSLBP2 (as determined by Western blotting) are indicated, with 100% representing the levels prior to addition of progesterone. At 7 h 70% of the oocytes had matured (as determined by the appearance of the white spot at the animal pole), and at 8 and 9 h all the selected oocytes had matured. No oocytes had matured at 6 h.
FIG. 2.
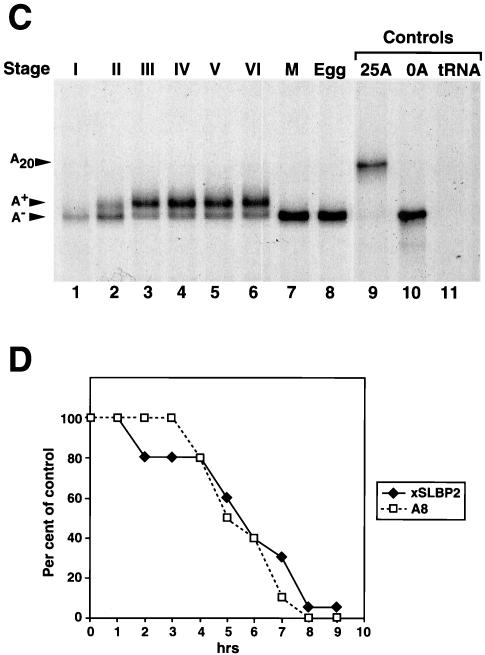
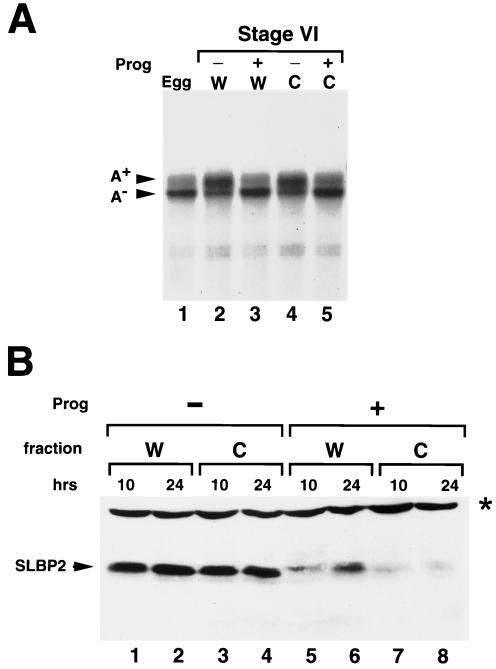
Deadenylation and xSLBP2 degradation at oocyte maturation does not require the nucleus. (A) Stage VI oocytes collected from a single frog were divided into two groups. One group (W) was incubated in OR-2 buffer with (+) progesterone (Prog) for 12 h (lane 3) or without (−) progesterone (lane 2). The second group of oocytes was enucleated (C); half of these oocytes were incubated in buffer (lane 4) without progesterone and the other half were treated with progesterone for 12 h (lane 5). A total of 20 oocytes were randomly selected from each of the four sets of oocytes, and RNAs were prepared. A total of 85% of the oocytes that had been treated with progesterone had matured after 12 h. One oocyte equivalent of RNA was analyzed by S1 nuclease mapping as described for Fig. 1C. Lane 1 shows the analysis of one egg equivalent of RNA from Xenopus eggs. (B) Stage VI oocytes from a single frog were treated as described for panel A. A total of 20 oocytes from each treatment were randomly selected after 10 h (lanes 1, 3, 5, and 7) or 24 h (lanes 2, 4, 6, and 8) of incubation with or without progesterone. Total cell protein was prepared and resolved by gel electrophoresis, and SLBP2 was detected by Western blotting. The oocytes in lanes 3, 4, 7, and 8 were enucleated, while the oocytes in lanes 1, 2, 5, and 6 were intact. The band labeled with a star represents a cross-reacting protein that remains constant during oocyte maturation and serves as an internal control.
The S1 probe used to determine luciferase RNA levels (see Fig. 7) was generated by linearization of the Luc-SL vector with DraII (which cleaves in the luciferase coding region). The 3′ end was labeled with the Klenow fragment of DNA polymerase I and [α-32P]dCTP. Total RNA was extracted (using TRIzol reagent) (Invitrogen) from oocyte lysates, and 1 oocyte equivalent of total RNA was hybridized to 5 ng of probe. Hybridization, S1 digestion, and detection were performed as previously described (25).
FIG. 7.
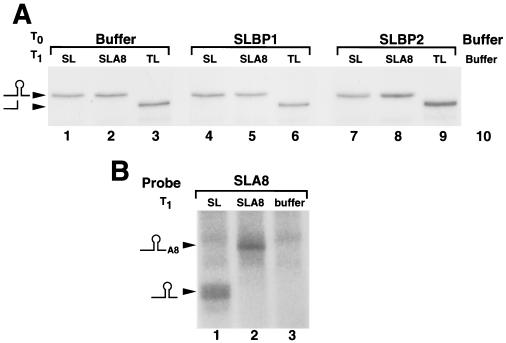
The different reporter mRNAs are equally stable, and the A8 tail is not lost after injection. (A) Oocytes were injected with buffer (lanes 1 to 3 and 10), xSLBP1 mRNA (lanes 4 to 6), or xSLBP2 mRNA (lanes 7 to 9) and were injected 16 h later with Luc-SL (lanes 1, 4, and 7), Luc-SLA8 (lanes 2, 5, and 8), Luc-TL mRNA (lanes 3, 6, and 9), or buffer (lane 10). RNA was prepared 16 h later, and one oocyte equivalent of RNA was analyzed by S1 nuclease mapping, using the Luc-SL gene labeled at the 3′ end of a restriction enzyme site in the luciferase coding region as described in Materials and Methods. These samples are from the collection of oocytes used as described for Fig. 5B and C. The probe protects the same fragment from the Luc-SL and Luc-SLA8 mRNA, which maps to the stem-loop (SL), and a 15-nt-shorter fragment from the Luc-TL mRNA, which differs in the loop. Since the probe is end labeled, the intensity of the band is proportional to the amount of mRNA in the sample. (B) RNA samples (used as described for panel A) from oocytes injected first with xSLBP1 mRNA and then with either Luc-SL (lane 1), Luc-SLA8 (lane 2), or buffer (lane 3) were analyzed using a probe which contains the SLA8 tail, with the end of the stem-loop 40 nt from the labeled end. The protected fragments were analyzed on a 10% polyacrylamide-7 M urea gel. The positions of the fragments ending at the stem-loop (40 nt) and the SLA8 tail (48 nt) are indicated.
For precise mapping of the 3′ end of luciferase mRNA (see Fig. 7), the terminal 230-nt sequence from the Luc-SL and Luc-SLA8 vector was amplified by PCR and a unique HindIII site was introduced in the forward primer. These amplified products were digested with HindIII and EcoRI and subcloned into homologous sites in pGEM-3Zf− to generate pGEM-Luc-SL and pGEM-Luc-SLA8. The S1 probes used to determine the integrity of the 3′ end of Luc-SL and Luc-SLA8 were generated by linearization of pGEM-Luc-SL and pGEM-Luc-SLA8 vectors with XbaI, which cuts 40 nt before the 3′ end of the histone mRNA, followed by labeling with the Klenow fragment of DNA polymerase I and [α-32P]dCTP. Hybridization was done at 56°C. S1 digestion was performed at 4°C as described above.
