Abstract
Tid1 is the mammalian counterpart of the Drosophila tumor suppressor Tid56 and is also a DnaJ protein containing a conserved J domain through which it interacts with the heat shock protein 70 (Hsp70) family of chaperone proteins. We generated a Tid1 conditional mutation in mice, and the subsequent global removal of the Tid1 protein was achieved by crossing these conditional knockout mice with general deletor mice. No Tid1−/− embryos were detected as early as embryonic day 7.5 (E7.5). Nonetheless, Tid1-deficient blastocysts were viable, hatched, formed an inner cell mass and trophectoderm, and implanted (E4.5), suggesting that the homozygous mutant embryos die between E4.5 and E7.5. To assess the function of Tid1 in embryonic cells, mouse embryonic fibroblasts with the homologous Tid1 floxed allele were produced. Tid1 removal in these cells led to massive cell death. The death of Tid1-deficient cells could be rescued by ectopic expression of wild-type Tid1 but not by expression of the Tid1 protein that had a mutated J domain and was thus incapable of binding to Hsp70. We propose that Tid1 is critical for early mammalian development, most likely for its function in sustaining embryonic-cell survival, which requires its association with Hsp70.
The Drosophila l(2)tid gene, Tid56, encodes the first and only member of the DnaJ cochaperone family that has been classified as a tumor suppressor (16, 17). The null mutation of the Tid56 gene not only keeps imaginal disks from differentiation but also leads to lethal tumorigenesis during the larval stage, suggesting that during embryogenesis Tid56 is involved in regulating cell differentiation, cell growth, and cell death (16, 17). The function of its mammalian counterpart, Tid1, in animals and in cells, however, has not been fully characterized. DnaJ proteins serve as cochaperones and regulatory factors for the heat shock protein 70 (Hsp70) family of molecular chaperones (24) and are characterized by a J domain, a highly conserved tetrahelical domain that binds to Hsp70 proteins to regulate their activities and provide substrate specificity (24). In fact, it has been shown that Tid1 interacts with Hsp70 proteins residing in cytosol (Hsp70/Hsc70) or in mitochondria (mtHsp70) through its DnaJ domain, indicating that Tid1 might exert its cellular function by modulating the activities and substrate-binding specificities of these Hsp70 proteins (25, 27).
Tid1 was originally isolated by yeast two-hybrid screening as a protein that interacts with the human papillomavirus E7 oncoprotein (22). Several reports have indicated that Tid1 functions as a regulator in intracellular signaling pathways related to cell survival, cell proliferation, and stress-induced cellular responses. For example, in agonist-induced programmed cell death, Tid1 has a role in both apoptotic and antiapoptotic processes (25). Tid1 has also been suggested to regulate intracellular signaling through its affinity with the RasGAP protein to modulate the Ras signaling pathway (27) and through its binding to Jak2 to regulate gamma interferon-dependent transcriptional activity (21). Additionally, it has been observed that Tid1 associates with human T-cell leukemia virus type 1 Tax and subsequently, by inhibiting IKKβ kinase activity, represses NF-κB activation induced by Tax, tumor necrosis factor alpha, and Bcl10 (6, 7).
To date, most information regarding the function of Tid1 has been generated from cell-based assays. To examine the function of Tid1 in vivo, we generated a conditional knockout mouse model of Tid1 and subsequently elucidated the role of Tid1 during early embryogenesis.
MATERIALS AND METHOD
Genomic DNA cloning of mouse Tid1
Mouse genomic DNA containing the Tid1 gene was isolated by PCR-based screening of a bacterial artificial chromosome library derived from 129/SvJ mouse embryonic stem (ES) cells (Incyte, St. Louis, Mo.). The primer sets used for screening were designed based on the mouse Tid1 sequence (GenBank accession no. AC079418): Tid99355U21 (5′-TGTCCCCGGGCCAAAATGGCT-3′) and Tid99562L21 (5′-TGGTGTTCCCCACGCTGGTTG-3′), and Tid94263U21 (5′-CCGCAGGAAGCAAGGATAGGC-3′) and Tid95069L21 (5′-CTCGGCGGTTCAGAGGGATCT-3′).
Disruption of the Tid1 gene in ES cells
A deletion allele of Tid1 was generated by introducing a double-stranded DNA linker, SalI-FRT-loxP-XhoI, into the XhoI site of the pPNT vector (28). The following PCR products from three regions were subcloned separately and later inserted into the modified pPNT vector: a 2.7-kb product of 5′-untranslated sequence, exon 1, and partial intron 1; a 0.8-kb product with partial intron 1, exon 2, and part of intron 2; and a 2.8-kb product containing partial intron 2, exon 3, and partial intron 3 (Fig. 1A). One extra loxP site was introduced by synthetic oligonucleotides in the middle of intron 1, and the second frt site was inserted into intron 2 as indicated in Fig. 1A. In the final targeting construct, the neomycin resistance (Neo) cassette of pPNT was flanked by two frt sites, and the exon 2-containing fragment of the Tid1 gene (0.8 kb) was flanked by two loxP sites. The orientation of the transcribed Neo cassette was in the opposite direction to that of Tid1. The insertion, orientation, and sequence fidelity of the inserted DNA fragments within the targeting vector were confirmed by DNA sequencing. Further details are available upon request.
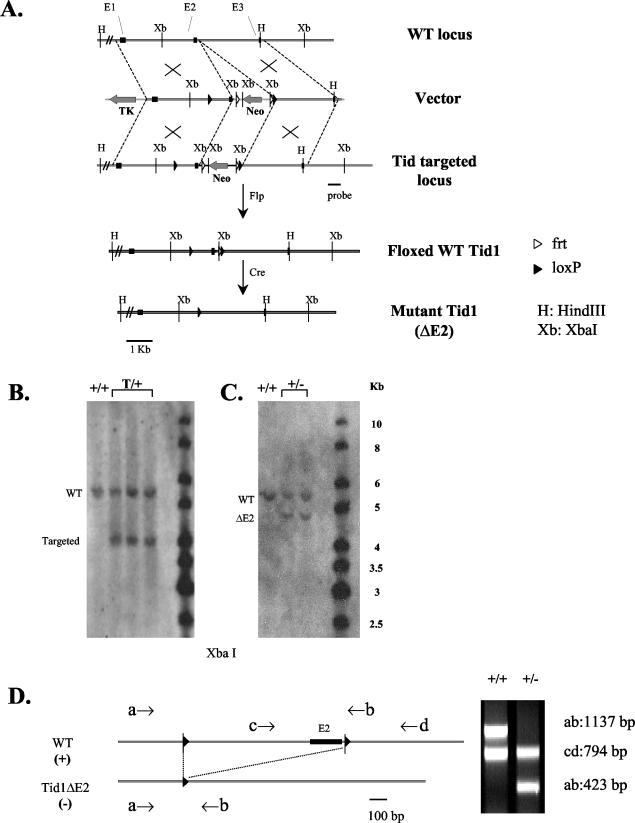
FIG. 1.