Analysis of xSLBP proteins
The stem-loop probe and the conditions used for the mobility shift assay were as previously described (25). Purification of the anti-xSLBP antibodies and the conditions used for the Western blotting analysis were as previously described (25, 33). Unlabeled and labeled RNAs ending in the stem-loop or the stem-loop followed by oligo(A) for the mobility shift experiments were transcribed (using T7 RNA polymerase) from appropriate templates.
In vitro transcription and translation
Transcription of capped and uncapped luciferase constructs (as well as of xSLBP1, xSLBP2, MS2, and MS2-hSLBP mRNAs) was performed as previously described (25). The synthesis of different SLBPs in vitro and the analysis of translation of the luciferase reporter mRNAs were done exactly as previously described, as were the protocols for microinjections, Western blotting, and luciferase assays (25).
RESULT
Over 25 years ago, it was reported that histone mRNAs in Xenopus oocytes, unlike histone mRNAs in other systems, contain poly(A) tails, as determined on the basis of their ability to bind to poly(dT) cellulose and as assayed by in vitro translation (15, 22, 23). Several years later it became apparent that the A tails were actually added onto the stem-loop sequence at the 3′ end of histone mRNAs (2) and that the A tails were quite short. Histone mRNAs cloned from cDNA libraries prepared from Xenopus ovary mRNA primed with oligo(dT) contain histone mRNAs encoding all five histone proteins with an A tail following the stem-loop whose length varies but is likely determined partly by the primers used to construct the library (40; Z. F. Wang and W. F. Marzluff, unpublished data). When we analyzed histone mRNAs by Northern blotting (even on high-agarose-percentage gels), however, the histone mRNAs from oocytes and eggs migrated identically (not shown). Since histone mRNAs are only 500 nt long, poly(A) tails of 25 to 50 nt would have been readily apparent.
Oocyte histone mRNA has an A tail of 6 to 10 nt
To quantitatively analyze the 3′ end of Xenopus histone mRNAs, we developed an S1 nuclease protection assay that can measure the length of the A tail on the 3′ end of histone mRNA (Fig. 1A). We prepared a 3′-labeled probe ending in the stem-loop followed by 20 As and 34 additional nucleotides. The probe was labeled at a site 130 nt from the end of the stem-loop, allowing us to precisely resolve even very short oligo(A) tails. For control mRNAs, we synthesized in vitro an mRNA ending at the stem-loop and one ending at the stem-loop with a 25-A extension. The probe containing 20 Ts will protect a 130-nt fragment from the wild-type histone mRNA and a 150-nt fragment from the mRNA ending in 20 As. A major challenge in the S1 mapping experiments is maintaining the integrity of the oligo(A-T) hybrids at the 3′ end of the mRNA during the experiment. To enhance annealing of the oligo(A-T) sequence, we used the minimal amount of enzyme necessary to degrade the free probe and performed the S1 digestions on ice (35). For each set of S1 nuclease assays we included the two control mRNAs. This approach allowed us to control for the loss of the A-T tail by S1 digestion of the relatively unstable oligo(A-T) hybrid, which occurred to different extents among the sets of samples.
We used this assay to determine the 3′ end of the Xenopus histone H2a mRNA during oogenesis. The control mRNA ending at the stem-loop protected a 130-nt fragment as expected, while the control mRNA ending in 25 As protected a major fragment of 150 nt (protecting the 20 Ts in the probe; Fig. 1B, lanes 3 and 4, respectively). The polyadenylated control mRNA displays a distribution of slightly smaller fragments as well as a small amount of the 130-nt protected fragment (Fig. 1B, lane 4). The 130-nt fragment could result either from “breathing” of the A-T hybrid, which allows the S1 nuclease to cleave the probe in the A stretch (resulting in subsequent complete removal of the A-T hybrid region), or from a small amount of the control mRNA that was not adenylated. We assayed total RNA from stage III and stage VI oocytes and matured oocytes. The oocyte histone mRNAs protected fragments longer than 130 nt. The most abundant fragments were 5 to 8 nt longer than those seen with the mature histone mRNA, with protected fragments up to 10 to 11 nt in length detected. Only a small proportion of the protected fragments were 130 nt, consistent with the great majority (if not all) of the mRNA ending in an oligo(A) tail (Fig. 1B, lanes 5 and 6). In contrast, RNA from in vitro-matured oocytes protected a major fragment of 130 nt, similar to the results seen with the standard synthetic histone mRNA (Fig. 1B, lane 7). These results are in general agreement with the results of Ballantine and Woodland (2).
We determined the state of the 3′ end of the Xenopus histone mRNAs during oogenesis and early development. In stage I oocytes, most of the histone mRNAs do not contain A tails (Fig. 1C, lane 1). The mRNA from stage II oocytes was partially adenylated. In stages III and VI of oogenesis, the great majority of the histone mRNAs contained a short A tail (Fig. 1C, lanes 3 to 6). Since RNAs from the same number of oocytes were analyzed for each lane, this assay allows us to determine the relative amounts of histone mRNAs in different stages of oogenesis. Note that as is true for most other maternal mRNAs (7) and as previously reported for histone mRNAs (38), most of the histone mRNA accumulates very early in oogenesis and the steady-state level of histone mRNA remains constant after stage III of oogenesis.
Different stage II oocyte preparations gave different proportions of adenylated histone mRNAs (the distinction between oocyte stages is somewhat subjective during the sorting of the oocytes), but all preparations of stage I oocytes contained less histone mRNA with little or no adenylated mRNA. We conclude that in very early oogenesis (stages I and II), most of the histone mRNAs do not contain A tails and that by stage III the great majority (if not all) of the histone mRNAs contain A tails.
To determine whether the A tails were lost during oocyte maturation, we treated stage VI oocytes with progesterone and compared the histone mRNA levels in stage VI oocytes to those of oocytes which had matured after progesterone treatment (Fig. 1C, lanes 6 and 7). We also analyzed the histone mRNA from eggs in a parallel assay (Fig. 1C, lane 8). The histone mRNA from oocytes matured in vitro with progesterone (as well as the histone mRNA from eggs) was not adenylated. Therefore, treatment of the oocytes with progesterone recapitulated the loss of the oligo(A) tails from histone mRNA.
xSLBP2, which inhibits histone mRNA translation, is also degraded during oocyte maturation (33). To determine the relative timings of these two changes, we analyzed the amounts of xSLBP2 by Western blotting and the proportion of histone mRNAs containing the oligo(A) tails by S1 nuclease assay at 1-h intervals after progesterone treatment. The removal of the oligo(A) tails and the destruction of xSLBP2 occurred in parallel (Fig. 1D). This assay has limited temporal resolution, due to the lack of precise synchrony with which oocytes respond to progesterone and to the absence of phenotypic markers for the progression of maturation. Although the removal of the oligo(A) tails and the destruction of xSLBP2 could not be temporally separated, both had occurred extensively prior to the appearance of the white spot on the animal pole that is indicative of germinal vesicle breakdown and completion of oocyte maturation.
Deadenylation of histone mRNA and degradation of xSLBP2 at maturation can occur in the absence of the nucleus
Messages that lack a cytoplasmic polyadenylation element (CPE) undergo default deadenylation at maturation, and their translational activity is inhibited (8, 31). An mRNA deadenylase called DAN has been purified from Xenopus oocytes, and there is a human orthologue called PARN. Removal of the nucleus or injection of antibodies against DAN (PARN) inhibits default deadenylation (14). To test whether removal of the histone mRNA oligo(A) tract at maturation requires the nucleus (like default deadenylation of non-CPE-containing mRNAs), enucleated oocytes were treated with progesterone to induce maturation. When enucleated oocytes were treated with progesterone, the histone mRNAs lost their oligo(A) tract to the same extent as whole oocytes treated with progesterone (Fig. 2A; compare lanes 3 and 5).
We also tested whether enucleated oocytes treated with progesterone degraded xSLBP2. Treatment of stage VI oocytes with progesterone resulted in a loss of xSLBP2 as analyzed by Western blotting (Fig. 2B, lanes 5 and 6). Identical results were obtained when the oocytes were enucleated prior to treatment with progesterone (Fig. 2B, lanes 7 and 8). We conclude that removal of the histone mRNA oligo(A) tail and the degradation of xSLBP2 can be mediated solely by cytoplasmic factors and is not dependent on germinal vesicle breakdown.
The presence of the oligo(A) tail does not affect binding of either xSLBP
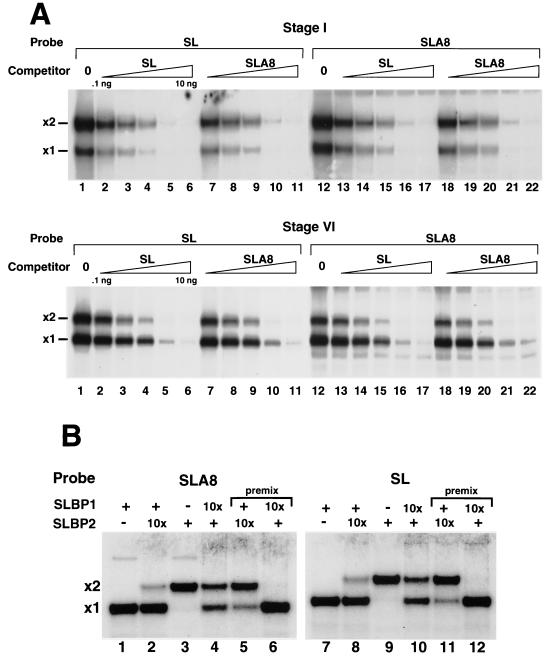
One function of the oligo(A) tail could be to help determine which xSLBP is associated with the histone mRNA. A second possibility is that another (or multiple) factor(s) could stably associate with the oligo(A) tail. Note that these tails are not long enough to be bound by the poly(A) binding protein (PABP), so it is unlikely that PABP binds to the oocyte histone mRNA via the poly(A) tract (24). To determine whether the two xSLBPs show different relative affinities for the histone stem-loop and the stem-loop with an oligo(A) tail, we performed mobility shift assays. We used extracts from stage I oocytes, which primarily form complexes containing xSLBP2 (33), and extracts from stage VI oocytes, which form complexes containing both xSLBP1 and xSLBP2, with xSLBP1 complexes being the more abundant species. We incubated each extract with either a radiolabeled stem-loop probe or with the stem-loop with 8 As (SLA8) attached. We then tested the ability of stem-loop and SLA8 unlabeled RNAs to compete for the binding of each xSLBP. Note that these assays are done using oocyte extracts, so any modifications of the xSLBPs that could alter their affinity for the stem-loop (which might occur at different stages of oogenesis) are likely to be retained. The two SLBPs bound the two probes with similar affinities, and the stem-loop and SLA8 unlabeled RNAs were equally effective in competing for the binding to either the SL or the SLA8 probe (Fig. 3A). Similar results were previously reported for the wild-type probe when the proteins were expressed in reticulocyte lysates (13).
FIG. 3.
xSLBP1 and xSLBP2 bind equally well to the stem-loop and the oligoadenylated stem-loop. (A) Extracts from stage I (top panel) or stage VI (bottom panel) oocytes were incubated with the stem-loop (SL) probe (lanes 1 to 11) or the radiolabeled oligoadenylated stem-loop probe (lanes 12 to 22). Increasing amounts of competitor stem-loop (0.1 to 10 ng; lanes 2 to 6 and lanes 13 to 17) or oligoadenylated stem-loop (0.1 to 10 ng; lanes 7 to 11 and lanes 18 to 22) were mixed with the probe prior to addition of the oocyte lysate. Complexes were resolved by electrophoresis on 8% polyacrylamide gels under native conditions and detected by autoradiography. The positions of the complexes with xSLBP1 (x1) and xSLBP2 (x2) are indicated. Lanes 1 and 12 were analyzed without an added competitor. (B) xSLBP1 and xSLBP2 were synthesized in reticulocyte lysates in the presence of [35S]methionine, and the amounts of each protein were estimated by autoradiography of the in vitro-synthesized proteins. The amounts of xSLBP1 and xSLBP2 required to completely shift the stem-loop probe or the SLA8 probe were determined. Mobility shift assays were performed with (+) or without (−) radiolabeled SLA8 probe (lanes 1 to 6) or the stem-loop probe (lanes 7 to 12). The same amount of xSLBP1 or xSLBP2 was incubated with the stem-loop probe (lanes 1, 3, 7, and 9). After incubation for 5 min at 4°C, a 10-fold excess (10x) of the other xSLBP was added (lanes 2, 4, 8, and 10) and the reaction mixture was incubated for 60 min prior to analysis. For lanes 5, 6, 11, and 12, the two proteins were mixed prior to the addition of the probe (premix).
We also tested the ability of the SLA25 probe to bind to proteins in the oocyte extracts. This probe, which contains 25 As after the stem-loop, can be bound by PABP. We observed complexes which resulted from both PABP and SLBP binding to the same probe (data not shown). Thus, the A8 tail is not long enough to bind PABP.
The SLBPs do not rapidly exchange in vitro
A striking finding in the previous work of Wang et al. was that xSLBP1 was bound to histone mRNA early in oogenesis whereas xSLBP2 was bound to the histone mRNA late in oogenesis (33). When the amounts of active xSLBPs are determined by mobility shift assays, however, xSLBP2 is the major SLBP early in oogenesis and xSLBP1 is the major SLBP late in oogenesis (33). Since it is possible that one function of the poly(A) tail is to alter the exchangeability of the SLBPs, we tested whether the xSLBPs would interchange in vitro on either the wild-type or the oligo(A) stem-loop probe.
We synthesized the xSLBPs in vitro using rabbit reticulocyte lysates and labeled the proteins with [35S]methionine to allow us to estimate the amount of protein (there is no mammalian SLBP in reticulocyte lysates; see reference 34). We incubated the probe with an excess of one of the SLBPs under conditions in which the entire radiolabeled probe formed a complex with an SLBP after incubation for 5 min at 4°C. An approximately 10-fold excess of the second SLBP was then added to the extract and incubated for an additional 60 min on ice. As controls the proteins were mixed together prior to addition of the probe.