Targeted disruption of the mouse Tid1 gene. (A) Partial restriction maps of the Tid1 locus, targeting vector, targeted locus, and targeted locus after Cre-mediated Tid1 deletion. The first three exons are indicated by solid boxes. WT, wild type; TK, thymidine kinase. (B) Genotyping ES cell clones by Southern blot hybridization of genomic DNA (restriction digestion with XbaI enzyme) with the probe shown in panel A. The genotype of each clone is indicated. T, targeted. (C) Genotyping mice derived from the cross between Tid1-targeted mice and general deletor mice by Southern blot analysis as described for panel B. (D) Genotyping Tid1+/+ and Tid1+/− mice by PCR. The PCR primers used for genotyping are indicated by arrows and are depicted along with maps of the corresponding alleles. On the right are the patterns of electrophoresed PCR products for Tid1+/+ and Tid1+/− mice.
The NotI-linearized targeting vector was introduced by electroporation into ES cells derived from the 129/SvJ mouse strain (Incyte, St. Louis, Mo.). Colonies resistant to 200 μg of G418 (GIBCO-BRL, Carlsbad, Calif.)/ml and 1 μM ganciclovir (Calbiochem, San Diego, Calif.) were picked and screened for homologous recombination first by PCR with the upstream Neo-specific primer Tidpcrp9997U30 (5′-CCAGTCCCTTCCCGCTTCAGTGACAACGTC-3′) and the downstream Tid1-specific primer Tidpcrp14009L30 (5′-TCCCGAGATCAGCTATCAATATGAATTACC-3′). Individual ES clones that passed the first screen were cultured and analyzed by Southern blotting and DNA sequencing in a second screen for clones containing two intact loxP sites flanking exon 2 of the Tid1 gene (Tid1 floxed) (Fig. 1B). Three clones of ES cells contained the targeting vector with the desired genomic integration site that had been introduced via homologous recombination.
Generation of Tid1 mutant and floxed mice
Three clones of targeted ES cells were injected separately into C57BL/6 blastocysts and later implanted into C57BL/6 pseudopregnant females (Jackson Laboratories, Bar Harbor, Maine). One highly chimeric male was produced and backcrossed to C57BL/6 females. The offspring were genotyped by Southern blotting and PCR analyses (details below) to detect germ line transmission from the chimeric male (Fig. 1B). To delete the Neo cassette in the Tid1+/targeted mice for generating mice with a functional Tid1 floxed allele, Tid1+/targeted mice were crossed with transgenic B6/J-TgN(ACTFLPe) mice (Jackson Laboratories) carrying an ACTB-FLPe gene (Fig. 1A and data not shown). Deletion of the Tid1 gene in Tid1 floxed mice was achieved by crossing Tid1+/targeted mice with transgenic FVB/N-TgN(ACTB-Cre) mice (Jackson Laboratories) carrying the ACTB-Cre gene (Fig. 1A, C, and D). The resulting Tid1+/− mice were maintained in a 129/SvJ, C57BL/6, and FVB/N mixed background and were later intercrossed to generate Tid1−/− embryos for subsequent studies. All animal protocols were in full compliance with the guidelines for animal care and were approved by the Animal Care Committee of the Scripps Research Institute.
Southern and PCR analyses for genotyping
For genotyping ES cell lines to identify targeted clones, 10 μg of genomic DNA was digested with XbaI, separated by electrophoresis through 0.7% agarose in 1× TAE buffer, blotted onto a nylon membrane (Nytran SuPerCharge; Schleicher & Schuell, Keene, N.H.), and probed with a 0.5-kb PCR fragment (Fig. 1A) that hybridizes downstream of the targeted region of Tid1. The 5.6-kb band corresponds to the wild-type allele, and the 4.0-kb band corresponds to the targeted allele (Fig. 1B). For genotyping weaned pups, genomic DNA was purified from tail biopsy specimens and analyzed by either Southern blotting or PCR analysis. For Southern analysis, 10 or 20 μg of DNA was digested with XbaI, separated by electrophoresis, blotted, and probed with the PCR product described above (Fig. 1A). The 5.6-kb fragment corresponds to the wild-type allele, and the 4.8-kb fragment corresponds to the mutant allele (ΔExon2) (Fig. 1C). For PCR analysis, genomic DNA was used as the template in a reaction where wild-type and mutant alleles were detected simultaneously. Four primers were designed to differentiate between the presence of wild-type and mutant Tid1 alleles. Primer a (5′-TCTAATGTCTTCTCCGGGAGT-3′), primer b (5′-AGTGACCAGACCAGCAATCTT-3′), and primer d (5′-TTTCCCAAAAGACCTGAACTC-3′) hybridized to both wild-type and Tid1/ΔExon2 alleles. Primer c (5′-GAATCAAATTCAGGGCCTTGT-3′) hybridized to the wild-type allele only and did not hybridize to the Tid1/ΔExon2 allele (Fig. 1D). Primers a and b amplified a 1,137-bp fragment from the wild-type allele and a 423-bp fragment from the Tid1/ΔExon2 allele (Fig. 1D). Primers c and d amplified a 794-bp fragment from the wild-type allele but did not amplify any product from the Tid1/ΔExon2 allele (Fig. 1D).
Embryos at different embryonic stages were genotyped by PCR analysis of DNA purified from either the yolk sac or the whole embryo. Tissues were digested overnight at 55°C in 300 μl of lysis buffer (50 mM Tris-HCI, pH 8.0-100 mM EDTA-0.5% sodium dodecyl sulfate [SDS]-500 μg of proteinase K/ml). DNA was purified by phenol-chloroform extraction, precipitated with isopropanol, and dissolved in 100 μl of 50 mM Tris-HCl (pH 8.0). This purified DNA (2 μl) was used for PCR-based genotyping as described above.
Blastocysts and blastocyst outgrowths were genotyped by nested PCR. The outgrowths were washed once with phosphate-buffered saline (PBS). Then, either blastocysts or outgrowths were lysed in 20 μl of PCR lysis buffer (10 mM Tris-HCl, pH 8.3-50 mM KCl-2.5 mM MgCl2-0.1 μg of gelatin/ml-0.45% NP-40-0.45% Tween 20-500 μg of proteinase K/ml) at 55°C for 1 h and then at 95°C for 10 min. Purified DNA (2 μl) was later used in the first PCR amplification with primers e (5′-CCAAGTATCGAGGTCTAATGT-3′) and f (5′-TCAGCTACAGTGTACCCCTAT-3′), which are located upstream and downstream from primers a and d, respectively, and a PCR product of ∼2 kb was generated (data not shown). The PCR conditions were the same as described above, except that the annealing temperature was 60°C instead of 55°C. This PCR product (2 μl) served as the template in a subsequent PCR amplification with primer sets ab and cd.
Timed pregnancies
For monitoring timed pregnancies, 6- to 10-week-old Tid1+/− females were mated with Tid1+/− males. The females were examined each morning for the presence of a vaginal plug (14). The females were sacrificed at E7.5, E8.5, E9.5, E10.5, E12.5, or E13.5 to isolate embryos. The isolated embryos were genotyped by PCR as described above.