When we formed a complex with xSLBP1 on either the SLA8 (Fig. 3B, lane 1) or the stem-loop (Fig. 3B, lane 7) probe, all of the probe formed a single complex. When we mixed an excess of xSLBP2 with xSLBP1 prior to addition to the probe, about 90% of the complexes formed contained xSLBP2 with both probes (Fig. 3B, lanes 5 and 11). However, when the same amount of excess xSLBP2 was added after formation of the complex with xSLBP1, there was a less than 10% exchange of the proteins in 60 min at 4°C (Fig. 3B, lanes 2 and 8). When the experiment was reversed (forming the complexes on xSLBP2 first), the addition of excess xSLBP1 resulted in partial exchange during the 60-min incubation. Partial exchange was observed on both probes (Fig. 3B, lanes 4 and 10). When the proteins were premixed in the same amounts used in the exchange experiments, the xSLBP1 complex was the only complex detected with either probe (Fig. 3B, lanes 6 and 12). These results demonstrate that the proteins do not exchange rapidly, in agreement with previous results that the RNA does not rapidly dissociate from SLBP (37). The presence of the oligo(A) sequence did not affect the exchange of the SLBPs in vitro.
Effect of the oligo(A) tract on translation in vitro
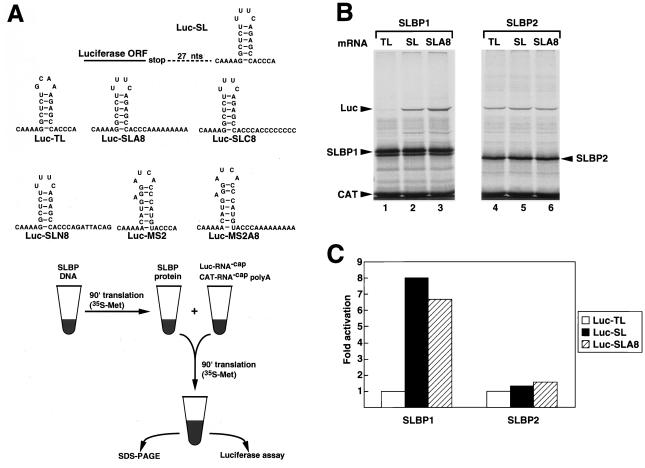
We have previously shown that xSLBP1 actively supports translation of a luciferase reporter mRNA ending in a stem-loop (both in reticulocyte lysates and in stage VI oocytes) whereas xSLBP2 does not (25). An MS2-SLBP fusion protein will also stimulate translation of luciferase reporter mRNAs that end in the histone stem-loop or the MS2 binding site (25). To determine whether the short A tail had any effect on the translation of the reporter mRNA ending in the stem-loop, we constructed a luciferase mRNA that ended in a stem-loop followed by 8 As (Luc-SLA8). As controls we constructed reporter mRNAs that ended in 8 Cs (Luc-SLC8) or in a random 8-nt sequence (Luc-SLN8). We also used mRNAs ending in the MS2 binding site (Luc-MS2) or in the MS2 site followed by 8 As (Luc-MS2A8). The constructs are shown in Fig. 4A.
FIG. 4.
xSLBP1 stimulates the translation of the Luc-SL and Luc-SLA8 mRNAs in vitro. (A) The seven reporter mRNAs used in the various translation experiments are represented at the top. A diagram of the in vitro translation assay (taken from reference 25) is shown in the lower panel. SLBP1 or SLBP2 was synthesized from capped synthetic mRNA in a reticulocyte lysate in the presence of [35S]methionine. Aliquots of lysates containing xSLBP1 or xSLBP2 were added to fresh lysate together with luciferase reporter mRNA and an uncapped polyadenylated CAT mRNA and incubated for 90 min. (B) The uncapped Luc-TL (lanes 1 and 4), Luc-SL (lanes 2 and 5), or Luc-SLA8 (lanes 3 and 6) mRNAs were added to reticulocyte lysates containing xSLBP1 (lanes 1 to 3) or xSLBP2 (lanes 4 to 6) together with uncapped polyadenylated CAT mRNA. The translation products were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography. There was a slight increase in the translation of the Luc-TL mRNA in the presence of xSLBP2 in the experiment whose results are shown, but this was not observed in multiple other experiments; in those experiments, there was essentially no effect seen on translation of either the Luc-TL or Luc-SL mRNAs by SLBP2 in the reticulocyte lysate (25). SL, stem-loop. (C) The reaction mixtures described for panel B were analyzed for luciferase activity; the activity levels are expressed relative to the Luc-TL mRNA levels.
In the in vitro assay, either xSLBP1 or xSLBP2 is first synthesized in a reticulocyte lysate from synthetic capped mRNA. Lysate containing the SLBP is then mixed with fresh lysate in the presence of an uncapped luciferase reporter mRNA and a competitor uncapped polyadenylated chloramphenicol acetyltransferase (CAT) mRNA (Fig. 4A). The CAT protein produced from this mRNA serves as an internal standard, and the luciferase expression is normalized to the expression of CAT protein within each experiment. This assay measures the ability of the reporter mRNA to compete effectively for initiation factors in the reticulocyte lysate. Previously we had shown that xSLBP1, but not xSLBP2, stimulates translation of the reporter Luc-SL mRNA but not of the Luc-TL mRNA containing a GCAA TL instead of the UUUC loop (25).
We compared the effect of the presence of xSLBP1 and xSLBP2 on translation of the Luc-SL, Luc-SLA8, and Luc-TL mRNAs in vitro. When xSLBP1 was synthesized in the lysate prior to the addition of the luciferase mRNAs, there was a six- to eightfold increase in the translation efficiency of both the Luc-SL and Luc-SLA8 mRNAs (Fig. 4B, lanes 2 and 3, and Fig. 4C) compared to the results seen with translation of Luc-TL mRNA (Fig. 4B, lane 1, and Fig. 4C), as determined both by luciferase assay and by autoradiography of the luciferase protein. Addition of a lysate containing xSLBP2 did not affect translation of any of the reporter mRNAs (Fig. 4B, lanes 4 to 6, and Fig. 4C). We conclude that the oligo(A) tail does not affect the ability of xSLBP1 to activate translation of the Luc-SLA8 mRNA in the reticulocyte lysate.
The oligo(A) tail reduces activation of translation of the Luc-SLA8 mRNA in Xenopus oocytes
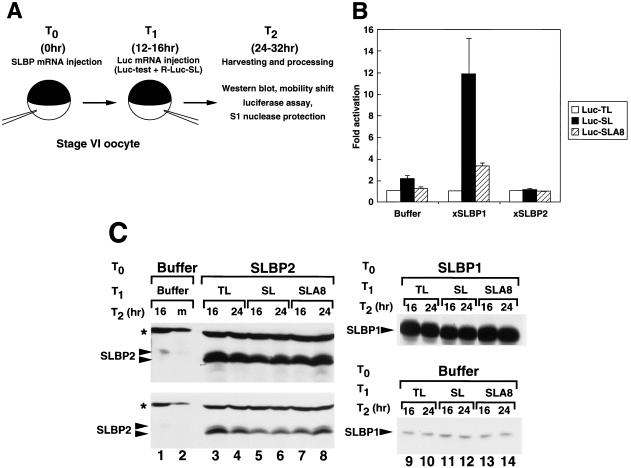
To assay translation of the reporter mRNAs in vivo (Fig. 5A) (25), we first injected a synthetic mRNA encoding an SLBP protein into stage VI oocytes. At 16 h later we injected equal amounts of a capped synthetic firefly luciferase mRNA (Luc-test) together with an internal standard capped Renilla luciferase mRNA ending in the histone stem-loop (R-Luc-SL). At different times after injection of the reporter mRNAs we harvested the oocytes and measured the firefly and Renilla luciferase activity levels. Using this assay we previously showed that xSLBP1 (but not xSLBP2) activates translation of the Luc-SL mRNA in vivo; the in vitro and in vivo assays gave essentially identical results (25).