Isolation of E3.5 embryos and in vitro culturing
Three- to 9-week-old Tid1+/− females were superovulated by injection intraperitoneally with pregnant mare's serum gonadotropin (5 IU per animal; Sigma-Aldrich Co. or Professional Compounding Centers of America, Houston, Tex.), followed by injection with 5 IU of human chorionic gonadotropin (Sigma-Aldrich Co. or Serono Laboratories, Randolph, Mass.), and 48 h later were mated with Tid1+/− males (18). At E3.5, the treated females were sacrificed, and embryos were isolated from their oviducts and cultured for 3 days in ES medium. For blastocyst outgrowth, individual blastocysts were placed on gelatin-coated 96-well plates with 0.2 ml of ES cell medium supplemented with leukemia inhibitory factor and were cultured again for 3 days (18). Outgrowths were photographed, washed with PBS, and lysed for genotyping by PCR as described above.
Antibody generation and immunoblot analysis
Antiserum against mouse Tid1 was generated in a New Zealand White rabbit (Animal Center of The Scripps Research Institute, La Jolla, Calif.) by using the synthetic oligopeptide (IQTDQKIRLTGKGIPRINC) derived from the mouse Tid1 amino acid sequence. The anti-MEK7 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). For immunoblots, cells were lysed for 10 min on ice in 50 mM Tris-HCl (pH 7.5), 10 mM EDTA, 1% SDS, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 15 μg of aprotinin/ml, and 2 μg of pepstatin/ml. Protein lysates were cleared by centrifugation for 20 min at 16,000 × g and 4°C, and protein concentrations were measured by the Bradford assay (Bio-Rad, Hercules, Calif.). Cell lysates (10 to 30 μg) were fractionated by SDS-polyacrylamide gel electrophoresis, and the proteins were transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked for 10 min in blocking buffer (10 mM Tris-HCl, pH 8.0-50 mM NaCl-0.2% Tween 20-5% nonfat dry milk), incubated with primary antibody for 2 h in blocking buffer (with 0.5% nonfat milk), and incubated in blocking buffer with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG; Pierce Biotechnology, Rockford, Ill.).
Generation of MEFs (14)
Mouse embryonic fibroblasts (MEFs) with the genotypes Tid1+/+, Tid1+/−, Tid1f/−, Tid1f/+, and Tid1f/f were generated by crossing Tid1f/+ mice with Tid1f/+ or Tid1+/− mice, and the resulting E13.5 embryos were collected. The yolk sac of each embryo was isolated and used for genotyping. Individual embryos were minced and then treated with trypsin for 10 min. After being washed with Dulbecco's modified Eagle's medium (DMEM) containing 15% fetal bovine serum, the MEFs were cultured in DMEM supplemented with 15% fetal bovine serum on 12-well plates coated with 0.1% gelatin.
Construction, preparation, and infection of recombinant adenovirus
The AdEasy vector system from Qbiogene, Inc. (Carlsbad, Calif.), was used according to the manufacturer's protocol to generate recombinant adenoviruses encoding human Tid1-L, Tid1-S, Tid1-S (H121Q), MEK7, or BAG1. Recombinant adenovirus encoding Cre recombinase under the control of the cytomegalovirus immediate-early promoter (Ad-Cre) was a gift from Shuang Huang, The Scripps Research Institute (La Jolla, Calif.). 293 cells were cultured in DMEM containing 10% fetal calf serum and used for producing recombinant adenoviruses. High-titer stocks of individual adenovirus particles were prepared by equilibrium centrifugation in CsCl. High-titer stocks contained 1 × 1012 to 10 × 1012 virus particles/ml, determined by using bovine serum albumin as a standard, with 1 μg of viral protein considered to be 4 × 109 virus particles. For adenovirus infection of MEFs, a multiplicity of infection of 5 × 104 was used to reach a 100% infection rate.
Immunofluorescence
MEFs were seeded into 12-well plates containing collagen-coated glass coverslips and allowed to adhere overnight. The slides were washed with PBS and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. After three washes with PBS, the cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature, rinsed, and blocked in 10% normal goat serum (Vector Laboratories, Burlingame, Calif.) for 1 h at room temperature. Next, the slides were incubated with 10 μg of affinity-purified murine Tid1 antibody in 2% goat serum/ml for 1 h at room temperature. Subsequently, the cells were incubated for 30 min with biotinylated goat anti-rabbit IgG (Vector Laboratories), rinsed, and incubated with streptavidin (1:1,000 dilution) conjugated with Alexa Fluor 568 (Molecular Probes Inc.) for 30 min at room temperature. Free streptavidin and biotin were blocked by using the avidin/biotin blocking kit (Vector Laboratories). Slides were incubated with 5 mg of the anti-mtHsp70 monoclonal antibody (Affinity Bioreagents, Golden, Colo.)/ml for 1 h, rinsed, and incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes Inc.) for 30 min. The nuclei of cells were stained with 2 μM ToPro 3 staining dye (Molecular Probes Inc.) for 20 min. The cells were viewed and photographed with the Bio-Rad MRC1024 laser scanning confocal microscope attached to a Zeiss Axiovert S100TV microscope. Images were collected on the confocal microscope using Bio-Rad's LaserSharp (version 3.2) software. For mitochondrial staining, live cells adhering to coverslips were fed with 0.5 μM Mitotracker Green FM (Molecular Probes Inc.) in fresh medium for 30 min at 37°C. Subsequently, the cells were fixed and permeabilized as described above. The cells were stained with anti-apoptosis-inducing factor (AIF), anti-cytochrome c (Santa Cruz Biotechnology), anti-Tid1, or anti-mtHsp70, respectively.
cDNA microarray
cDNA microarrays containing ∼13,000 mouse cDNA clones in three physically different slides were used. The cDNA clones were obtained commercially from Research Genetics (IMAGE consortium) and Incyte Genomics, as well as internal sources. Each clone was printed as two independent spots on a given chip, on the left and right sides. Printing, hybridization, scanning, and image analysis have been previously described (2).
Statistical analysis of microarray data
From Tid1f/+ and Tid1f/− MEFs on days 4, 5, and 6 after Ad-Cre treatment, total RNAs were collected and hybridized to cDNA microarrays. Intensity data were multiplicatively normalized to the 75th percentile of each microarray, which was arbitrarily set to the value of 100. The normalized data were analyzed by the t test comparing the Tid1f/+ and Tid1f/− MEFs on days 4, 5, and 6. The normalized data were also analyzed by using the analysis of variance model, considering that day {4, 5, or 6} and type {Tid1f/+ and Tid1f/− MEFs} are two factors with interaction. By using this model, P values were generated for each gene on the microarray for the effects of day, type, and day-and-type interaction. Ratios were calculated by using median values for each group. The P value of 0.05 was used as the cutoff for selecting genes with significant changes.
Reverse transcription-PCR (RT-PCR)
Total RNAs were isolated from Tid1f/+ and Tid1f/− MEFs on day 6 after the Ad-Cre treatment. First-strand cDNAs were synthesized from these RNAs (1 μg) with the SuperScript Preamplification system kit (Invitrogen, Carlsbad, Calif.). First-strand cDNA (0.1 μg) was then used for PCR with specific oligonucleotide primers designed for detecting messages of BAG1, MEK7, or GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The individual primer sets for GAPDH, BAG1, and MEK7 were as follows: GAPDH sense (5′-TGCACCACCAACTGCTTAGC-3′) and antisense (5′-GGCATGGACTGTGGTCATGAG-3′); BAG1 sense (5′-CGTGATCGTCACCCACAGCAA-3′) and antisense (5′-CAACAAGCTGAGCCAAGTCTT-3′); and MEK7 sense (5′-CGCATCGACCCTCCAGATCC-3′) and antisense (5′-CTGTCCTGTTGCCAGCTCCAC-3′). The numbers of PCR cycles for amplifying specific gene products were 26 (BAG1), 28 (MEK7), and 22 (GAPDH). The PCR products were separated by electrophoresis through 4% agarose and visualized by ethidium bromide staining.
RESULT
Generation of Tid1 floxed and mutant mice
To determine the pathophysiological function of Tid1 both during early development and in adults, we used a conditional knockout strategy to generate mouse ES cell lines with a mutant Tid1 allele. Briefly, a targeting vector was constructed by inserting two loxP sequences flanking exon 2 of the Tid1 gene (Fig. 1A and B). A Cre-mediated excision of this Tid1 floxed allele deleted exon 2 of the Tid1 gene and also created a frameshift, disrupting the translation of the key functional domains of Tid1, including the DnaJ domain, a glycine- and phenylalanine (G/F)-rich region, a cysteine-rich part resembling a zinc finger and consisting of four CXXCXGXG repeats, and a nonhomologous C-terminal domain (24). Therefore, deleting exon 2 of Tid1 would most likely lead to a null phenotype. We produced the Tid1 floxed mice, which developed to adulthood normally and appeared to be healthy for at least 12 months after birth.
Loss of Tid1 resulted in embryonic lethality between E4.5 and E7.5, but Tid1−/− blastocysts are viable
Deletion of the Tid1 gene in all tissues was achieved by crossing Tid1+/targeted mice with a general deletor mouse strain [FVB/N-TgN(ACTB-Cre)] carrying the ACTB-Cre gene (Fig. 1C and D), and the offspring of this cross that had the Tid1+/− genotype were later intercrossed. The resultant 121 weaned pups were genotyped, and no Tid1−/− mice were found, indicating that animals lacking functional Tid1 are not viable. Heterozygous mutant and wild-type mice were born at an ∼2:1 ratio (Table 1) and grew normally with no discernible phenotypic differences for >9 months.
TABLE 1.
Genotypes of progeny from heterozygous matingsa
| Stage | No. with genotype
|
No. of empty implantations | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| At weaning | 41 | 80 | 0 | |
| E13.5 | 10 | 17 | 0 | 7 |
| E12.5 | 7 | 9 | 0 | 5 |
| E10.5 | 18 | 34 | 0 | 19 |
| E9.5 | 3 | 6 | 0 | 2 |
| E8.5 | 5 | 18 | 0 | 10 |
| E7.5 | 5 | 12 | 0 | 9 |
| Blastocysts | 14 | 26 | 9 | |
Genotypes were determined by PCR.
To determine the stage of embryonic development at which the homozygous mutant embryos die, we isolated and genotyped embryos at E7.5, E8.5, E9.5, E10.5, E12.5, and E13.5 from heterozygote (Tid1+/−) intercrosses (Table 1). For each time point analyzed, no homozygous mutant embryos were recovered, suggesting that Tid1−/− embryos die before E7.5 (P < 0.01) (Table 1). The ratio between heterozygous and wild-type embryos is 2:1. Approximately 25% of the total implantation sites were empty, suggesting that implantation had occurred at these sites but failed to proceed beyond E7.5 (Table 1). Conceivably, the Tid1−/− embryos were responsible for these empty sites; if so, Tid1 null embryos could implant (E4.5) but were unable to survive in the uterus through E7.5.
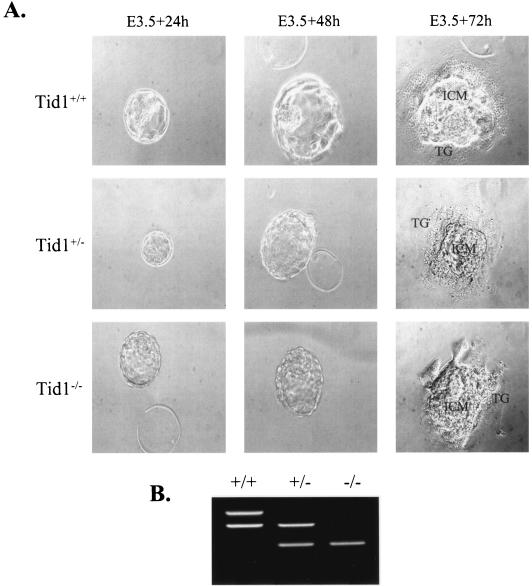
The failure of Tid1−/− embryos to develop past E7.5 suggested that at the early development phase Tid1 might be required for blastocyst formation. The E3.5 embryos from heterozygote intercrosses were isolated and allowed to develop for up to 3 days in vitro on gelatinized petri dishes (Fig. 2A). After that, viable blastocysts were genotyped (Fig. 2B). Eighteen percent (9 of 49) of these were homozygous mutants, 53% (26 of 49) were heterozygotes, and 29% (14 of 49) were wild type, which agrees with the Mendelian ratio of 1:2:1 (Table 1). Mutant blastocysts formed an inner cell mass and trophectoderm and were morphologically indistinguishable from wild-type and heterozygote outgrowths (Fig. 2A). These results suggest that Tid1 is not required for blastocyst establishment but is required for later embryonic development between E4.5 and E7.5.
FIG. 2.
Tid1−/− blastocysts were viable, hatched, and formed an inner cell mass and trophectoderm. (A) Blastocysts derived from Tid1+/− intercrosses were cultured in vitro for 3 days. In each outgrowth, the inner cell mass (ICM) and trophectoderm showed no differences. TG, trophoblast giant cell. (B) The genotype of each outgrowth was determined by PCR as described in the legend to Fig. 1D. The patterns of electrophoresed nested-PCR products are shown for ES cells with Tid1+/+, Tid1+/−, and Tid1−/− genotypes.