FIG. 5.
xSLBP1 does not activate translation of Luc-SLA8 mRNA in vivo. (A) A schematic of the assay is shown (25). Oocytes were injected with a synthetic mRNA encoding an SLBP protein and were injected 16 h later with a firefly luciferase reporter mRNA together with a Renilla luciferase internal control ending in the stem-loop. A total of 20 oocytes from each time point were pooled. The ratio of the firefly and Renilla luciferase activity levels was used to determine the relative translational activity levels of the various reporter mRNAs. The activity of the Luc-TL mRNA was set at 1. The same lysates were assayed for xSLBP proteins by Western blotting and for luciferase RNAs by S1 nuclease mapping. (B) Oocytes were injected with buffer, xSLBP1 mRNA, or xSLBP2 mRNA and were injected 16 h later with Luc-TL, Luc-SL, or Luc-SLA8 and the Renilla luciferase internal control. Oocytes were harvested 8, 16, or 24 h later, and the luciferase activity levels were determined. The luciferase activity was normalized to the Renilla luciferase level at each time point, and the average and standard deviations of the relative expression levels are shown. There was a steady increase in the absolute amount of luciferase activity during the incubation, and the absolute luciferase activity level was about 50% higher in the 24-h than in the 16-h samples. (C) The amounts of xSLBP2 (left panels) or xSLBP1 (right panels) in the 16- and 24-h extracts were determined by Western blotting. (Left panels) Lanes 1 and 2 show endogenous xSLBP2 levels in oocytes injected with buffer and oocytes matured with progesterone, respectively. A lighter exposure of the gel region is shown in the bottom panel. The star indicates a cross-reacting protein, which serves as an internal control (Fig. 2B). (Right panels) All of the samples in lanes 9 to 14 come from the same gel and were exposed for the same time. The exogenous xSLBP2 has a slightly faster mobility than the endogenous xSLBP2 due to variations among the results for different frogs (25). SL, stem-loop.
In the first experiments we determined the levels of luciferase activity at 8, 16, and 24 h after injection of the synthetic luciferase mRNAs containing different 3′ ends. In oocytes injected with buffer (Fig. 5B), there was a two- to threefold increase in luciferase activity from the Luc-SL mRNA (compared to the results seen with Luc-TL) due to the activity of endogenous xSLBP1 in the oocytes (25). However, the Luc-SLA8 was translated at levels very similar to those of the Luc-TL mRNA. When xSLBP1 mRNA was injected prior to injection of the luciferase mRNA, there was an 8- to 15-fold increase in luciferase activity from the Luc-SL mRNA after injection of the reporter mRNAs but only a 2- to 3-fold increase in luciferase activity from the Luc-SLA8 mRNA compared with the results seen with Luc-TL mRNA (Fig. 5B). The Luc-SL, Luc-SLA8, and Luc-TL synthetic mRNAs exhibited similar levels of activity after overexpression of xSLBP2 (Fig. 5B), since the excess xSLBP2 prevents activation of translation by the endogenous xSLBP1 (25). The relative expression levels of luciferase from the different mRNAs at 8, 16, and 24 h after injection of the reporter mRNAs were similar.
We measured (by Western blotting) the amount of xSLBP1 and xSLBP2 present in the oocytes injected with the synthetic SLBP mRNAs. The oocytes injected with the xSLBP2 or xSLBP1 mRNA expressed a large excess of the xSLBP both 16 and 24 h after injection of the reporter mRNAs (Fig. 5C) compared with the oocytes injected with buffer (Fig. 5C, lane 1, lanes 9 to 14, bottom panels).
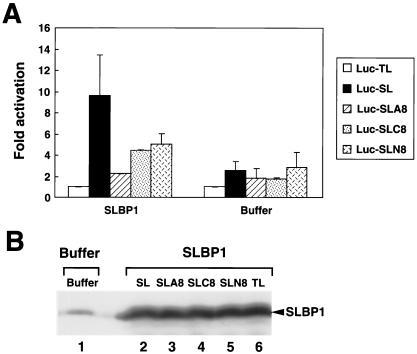
We repeated this experiment with an independent set of oocytes. As additional controls we also used reporter mRNAs ending in a stem-loop followed by 8 Cs (Luc-SLC8) or by a random nucleotide sequence (Luc-SLN8; Fig. 4A). The oocytes were harvested 24 h after injecting these reporter mRNAs. In these oocytes, expression of exogenous xSLBP1 results in an approximately sixfold increase in translation of the Luc-SL reporter mRNA compared to the results seen with the Luc-TL mRNA. In contrast, translation of the Luc-SLA8 mRNA was stimulated only twofold in the oocytes expressing xSLBP1 (Fig. 6A). However, the Luc-SLC8 and Luc-SLN8 RNAs were translated with only slightly reduced efficiency compared with the Luc-SL RNA. Expression of xSLBP2 in the oocytes prior to injection of the reporter mRNAs again reduced translation of the Luc-SL mRNA compared to the results seen with injection of buffer, and all five reporter mRNAs were translated to the same extent (Fig. 6A). In these oocytes injected with buffer there was a twofold activation of the Luc-SL (compared to the results seen with Luc-TL mRNA reporter) due to the presence of endogenous xSLBP1 in the oocyte. There was no activation of the Luc-SLA8 mRNA; this RNA was translated to the same extent as the Luc-TL mRNA (Fig. 6A), although Luc-SLC8 and Luc-SLN8 were both translated at rates similar to those seen with the Luc-SL RNA. The levels of the exogenous proteins were the same in all extracts regardless of which reporter mRNAs had been injected (Fig. 6B). We also tested the ability of all five of these capped RNA preparations to translate in the rabbit reticulocyte lysate, and they were all equally active (not shown, but see Fig. 8).
FIG. 6.
Failure to activate translation is specific for the oligo(A) tail. Oocytes were injected with either xSLBP1 mRNA or buffer. At 16 h later they were injected with one of the reporter mRNAs shown in Fig. 4A. After incubation for an additional 16 h, oocytes were harvested and luciferase activity levels were measured (A). The averages of the results of two independent experiments (with the standard deviations indicated) are shown. A portion of the extract was also analyzed for xSLBP1 levels by Western blotting in oocytes injected with buffer (panel B, lane 1) or with the indicated constructs (panel B, lanes 2 to 6). SL, stem-loop.
FIG. 8.