Tid1 is critical for embryonic-fibroblast survival
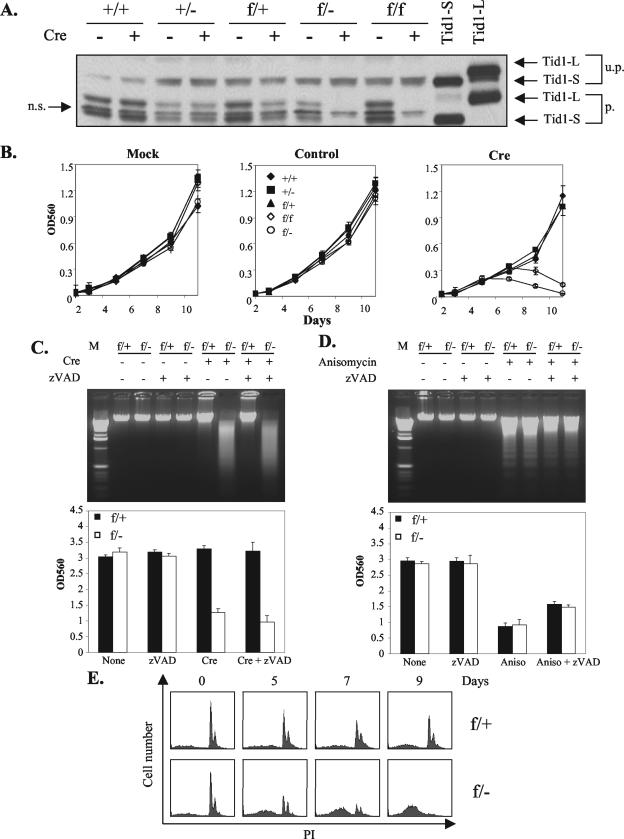
To explore the reason for the early death of Tid1-deficient embryos, we established MEFs with the genotypes Tid1+/+, Tid1+/−, Tid1f/f, Tid1f/+, and Tid1f/−. Treating these MEFs with recombinant adenovirus encoding Cre recombinase effectively and irreversibly disrupted the Tid1 floxed allele and abrogated consequent Tid1 protein production from the Tid1 floxed allele. As shown in Fig. 3A, after Ad-Cre treatment, both splicing isoforms of Tid1 protein, Tid1-S and Tid1-L (22, 25), were completely removed in Tid1f/− and Tid1f/f MEFs, and the amount of Tid1 protein produced from Tid1f/+ MEFs was about half the amount produced from Tid1+/+ MEFs (Fig. 3A). The earliest stage that Tid1 proteins became undetectable was on day 4 after the Ad-Cre treatment in Tid1f/− and Tid1f/f MEFs (data not shown). The growth of treated MEFs was normal in cells with either one or both Tid1 genes intact. On the other hand, the proliferation of Tid1-deficient MEFs was impaired, and the number of viable cells declined dramatically 5 to 6 days after Ad-Cre addition (Fig. 3B). Eventually, none of the Tid1-deficient MEFs survived the Ad-Cre treatment. For MEFs infected with the control adenovirus, no growth difference was observed for each MEF cell type (Fig. 3B).
FIG.3.
Tid1 gene product is required for cell survival. (A) MEFs with Tid1+/+, Tid1+/−, Tid1f/+, Tid1f/−, and Tid1f/f genotypes were infected with Ad-Cre (Cre) (+) or uninfected (−) as indicated. After 6 days, total cellular proteins from these cells were collected and separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting with an antibody against mouse Tid1. The recombinant Tid1L and Tid1S proteins generated from 293T cells transfected with expression plasmids encoding Tid1-L or Tid1-S (two splicing isoforms of Tid1) were used as controls. u.p., unprocessed; p., processed; n.s., nonspecific. (B) After infection with nothing (Mock), control adenovirus (control), or Ad-Cre (Cre) as indicated, the above-mentioned MEF cell lines were analyzed for growth and viability by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assays at the indicated time intervals. OD560, optical density at 560 nm. The error bars indicate standard deviations. (C) Tid1f/+ and Tid1f/− MEFs were treated as for panel A. After 9 days, genomic DNAs were purified from these cells and separated by agarose gel electrophoresis (top). Some MEFs were also treated with 50 μM zVAD (a caspase 3 inhibitor) as indicated. The relative amounts of viable cells for these treated MEFs were also assessed by MTT assays (bottom). (D) Tid1f/+ and Tid1f/− MEFs were treated with 5 μg of anisomycin/ml (50 μM zVAD) as indicated for 18 h, and their genomic DNAs (top) and viabilities (bottom) were analyzed as for panel C. The MTT assays were performed in triplicate and repeated at least three times (the values are means plus standard deviations). (E) Tid1f/+ and Tid1f/− MEFs were infected with Ad-Cre and collected at the indicated time intervals. The cells were fixed and stained with propidium iodide (PI) and analyzed by flow cytometry.
To characterize further the acute cell death triggered by Tid1 deletion, we subjected Tid1f/− and Tid1f/+ MEFs, after disruption of the Tid1 floxed allele, to DNA fragmentation and fluorescence-activated cell sorter analysis (Fig. 3C and E). The genomic DNA from Ad-Cre-treated Tid1f/− cells showed DNA degradation in the form of DNA smearing rather than DNA laddering (Fig. 3C). Fluorescence-activated cell sorter analysis also showed that with Ad-Cre infection, the Tid1f/− MEFs increased their subdiploid DNA contents (Fig. 3E). Both the DNA smearing and the increase in subdiploid DNA suggest that the Tid1-deficient MEFs were undergoing a process of cell death that is not typical apoptosis (Fig. 3C and E). On the other hand, the growth and viability of Tid1f/+ MEFs after Ad-Cre infection, with one Tid1 allele remaining intact, were not affected (Fig. 3B, C, and E). Moreover, in the presence of the caspase 3 inhibitor zVAD, the death of Tid1-deficient cells was not reduced (Fig. 3C), and the cell death triggered by the apoptosis inducer aniosmycin was partially blocked in both Tid1f/+ and Tid1f/− cells (Fig. 3D). These results suggest that cell death caused by Tid1 removal is not mediated by a typical caspase 3-dependent apoptotic process.
A functional DnaJ domain of Tid1 is essential for viability of MEF cells
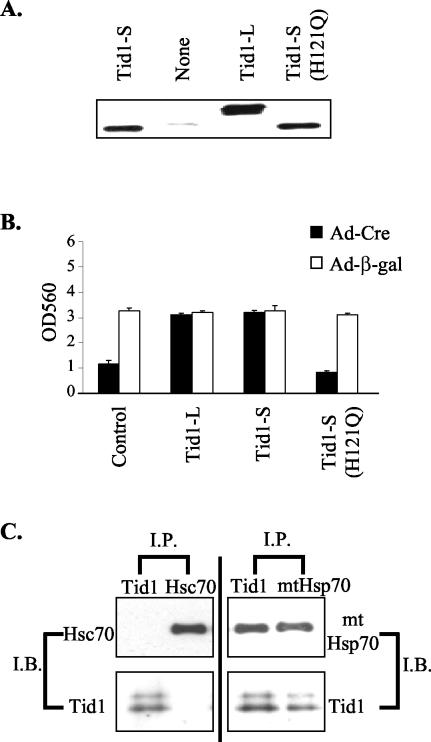
As a DnaJ protein family member, Tid1 interacts with Hsp70 proteins via its highly conserved DnaJ domain and subsequently activates the ATPase activity of Hsp70. To test whether the J domain in Tid1 was critical for the role of Tid1 in cell survival, we generated a Tid1 protein with an impaired J domain, Tid1-S(H121Q), by mutating one amino acid residue within the J domain that is critical for the interaction of Tid1 with Hsp 70 proteins (Fig. 4A) (25). Ad-Cre-treated Tid1f/− cells subsequently infected with adenovirus encoding either wild-type Tid1-S or Tid1-L could survive the loss of the functional Tid1 allele (Fig. 4B). In contrast, infection with Ad-Tid1-S(H121Q) adenovirus failed to compensate for the disruption of the normal Tid1 gene within Tid1f/− MEFs (Fig. 4B). These data suggest that Tid1 association with Hsp70 is critical for maintaining the survival of MEFs.
FIG. 4.
DnaJ domain-dependent interaction between Tid1 and Hsp70 is critical for cell survival. (A) Total proteins were collected from Tid1f/− MEFs infected with recombinant adenoviruses encoding human Tid1-L, Tid1-S, or Tid1-S (H121Q) as indicated and analyzed by Western blotting with the anti-Tid1 antibody. (B) Tid1f/− MEFs were first infected with Ad-Cre or Ad-β-gal adenovirus, and after 2 days, the treated cells were infected with adenoviruses encoding human Tid1-L, Tid1-S, or Tid1-S (H121Q) as indicated. Nine days after the first infection, the number of viable cells for these MEFs was determined by MTT assay. OD560, optical density at 560 nm. The error bars indicate standard deviations. (C) Endogenous Tid1-L and Tid1-S proteins coimmunoprecipitated with mtHsp70. Total protein from MEFs was used for immunoprecipitation (I.P.) experiments with antibody specific for Tid1, mtHsp70, or Hsc70. The resulting immunocomplexes were analyzed by immunoblotting (I.B.) with antibody against Tid1, mtHsp70, or Hsc70, as indicated.
The Tid1 protein has been shown to interact with both mitochondrial heat shock protein 70 (mtHsp70) and nonmitochondrial Hsp70 (25, 27). Nonetheless, Syken et al. (25) reported that in human osteosarcoma cells, U2OS, the Tid1 protein interacts only with mtHsp70 and not with cytosolic Hsp70 (Hsc70). To identify which Hsp70 Tid1 interacts with in MEFs, we used specific antibody against Tid1, mtHsp70, or the nonmitochondrial Hsp70 homolog, Hsc70, in coimmunoprecipitation-immunoblotting experiments. The antibody specific for mouse Tid1 immunoprecipitated both mature forms of Tid1 associated with mtHsp70 but did not immunoprecipitate Hsc70 from MEFs (Fig. 4C). The antibody specific for mtHsp70 protein immunoprecipitated mtHsp70 in complex with Tid1, but in immunoprecipitates prepared from cell extracts with the anti-Hsc70 antibody, no Tid1 was detected (Fig. 4C).
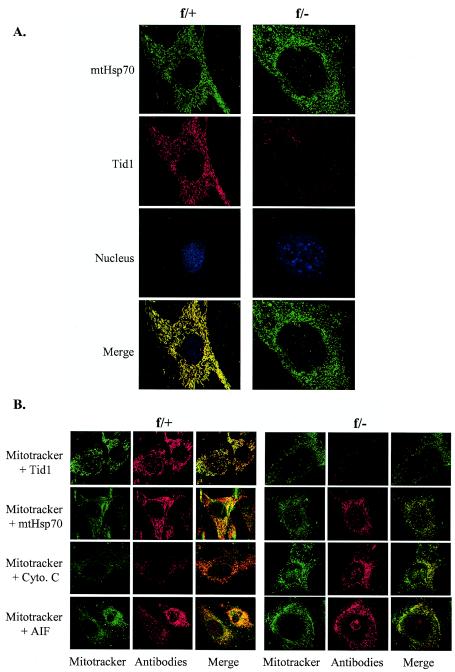
We further investigated the subcellular localization of Tid1 in MEFs by immunofluorescence microscopy. In Tid1f/+ MEFs, 5 days after Ad-Cre infection, endogenous Tid1 and mtHsp70 were separately stained by using antibody specific for either Tid1 or mtHsp70. Both Tid1 and mtHsp70 were detected mostly in the cytoplasm, colocalized, and displayed a short filamentous pattern (Fig. 5A). Not surprisingly, no Tid1 protein could be detected in Ad-Cre-treated Tid1f/− MEFs, and interestingly, the subcellular distribution of mtHsp70 turned punctate (Fig. 5A). The change of the subcellular distribution of mtHsp70 from short filament to punctate was not dependent on the genotype of the MEFs, since before the deletion of Tid1, mtHsp70 and Tid1 colocalized in a short-string pattern within the cytosol of Tid1f/− MEFs (data not shown). These data suggest that the Tid1 protein in MEFs is located within the cytoplasm and that mtHsp70 is the major Hsp70 in complex with Tid1. To further examine the subcellular localization of Tid1 or mtHsp70 in MEFs, Mitotracker, a mitochondrion-specific dye, was used. In Tid1f/+ MEFs 5 days after Ad-Cre infection, most of the Tid1 costained with the Mitotracker, demonstrating that Tid1 proteins were mainly restricted within the mitochondria (Fig. 5B).
FIG.5.
Tid1 and mtHsp70 proteins colocalize within mitochondria. (A) Immunofluorescence staining of Tid1 and mtHsp70 proteins in Tid1f/+ and Tid1f/− MEFs 5 days after treatment with Ad-Cre. (B) The same Ad-Cre-treated Tid1f/+ and Tid1f/− MEFs were first stained with Mitotracker and subsequently double stained with antibody against Tid1, mtHsp70, cytochrome c (Cyto. C), or AIF as indicated.
Similar patterns were observed when Tid1f/+ MEFs were double-stained with Mitotracker and mtHsp70, Mitotracker and cytochrome c, or Mitotracker and AIF (Fig. 5B). In Ad-Cre-treated Tid1f/− MEFs, the intensity of Mitotracker staining was attenuated compared to that of Tid1f/+ MEFs (Fig. 5B). Interestingly, some AIF proteins were translocated from the mitochondria to the nucleus, suggesting that the Tid1f/− MEFs, with Tid1 deleted, were undergoing the process of cell death (Fig. 5B).
Decreased expression of BAG1 and MEK7 genes is partially responsible for cell death resulting from Tid1 removal
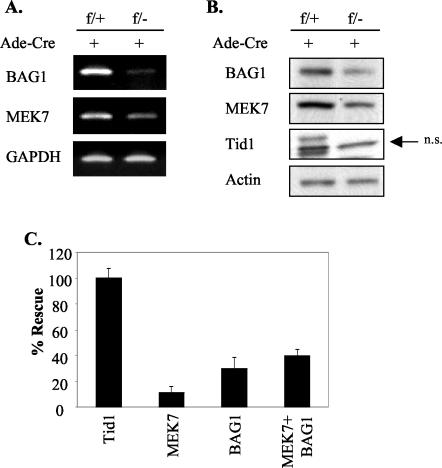
The role of Tid1 in sustaining cell survival most likely occurs through its association with mtHsp70 and the consequent biological effect of this interaction. It has been demonstrated that mitochondrial Hsp70 (mtHsp70/mortalin/Grp75) influences cell mortality by modulating the p53-mediated gene expression of two downstream proteins, MDM2 and p21 (30). Thus, the impact on the expression levels of the p53, MDM2, and p21 proteins was assessed after Tid1 removal in MEFs. We detected no significant alteration in the expression of these proteins (data not shown). Next, by cDNA microarray, we examined 13,000 known genes to determine the gene expression profiles altered by Tid1 removal. Total RNAs from both Ad-Cre-infected Tid1f/+ and Tid1f/− MEFs were purified and examined by cDNA microarray-based analysis. After statistical analysis, we found a total of 91 genes that showed a >2-fold change caused by disrupting both Tid1 alleles (data not shown). Among these, we found that the expression of the BAG1 and MEK7 genes, important for cell growth and survival (8, 26), decreased >2.5-fold in cells without the Tid1 protein. Semiquantitative RT-PCR analysis was used to confirm the results from the cDNA microarray (Fig. 6A). Additionally, by immunoblot assay, the decreased expression of BAG1 and MEK7 proteins after Tid1 deletion was established by comparing their expression levels in Tid1f/+ and Tid1f/− MEFs after Ad-Cre treatment. The expression of the internal control, β-actin, was not altered in these two treated MEFs (Fig. 6B). For determining the contribution of BAG1 and MEK7 in Tid1-dependent cell survival, adenoviruses expressing human recombinant BAG1 or MEK7 were constructed and introduced, separately or jointly, into Tid1f/− MEFs 2 days after the infection of Ad-Cre. Supplementary MEK7 or BAG1 prevented 12 or 31%, respectively, of cell death caused by the loss of Tid1 in Tid1f/− cells (Fig. 6C). Combined treatment with Ad-MEK7 and Ad-BAG1 had an additive effect on rescuing cell death in Tid1f/− cells with Tid1 deleted, suggesting that the survival signals generated from MEK7 or BAG1 do not overlap in Tid1-deficient MEFs. These results indicate that the decreased expression of BAG1 and MEK7 partially accounts for the cell death induced by removing endogenous Tid1 protein from MEFs.