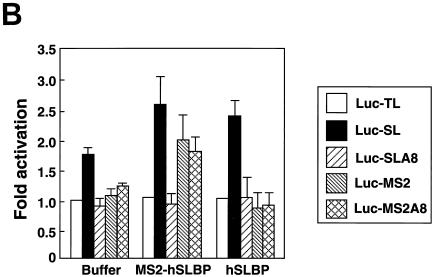
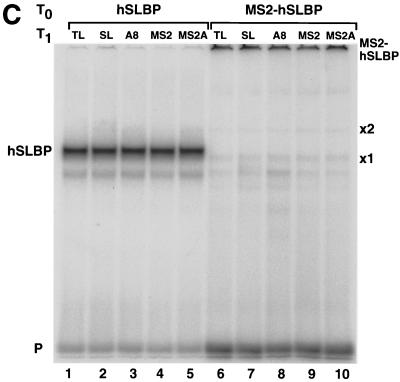
The MS2-hSLBP protein activates translation of the Luc-MS2A8 mRNA but not the Luc-SLA8 mRNA. (A) Capped luciferase reporter mRNAs (Luc-TL, lane 1; Luc-SL, lane 2; Luc-SLA8, lane 3; Luc-pA, lane 4; Luc-MS2, lane 5; Luc-MS2A8, lane 6) were mixed with equal amounts of the R-Luc-SL mRNA and translated in a reticulocyte lysate in the presence of [35S]methionine. The products were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography. The constructs are those shown in Fig. 4A (except for Luc-pA, which contains a 50-nt poly(A) tail) (25). SL, stem-loop; F/Luc, firefly luciferase; R/Luc, Renilla luciferase. (B) Xenopus oocytes were injected with buffer or with synthetic mRNAs encoding hSLBP, a MS2-hSLBP fusion protein, or the MS2 protein. At 16 h later the reporter mRNAs (Luc-TL, Luc-SL, Luc-SLA8, Luc-SLMS2, and Luc-MS2A8) analyzed as described for panel A were injected, and the oocytes were incubated for an additional 16 h. The oocytes were harvested, and luciferase activity levels were determined, with the activity of Luc-TL set at 1. The averages of the results of two independent experiments using different frogs are shown. (C) The extracts (described for panel B) from oocytes containing the reporter mRNAs indicated at the top of each lane and expressing either human SLBP (lanes 1 to 5) or the MS2-hSLBP fusion protein (lanes 6 to 10) were incubated with the radiolabeled stem-loop probe, and the complexes were resolved by native gel electrophoresis. The positions of the endogenous xSLBP1 and xSLBP2 complexes are indicated by x1 and x2. Because of the large size of the MS2-hSLBP fusion protein, the complex migrates near the top of the gel.
The reporter mRNAs are stable and the 3′ ends remain intact after injection
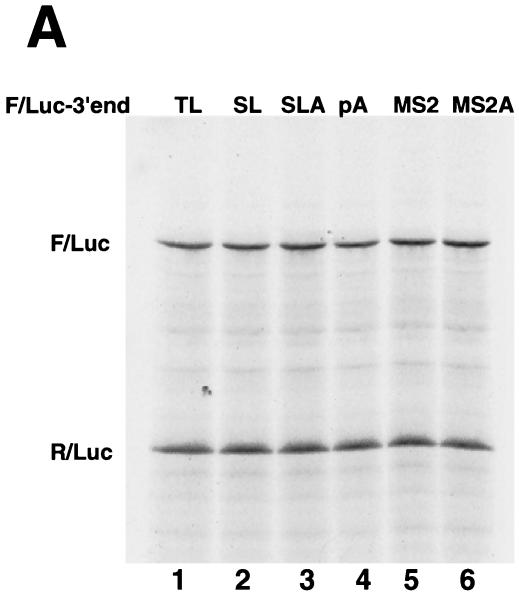
To rule out the possibility that these results were due to different levels of stability of the reporter mRNAs, we performed S1 protection assays using total RNA from oocytes harvested 16 h after luciferase mRNA injection. Two different probes were used in the S1 nuclease assays. We used a 3′ end-labeled DNA probe that was complementary to the Luc-SL construct to measure the total amount of each of the reporter mRNAs. This probe hybridizes to all the luciferase mRNAs, and the protected fragments are identical for the Luc-SL and Luc-SLA8 mRNAs and 15 nt shorter for the Luc-TL mRNA (which diverges from the Luc-SL mRNA at the start of the loop) (Fig. 7A). Since there is a single radioactive phosphate in the probe, the intensity of the protected fragment is directly proportional to the concentration of the mRNA. Similar amounts of all three mRNAs were present in the oocytes harvested 16 h after injection of the reporter mRNAs, regardless of which xSLBP was expressed in the oocytes (Fig. 7A). These data demonstrate that the stability of the injected luciferase mRNA is not affected by the nature of its 3′ end or by the amount of either xSLBP. Thus, the differences in translation efficiency levels are not due to differences in mRNA stability.
The probe used for the S1 protection assay in Fig. 7A would not detect any change that might occur on the oligo(A) tail of the Luc-SLA8 mRNA, since it extended only to the end of the stem-loop. To verify the integrity of the 3′ end of the reporter mRNAs 16 h after injection, we designed a short probe that contained the 3′ end of the Luc-SLA8 mRNA (Fig. 7B). To facilitate resolution of the small changes in length, this probe protected only 40 nt prior to the end of the stem-loop. We analyzed the RNA from oocytes that overexpressed xSLBP1 and that were injected with Luc-SL and Luc-SLA8 mRNAs. At 16 h after injection of the reporter mRNAs, the 3′ end of the Luc-SLA8 mRNA still contained the eight adenosines (Fig. 7B, lane 2) whereas the Luc-SL mRNA did not have any adenosines added to the 3′ end (Fig. 7B, lane 1). These protected fragments were absent from oocytes that did not have reporter mRNA injected (Fig. 7B, lane 3). Since the same probe was used in lanes 1 to 3, this experiment also confirms that the amounts of the Luc-SL and Luc-SLA8 mRNA were similar in the injected oocytes.
Translation repression in stage VI Xenopus oocytes requires the stem-loop and the oligo(A) tail
Previously we showed that an MS2-SLBP fusion protein can activate translation of a reporter mRNA that ends in an MS2 binding site as well as of an RNA that ends in the stem-loop (25). We compared the ability of the MS2-SLBP fusion protein to activate translation of reporter mRNAs that end either in the MS2 site or the MS2 site followed by 8As (Luc-MS2A8) with its ability to activate translation of the Luc-SL and Luc-SLA8 mRNAs. We synthesized five capped reporter mRNAs (encoding firefly luciferase) with different 3′ ends and mixed them with the internal control mRNA (encoding Renilla luciferase) ending in the stem-loop. All of these mRNAs were translated to the same extent in the reticulocyte lysate in the absence of SLBP (Fig. 8A).