FIG. 6.
Decreased expression of BAG1 and MEK7 genes is partially responsible for cell death resulting from cellular Tid1 removal. (A) Tid1f/+ and Tid1f/− MEFs were infected (+) with Ad-Cre, and after 6 days, total RNAs from these MEFs were collected and analyzed by semiquantitative RT-PCR with the specific primer set for BAG1, MEK7, or GAPDH. The PCR products were separated through 4% agarose. (B) Total cellular extracts were prepared from these Ad-Cre-treated Tid1f/+ and Tid1f/− cells and analyzed by immunoblotting with anti-Tid1, anti-BAG1, anti-MEK7, or anti β-actin antibodies as indicated. n.s., nonspecific. (C) Tid1f/− MEFs were infected with Ad-Cre on day 1 and infected with human recombinant Tid1, BAG1, or MEK7 adenovirus on day 3. The number of viable cells on day 9 for each treated cell type was measured by MTT assay. The rescue rate for cells treated with the combination of Ad-Cre and Ad-Tid1 was set as 100%, and 0% was set for cells treated with Ad-Cre only. The error bars indicate standard deviations.
DISCUSSION
In this study, we generated a null mutation of the Tid1 gene by homologous recombination in mouse ES cells. The Tid1 mutation does not affect the differentiation of the diploid trophoblast in the inner cell mass tissue of the early embryonic blastocyst and yet was lethal before E7.5 of gestation. Interestingly, no hyperproliferative phenotype was observed in the Tid1-null embryos of mice, as was observed in the Tid 56 mutant embryo of Drosophila (16). This phenotypic divergence between mammal and insect might be caused by a developmental role of Tid1 in mammals different from that of its Drosophila counterpart, Tid56. However, we cannot exclude the possibility that in mammals a protection mechanism exists that aborts embryogenesis once the Tid1 mutant embryo becomes tumorigenic. If this is the case, the window to identify those embryos would be very small (between E4.5 and E7.5), and within that period of time, the embryos would be too tiny to isolate and to genotype. The other possibility is that Tid1 is vital for certain crucial cell types during early embryonic development and that disruption of the Tid1 gene in these cells causes embryonic lethality, preventing the observation of the anticancer function of Tid1 for other cell types generated in the later stages of embryogenesis and/or in the adult animal. Early lethality has also been observed in mouse embryos deficient in other tumor suppressors, such as Brca1, Brca2, and Rb, due to their roles in cellular proliferation and differentiation during early development (11, 12, 19, 31). Therefore, it is not unexpected that Tid1, as a putative tumor suppressor, has a role in embryonic development.
Tid1 binds to mtHsp70/mortalin, whose function has been suggested in orchestrating protein transfer across mitochondrial membranes, as well as in cell survival, for its inhibitory effect on p53 (30). However, Tid1 deletion in MEFs does not affect p53-dependent transcription of MDM2 and p21. Possibly, the Tid1/mtHsp70 complex is not involved in modulating p53 activity, at least during early embryogenesis or in embryonic fibroblasts. Nevertheless, we found that Tid1 removal significantly reduces transcription and consequently lowers the level of protein expression of two molecules, BAG1 and MEK7, whose functions in sustaining cell survival have been demonstrated (8, 26). BAG1 is a multifunctional protein that has affinity with a wide variety of cellular targets for regulating cell survival pathways, and its expression is frequently altered in malignant cells (26). MEK7 is one of the MAP2K proteins of the signal pathway of c-Jun N-terminal protein kinase (JNK), which is a critical mediator for transducing message from activated MAP3K to MAPK (JNK) (8). The JNK signaling pathway is critical in regulating both the apoptotic and the antiapoptotic processes (8); thus, it is logical that down-regulation of MEK7 expression is partially responsible for the death of Tid1-deficient MEFs. Tid1 has also been shown to interact with RasGAP and Jak2 kinase to regulate growth control pathways (21, 27), possibly through its role as a cochaperone for Hsp70 (21, 27). These pathways are both important for delivering antiapoptotic or growth signals, resulting in hyperproliferation and, sometimes, cancerous cellular phenotypes (4, 29). Key downstream effectors for these pathways are transcriptional machineries which, after activation, augment the expression of mitogenic genes (4, 29). It will be interesting to see whether the transcription of the BAG1 and MEK7 genes is affected by these two signaling pathways in a Tid1-dependent manner.
More than two dozen tumor suppressor genes are known in Drosophila (9, 10). A number of these genes have human homologues whose tumor suppressor functions have been well documented, such as PTEN, RB, and p16INK4a (3, 20, 23). The anticancer roles of the mammalian counterparts of some of these fly tumor suppressor genes, such as that of Tid56 (Tid1), are not clear, although the loss of Tid1 expression has been implicated in basal cell carcinoma (5). Several reports have associated the functions of Tid1 in processing oncoproteins, for modulating transformation signals, and in regulating apoptotic and antiapoptotic signaling pathways (6, 7, 21, 22, 27). Additionally, overexpression of Tid1 reduces the uncontrolled growth of cancer cells (7). Decreasing cellular Tid1 should promote the proliferation or the malignancy of cells, but we see an opposite effect in embryonic fibroblasts (Fig. 3B). One possible reason is that Tid1 has different functions in different cell types or at different stages of development. This hypothesis is supported by our recent observation that mice with a hepatocyte-specific Tid1 deletion are viable and appear healthy at 4 weeks of age (unpublished data). The other possibility is that loss of Tid1 might lead to an epigenetic condition that results in cell death, excluding those rare cells that undergo additional mutations and develop into cancer. A precedent in carcinogenesis for this kind of inconsistency is the discovery of c-myc: in primary cells, overexpression of c-myc leads to programmed cell death (15); however, for rare cells that contain additional mutations, such as in bcl2, bax, or ras, elevated c-myc promotes their proliferation and survival and induces them to malignancy (1, 13).
To study the pathophysiological function of Tid1 in adult animals and to investigate the role that the loss of this (potential) tumor suppressor gene plays in tumor initiation and progression in vivo, we will take advantage of the conditional nature of Tid1 floxed mice to circumvent the early embryonic death caused by global deletion of Tid1. We are currently breeding our conditional knockout model with various Cre transgenic lines to generate mice in which deletion of the Tid1 gene is restricted to certain types of tissues or selected cell lineages, as well as generating mice in which removal of the Tid1 protein is inducible in adult animals.
In summary, our results demonstrate that Tid1 is a crucial element in early embryogenesis and for sustaining embryonic-cell survival. We also provide evidence that the generation of Tid1-dependent survival signals requires interaction between Tid1 and the Hsp70 chaperone.
REFERENCE
- 1.Aunoble, B., R. Sanches, E. Didier, and Y. J. Bignon. 2000. Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer. Int. J. Oncol. 16:567-576. [DOI] [PubMed] [Google Scholar]
- 2.Bonaventure, P., H. Guo, B. Tian, X. Liu, A. Bittner, B. Roland, R. Salunga, X. J. Ma, F. Kamme, B. Meurers, et al. 2002. Nuclei and subnuclei gene expression profiling in mammalian brain. Brain Res. 943:38-47. [DOI] [PubMed] [Google Scholar]
- 3.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 17:1395-1413. [DOI] [PubMed] [Google Scholar]
- 5.Canamasas, I., A. Debes, P. G. Natali, and U. Kurzik-Dumke. 2003. Understanding human cancer using drosophila: Tid47, a cytosolic product of the DnaJ-like tumor suppressor gene l(2)tid, is a novel molecular partner of patched related to skin cancer. J. Biol. Chem. 278:30952-30960. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, H., C. Cenciarelli, M. Tao, W. P. Parks, and C. Cheng-Mayer. 2002. HTLV-1 Tax-associated hTid-1, a human DnaJ protein, is a repressor of IκB kinase beta subunit. J. Biol. Chem. 277:20605-20610. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, H., C. Cenciarelli, Z. Shao, M. Vidal, W. P. Parks, M. Pagano, and C. Cheng-Mayer. 2001. Human T cell leukemia virus type 1 Tax associates with a molecular chaperone complex containing hTid-1 and Hsp70. Curr. Biol. 11:1771-1775. [DOI] [PubMed] [Google Scholar]
- 8.Dhanasekaran, N., and R. E. Premkumar. 1998. Signaling by dual specificity kinases. Oncogene 17:1447-1455. [DOI] [PubMed] [Google Scholar]
- 9.Gateff, E. 1994. Tumor suppressor and overgrowth suppressor genes of Drosophila melanogaster: developmental aspects. Int. J. Dev. Biol. 38:565-590. [PubMed] [Google Scholar]
- 10.Gateff, E., and B. M. Mechler. 1989. Tumor-suppressor genes of Drosophila melanogaster. Crit Rev. Oncog. 1:221-245. [PubMed] [Google Scholar]
- 11.Gowen, L. C., B. L. Johnson, A. M. Latour, K. K. Sulik, and B. H. Koller. 1996. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat. Genet. 12:191-194. [DOI] [PubMed] [Google Scholar]
- 12.Hakem, R., J. L. de la Pompa, C. Sirard, R. Mo, M. Woo, A. Hakem, A. Wakeham, J. Potter, A. Reitmair, F. Billia, E. Firpo, C. C. Hui, J. Roberts, J. Rossant, and T. W. Mak. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85:1009-1023. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, B., A. Amanullah, M. Shafarenko, and D. A. Liebermann. 2002. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene 21:3414-3421. [DOI] [PubMed] [Google Scholar]
- 14.Hogan, B., F. Costantini, R. Beddington, and E. Lacy. 1995. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Klefstrom, J., I. Vastrik, E. Saksela, J. Valle, M. Eilers, and K. Alitalo. 1994. c-Myc induces cellular susceptibility to the cytotoxic action of TNF-alpha. EMBO J. 13:5442-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurzik-Dumke, U., D. Gundacker, M. Renthrop, and E. Gateff. 1995. Tumor suppression in Drosophila is causally related to the function of the lethal(2) tumorous imaginal discs gene, a dnaJ homolog. Dev. Genet. 16:64-76. [DOI] [PubMed] [Google Scholar]
- 17.Kurzik-Dumke, U., A. Debes, M. Kaymer, and P. Dienes. 1998. Mitochondrial localization and temporal expression of the Drosophila melanogaster DnaJ homologous tumor suppressor Tid50. Cell Stress Chaperones 3:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, B., J. C. Ruiz, and K. T. Chun. 2002. CUL-4A is critical for early embryonic development. Mol. Cell. Biol. 22:4997-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig, T., D. L. Chapman, V. E. Papaioannou, and A. Efstratiadis. 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11:1226-1241. [DOI] [PubMed] [Google Scholar]
- 20.Mayo, L. D., and D. B. Donner. 2002. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem. Sci. 27:462-467. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar, S., B. P. Pollack, K. T. Lin, S. V. Kotenko, J. R. Cook, A. Lewis, and S. Pestka. 2001. hTid-1, a human DnaJ protein, modulates the interferon signaling pathway. J. Biol. Chem. 276:49034-49042. [DOI] [PubMed] [Google Scholar]
- 22.Schilling, B., T. De Medina, J. Syken, M. Vidal, and K. Munger. 1998. A novel human DnaJ protein, hTid-1, a homolog of the Drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology 247:74-85. [DOI] [PubMed] [Google Scholar]
- 23.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 24.Silver, P. A., and J. C. Way. 1993. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 74:5-6. [DOI] [PubMed] [Google Scholar]
- 25.Syken, J., T. De Medina, and K. Munger. 1999. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc. Natl. Acad. Sci. USA 96:8499-8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayama, S., and J. C. Reed. 2001. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3:E237-E241. [DOI] [PubMed] [Google Scholar]
- 27.Trentin, G. A., X. Yin, S. Tahir, S. Lhotak, J. Farhang-Fallah, Y. Li, and M. Rozakis-Adcock. 2001. A mouse homologue of the Drosophila tumor suppressor l(2)tid gene defines a novel Ras GTPase-activating protein (RasGAP)-binding protein. J. Biol. Chem. 276:13087-13095. [DOI] [PubMed] [Google Scholar]
- 28.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 29.Verma, A., S. Kambhampati, S. Parmar, and L. C. Platanias. 2003. Jak family of kinases in cancer. Cancer Metastasis Rev. 22:423-434. [DOI] [PubMed] [Google Scholar]
- 30.Wadhwa, R., S. Takano, M. Robert, A. Yoshida, H. Nomura, R. R. Reddel, Y. Mitsui, and S. C. Kaul. 1998. Inactivation of tumor suppressor p53 by mot-2, a hsp70 family member. J. Biol. Chem. 273:29586-29591. [DOI] [PubMed] [Google Scholar]
- 31.Williams, B. O., E. M. Schmitt, L. Remington, R. T. Bronson, D. M. Albert, R. A. Weinberg, and T. Jacks. 1994. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 13:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]