These same RNA preparations were injected into Xenopus oocytes expressing either human SLBP, MS2 protein, or the MS2-SLBP fusion protein (Fig. 8B). Both the human SLBP and the MS2-SLBP fusion protein were overexpressed relative to the endogenous SLBPs, as measured by the mobility shift assay (Fig. 8C). The human SLBP activated translation of the Luc-SL mRNA but not that of the Luc-SLA8 mRNA or that of the mRNAs ending in the MS2 binding site. We tested whether placing an oligo(A) tail on the reporter mRNA ending in the MS2 site would affect activation of translation by the MS2-SLBP fusion protein. The MS2-SLBP fusion protein activated the levels of translation of the Luc-MS2 and the Luc-SL mRNAs to the same extent. However, the MS2-SLBP protein gave different results with the mRNAs that ended in 8 As. Like the hSLBP, the MS2-SLBP did not activate translation of the Luc-SLA8 mRNA. However, the MS2-SLBP did activate translation of the Luc-MS2A8 mRNA significantly more than it supported translation of the Luc-SLA8 mRNA. This result suggests that the inactivation of the Luc-SLA8 mRNA requires that the adenosines be attached to the histone stem-loop and suggests that there is a direct interaction between the stem-loop, an SLBP, and the eight adenosines and any associated factor(s) required for translational repression.
DISCUSSION
During oogenesis and early embryogenesis most of the regulation of gene activity occurs at the level of translation. A number of maternal mRNAs are stored in the Xenopus oocyte in an inactive form, and their translation is ultimately controlled in a precise temporal and spatial pattern (11) during oocyte maturation and early embryogenesis. These mRNAs include the mRNAs encoding essential proteins for cell cycle progression (3, 29). Histone proteins are critical components of the chromosome, and their assembly into newly replicating chromatin must occur appropriately during each S phase. There is not only storage of histone mRNA during oogenesis but also storage of histone proteins complexed with specific histone binding proteins, nucleoplasmin, and N1/N2 (5). Like the histone mRNAs, most of the histone protein stored in the oocyte is also synthesized prior to stage III, after which time the histone mRNA is translationally inactive (39). Translation of histone mRNAs is reactivated at oocyte maturation (1).
Translational regulation of histone mRNA
A major component of the regulation of translation of histone mRNA during Xenopus oogenesis is the presence of xSLBP1 and xSLBP2, two different proteins that bind the 3′ end of these mRNAs (33). xSLBP1 stimulates translation in vivo and in vitro of reporter mRNAs that end with the 3′ end of histone mRNA (25) and likely is required for efficient translation of histone mRNAs in vivo. xSLBP2, which is oocyte specific, does not support translation of histone mRNA and probably is involved in translational repression of histone mRNA.
Oocyte histone mRNAs differ from other histone mRNAs in that there is an oligo(A) tail added to their 3′ end. This tail is too short to bind a molecule of PABP (24). The oligo(A) tail is added to the histone mRNA early in oogenesis and is associated with the translationally inactive histone mRNA. In vitro both xSLBP1 and xSLBP2 bind to the oligoadenylated histone mRNAs with similar affinities, and these proteins do not readily exchange. However, in vivo it is likely that other factors bind to the oligoadenylated 3′ end and it is possible that these factors interact with xSLBP2. Other than xSLBP2, the protein components of the stored histone mRNA are not known. When stage VI oocytes were matured in vitro, we did not observe degradation of exogenously expressed xSLBP2 whether or not it was bound to histone mRNP or detect removal of the oligo(A) tail from injected in vitro-synthesized histone mRNAs (unpublished results). These results suggest that the xSLBP2/histone mRNA complex must be stored in a specific compartment or particle in the oocyte, which allows degradation of xSLBP2 and removal of the oligo(A) tail after oocyte activation. Exogenous xSLBP2 and/or histone mRNA may not be sequestered properly in the oocyte for degradation and deadenylation to occur at maturation.
A second possible function of the oligo(A) tail is to stabilize the histone mRNA for the long-term storage during oogenesis. We have recently characterized a mammalian 3′ exonuclease that specifically binds the 3′ end of histone mRNA (6), and this exonuclease would not bind to the oligoadenylated form of histone mRNA.
Strikingly, the oligoadenylated histone mRNA is not efficiently translated in Xenopus oocytes (even in the presence of a large excess of xSLBP1). This is not a result of a lower affinity of the xSLBP1 for the oligoadenylated histone mRNA, since xSLBP1 binds well to the oligoadenylated stem-loop and is equally active in stimulating translation of wild-type histone mRNA and oligoadenylated histone mRNAs in a reticulocyte lysate. The minimal xSLBP1 protein required for translation activation (a deletion of the first 68 amino acids and of the C-terminal domain; see reference 25) was also inactive in translation of the oligoadenylated histone mRNAs (data not shown). This result strongly suggests that there are oocyte-specific factors that interact with the oligoadenylated histone mRNA, inhibiting its translation by preventing xSLBP1 from interacting with its target in the initiation machinery. Activation of histone mRNA translation when oocytes are treated with progesterone requires both the removal of the xSLBP2 from the stored histone mRNA and the removal of the oligo(A) tail. Both of these events occur in enucleated oocytes that have been treated with progesterone. It is likely that a result of progesterone treatment is activation of the pathway leading to both xSLBP2 removal from the histone mRNA and destruction of xSLBP2 (as well as removal of the oligo(A) tail from the histone mRNA). It is reasonable to suspect that these events are mediated by the same signal.
Translation regulation of other mRNAs
Extensive progress has been made recently in understanding the mechanism of translational regulation of the c-mos mRNA required for initiation of oocyte maturation (26, 28) as well as the regulation of translation of cyclin B mRNA (3, 29). Translation of these polyadenylated mRNAs is regulated by alterations in the length of the poly(A) tail and by proteins that interact with the untranslated regions (4, 20). c-mos mRNA is one of the earliest mRNAs whose translation is altered in response to progesterone treatment (26). Activation of c-mos translation requires the Eg2 kinase which is activated by treatment with progesterone (19, 20). c-mos translation activation also requires the CPE located in the 3′ untranslated region of c-mos mRNA. CPEB, the protein that binds the CPE, is phosphorylated upon progesterone stimulation. Before oocyte maturation CPEB is linked to eIF-4E, the cap-binding protein, via the maskin protein. Maskin prevents the assembly of the translation preinitiation complex by impeding the interaction between eIF-4E and eIF-4G. Phosphorylation of CPEB in response to progesterone stimulation causes the release of CPEB-bound maskin from eIF-4E and promotes the recruitment of CPSF to c-mos mRNA (27). CPSF then recruits a poly(A) polymerase, possibly a novel poly(A) polymerase (32), that lengthens the poly(A) tail. These changes in translation of polyadenylated mRNAs important for cell-cycle regulation are occurring at the same time as the changes in the histone mRNA.
A second consequence of treatment of oocytes with progesterone is the shortening of the poly(A) tail on some of the oocyte mRNAs, resulting in translation arrest (8). The shortening of the poly(A) tail requires the presence of PARN, a 3′ exonuclease specific for poly(A) tails (14). It has been reported that shortening of the poly(A) tails on oocyte mRNAs requires the breakdown of the germinal vesicle, since it does not occur in enucleated oocytes treated with progesterone (30). The results shown in Fig. 2 suggest that the mechanism for the removal of the oligo(A) tail from histone mRNA may be distinct from the removal of the poly(A) tails from other mRNAs. Whether the same signal transduction pathway triggers these events after progesterone treatment is not known. It is noteworthy that these two deadenylation reactions have opposite effects: translation of maternal adenylated mRNAs is inhibited, while histone mRNA translation is activated. Whether the signaling pathway that results in activation of histone mRNA translation requires the Eg2 kinase (the critical regulator of translation activation of polyadenylated mRNAs) is not known.
Acknowledgments
This work was supported by NIH grant GM58921 to W.F.M.
We thank Zbig Dominski and Eric Wagner for critical review of the manuscript.
REFERENCE
- 1.Adamson, E. D., and H. R. Woodland. 1977. Changes in the rate of histone synthesis during oocyte maturation and very early development of Xenopus laevis. Dev. Biol. 57:136-149. [DOI] [PubMed] [Google Scholar]
- 2.Ballantine, J. E., and H. R. Woodland. 1985. Polyadenylation of histone mRNA in Xenopus oocytes and embryos. FEBS Lett. 180:224-228. [DOI] [PubMed] [Google Scholar]
- 3.Barkoff, A. F., K. S. Dickson, N. K. Gray, and N. Wickens. 2000. Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 220:97-109. [DOI] [PubMed] [Google Scholar]
- 4.De Moor, C. H., and J. D. Richter. 2001. Translational control in vertebrate development. Int. Rev. Cytol. 203:567-608. [DOI] [PubMed] [Google Scholar]
- 5.Dilworth, S. M., S. J. Black, and R. A. Laskey. 1987. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell 51:1009-1018. [DOI] [PubMed] [Google Scholar]
- 6.Dominski, Z., X. Yang, H. Kaygun, and W. F. Marzluff. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12:295-305. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin, M. B., and E. Dworkin-Rastl. 1985. Changes in RNA titers and polyadenylation during oogenesis and oocyte maturation in Xenopus laevis. Dev. Biol. 112:451-457. [DOI] [PubMed] [Google Scholar]
- 8.Fox, C. A., and M. Wickens. 1990. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 4:2287-2298. [DOI] [PubMed] [Google Scholar]
- 9.Gallie, D. R., N. J. Lewis, and W. F. Marzluff. 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 24:1954-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graves, R. A., S. E. Wellman, I.-M. Chiu, and W. F. Marzluff. 1985. Differential expression of two clusters of mouse histone genes. J. Mol. Biol. 183:179-194. [DOI] [PubMed] [Google Scholar]
- 11.Groisman, I., Y. S. Huang, R. Mendez, Q. P. Cao, W. Theurkauf, and J. D. Richter. 2000. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103:435-447. [DOI] [PubMed] [Google Scholar]
- 12.Hake, L. E., and J. D. Richter. 1997. Translational regulation of maternal mRNA. Biochim. Biophys. Acta 1332:M31-M38. [DOI] [PubMed] [Google Scholar]
- 13.Ingledue, T. C., Z. Dominski, R. Sanchez, J. A. Erkmann, and W. F. Marzluff. 2000. Dual role for the RNA binding domain of Xenopus laevis SLBP1 in histone pre-mRNA processing. RNA 6:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Körner, C. G., M. Wormington, M. Muckenthaler, S. Schneider, E. Dehlin, and E. Wahle. 1998. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 17:5427-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levenson, R. G., and K. B. Marcu. 1976. On the existence of polyadenylated histone mRNA in Xenopus oocytes. Cell 9:311-322. [DOI] [PubMed] [Google Scholar]
- 16.Marzluff, W. F. 1992. Histone 3′ ends: essential and regulatory functions. Gene Expr. 2:93-97. [PMC free article] [PubMed] [Google Scholar]
- 17.Marzluff, W. F., and R. J. Duronio. 2002. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14:692-699. [DOI] [PubMed] [Google Scholar]
- 18.Marzluff, W. F., P. Gongidi, K. R. Woods, J. P. Jin, and L. Maltais. 2002. The human and mouse replication-dependent histone genes. Genomics 80:487-498. [PubMed] [Google Scholar]
- 19.Mendez, R., L. E. Hake, T. Andresson, L. E. Littlepage, J. V. Ruderman, and J. D. Richter. 2000. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404:302-307. [DOI] [PubMed] [Google Scholar]
- 20.Mendez, R., K. G. Murthy, K. Ryan, J. L. Manley, and J. D. Richter. 2000. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell 6:1253-1259. [DOI] [PubMed] [Google Scholar]
- 21.Newport, J., and M. A. Kirschner. 1982. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 3:687-696. [DOI] [PubMed] [Google Scholar]
- 22.Ruderman, J. V., and M. L. Pardue. 1977. Cell-free translation analysis of messenger RNA in echinoderm and amphibian early development. Dev. Biol. 60:48-68. [DOI] [PubMed] [Google Scholar]
- 23.Ruderman, J. V., and M. L. Pardue. 1978. A portion of all major classes of histone messenger RNA in amphibian oocytes is polyadenylated. J. Biol. Chem. 253:2018-2025. [PubMed] [Google Scholar]
- 24.Sachs, A. B., R. W. Davis, and R. B. Kornberg. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets, M. D., M. Wu, and M. Wickens. 1995. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature 374:511-516. [DOI] [PubMed] [Google Scholar]
- 27.Stebbins-Boaz, B., Q. Cao, C. H. De Moor, R. Mendez, and J. D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 4:1017-1027. [DOI] [PubMed] [Google Scholar]
- 28.Stebbins-Boaz, B., L. E. Hake, and J. D. Richter. 1996. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 15:2582-2592. [PMC free article] [PubMed] [Google Scholar]
- 29.Tay, J., R. Hodgman, and J. D. Richter. 2000. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 221:1-9. [DOI] [PubMed] [Google Scholar]
- 30.Varnum, S. M., C. A. Hurney, and W. M. Wormington. 1992. Maturation-specific deadenylation in Xenopus oocytes requires nuclear and cytoplasmic factors. Dev. Biol. 153:283-290. [DOI] [PubMed] [Google Scholar]
- 31.Varnum, S. M., and W. M. Wormington. 1990. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: a default mechanism for translational control. Genes Dev. 4:2278-2286. [DOI] [PubMed] [Google Scholar]
- 32.Wang, L. T., C. R. Eckmann, L. C. Kadyk, M. Wickens, and J. Kimble. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419:312-316. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Z.-F., T. C. Ingledue, Z. Dominski, R. Sanchez, and W. F. Marzluff. 1999. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol. Cell. Biol. 19:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Z.-F., M. L. Whitfield, T. I. Ingledue, Z. Dominski, and W. F. Marzluff. 1996. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 35.Wellman, S. E., P. J. Casano, D. R. Pilch, W. F. Marzluff, and D. B. Sittman. 1987. Characterization of mouse H3.3-like histone genes. Gene 59:29-39. [DOI] [PubMed] [Google Scholar]
- 36.Wickens, M. P., J. Kimble, and S. Strickland. 1996. Translational control of developmental decisions, p. 411-450. In J. W. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Williams, A. S., and W. F. Marzluff. 1995. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 23:654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodland, H. R. 1980. Histone synthesis during the development of Xenopus. FEBS Lett. 121:1-7. [DOI] [PubMed] [Google Scholar]
- 39.Woodland, H. R., and E. D. Adamson. 1977. The synthesis and storage of histones during oogenesis of Xenopus laevis. Dev. Biol. 57:118-135. [DOI] [PubMed] [Google Scholar]
- 40.Zernik, M., N. Heintz, I. Boime, and R. G. Roeder. 1980. Xenopus laevis histone genes: variant H1 genes are present in different clusters. Cell 22:807-815. [DOI] [PubMed] [Google Scholar]