Abstract
Interleukin-10 (IL-10) is a potent anti-inflammatory cytokine that suppresses the induction of proinflammatory cytokine genes, including the IL-12 p40 gene. Despite considerable effort examining the effect of IL-10 on specific transcription factors and signaling molecules, the mechanism by which IL-10 inhibits gene transcription has remained elusive. To provide a different perspective to this problem, we examined the effect of IL-10 on molecular events occurring at the endogenous IL-12 p40 locus in lipopolysaccharide-stimulated peritoneal macrophages. IL-10 abolished recruitment of RNA polymerase II to the p40 promoter. However, it only modestly reduced binding of C/EBPβ, as monitored by genomic footprinting and chromatin immunoprecipitation. It also had little effect on NF-κB complexes that are critical for p40 induction. A substantial reduction in nucleosome remodeling at the p40 promoter was observed, but the magnitude of this reduction appeared insufficient to account for the strong inhibition of transcription. Finally, a lipopolysaccharide-inducible DNase I hypersensitive site identified 10 kb upstream of the start site was unaffected by IL-10. Thus, despite a dramatic reduction in p40 transcription, several events required for activation of the endogenous p40 gene occurred relatively normally. These findings suggest that IL-10 blocks one or more events that occur after p40 locus decondensation and nucleosome remodeling and after, or in parallel with, the binding of a subset of p40 transcriptional activators.
The inflammatory response is an essential component of the host defense against microbial pathogens. However, when excessive or regulated improperly, inflammation can lead to harmful and even fatal consequences. Among the many biological agents that suppress inflammatory responses, the cytokine interleukin-10 (IL-10) is one of the most potent and significant. Numerous studies have shown that IL-10 treatment can decrease the severity of inflammatory processes in vivo (34). For example, IL-10 protects mice against endotoxin-induced lethality (19, 21, 32). Furthermore, IL-10−/− mice develop an inflammatory Crohn's-like disease and exhibit dysregulated inflammatory responses (25). These findings suggest that key functions of IL-10 are to maintain homeostasis of the immune system and to protect the host from excessive inflammation.
IL-10 is secreted primarily by activated macrophages and T cells and inhibits numerous events in macrophages and neutrophils, including major histocompatibility complex class II expression, oxidative burst and nitric oxide production, phagocytosis, and the production of proinflammatory cytokines (9, 13, 34). IL-10 carries out these functions by interacting with the IL-10 receptor, which induces signaling through the Janus kinase/signal transducer and activator of transcription (Jak/Stat) pathway (34). In Jak1−/− and Stat3−/− macrophages, IL-10 was unable to inhibit lipopolysaccharide (LPS)-induced gene expression, demonstrating essential roles for these proteins (40, 46). The suppressor of cytokine signaling-3 (SOCS3) protein has also been implicated in the IL-10 pathway (3), although more recent studies with SOCS3-deficient mice have demonstrated that this protein is not essential for the suppression of LPS-induced gene expression by IL-10 (26, 55).
Beyond the requirement for molecules that interact with the IL-10 receptor, little is known about the mechanism by which IL-10 inhibits the expression of proinflammatory cytokine genes (13, 34). The suppression of cytokine genes in macrophages appears to be indirect, as Stat protein binding sites that act in a negative manner have not been identified in the promoters for these genes. Previous studies demonstrated that IL-10 can both inhibit cytokine gene transcription and destabilize cytokine mRNAs (1, 4, 6, 7, 11, 12, 14, 23, 29, 38, 41, 44, 45, 47, 51, 52). Some studies have suggested that IL-10 prevents NF-κB activation by LPS and other stimuli (29, 38, 41, 44, 45, 52), but no effect on NF-κB was observed in other studies (3, 10-12, 47). Variable effects on the activation of AP-1 and mitogen-activated protein kinases have also been reported (3, 11-13, 16, 18, 24, 38, 52), but IL-10 had no effect on the activation of several other transcription factors (12, 47, 52). The variable effects on NF-κB, AP-1, and mitogen-activated protein kinases may in part be due to the use of macrophages from different sources, although in some cases conflicting results have been obtained when using the same stimuli and similar macrophage populations. Microarray experiments revealed that a number of genes are upregulated by IL-10 in macrophages, but few potential contributors to the inhibition of cytokine gene transcription have emerged (7, 27). The transcriptional coactivator c-Maf was identified in one study as an IL-10-inducible protein that may contribute to the inhibition of proinflammatory cytokine genes, but transcriptional inhibition by IL-10 was unaffected in c-Maf−/− macrophages (7).
IL-12 is a proinflammatory cytokine that serves as a critical bridge between innate and adaptive immunity by stimulating the development of T-helper 1 (Th1) cells (17, 48-50). Antigen-stimulated Th1 cells secrete cytokines that contribute to host defense against intracellular pathogens. The bioactive IL-12 p70 heterodimer contains p35 and p40 subunits, which are synthesized in macrophages and dendritic cells in response to intracellular pathogens and bacterial products, including LPS (48-50).
The murine IL-12 p40 promoter contains several transcription factor binding sites that contribute to gene induction in LPS-stimulated macrophages (31, 35, 36, 56). The three control elements that have been characterized most extensively bind NF-κB, C/EBP, and AP-1 family members. An analysis of mice lacking specific NF-κB family members revealed that c-Rel is selectively required for p40 induction in macrophages stimulated with LPS plus gamma interferon (IFN-γ) (42). The involvement of C/EBPβ in p40 induction was supported by a number of findings, including efficient association of C/EBPβ with the endogenous p40 promoter in chromatin immunoprecipitation (ChIP) experiments (5, 36). Transcription of the p40 gene is also regulated at the chromatin level. A positioned nucleosome spans the transcription factor binding sites at the p40 promoter and is selectively remodeled upon macrophage activation (54). Remodeling of this nucleosome is independent of c-Rel and may also be independent of C/EBP and AP-1 proteins, suggesting that remodeling complexes may be recruited by other inducible factors (53).
A previous study provided compelling evidence that the inhibition of IL-12 p40 expression by IL-10 in human peripheral blood mononuclear cells occurs at the level of transcription initiation (1). However, efforts to elucidate the mechanism of inhibition were unsuccessful, largely because inhibition of p40 promoter activity by IL-10 was not observed in transient- or stable-transfection assays with p40 promoter-reporter plasmids (1; L. Zhou and S. T. Smale, unpublished results). Another technical hurdle that has slowed efforts to dissect the mechanism of p40 inhibition by IL-10 is that the magnitude of inhibition is significantly smaller in transformed macrophage lines than in primary macrophages (Zhou and Smale, unpublished).
Because the strategies used to elucidate the IL-10 inhibition mechanism have thus far been unsuccessful, we used a different strategy to gain mechanistic insight. Specifically, we monitored the effect of IL-10 on events occurring at the endogenous p40 promoter in LPS-stimulated peritoneal macrophages. An attractive feature of this approach is that the experiments can be performed in primary macrophages, which support strong inhibition by IL-10 and yield results that may be more physiological than those obtained in transformed cell lines. Furthermore, by studying the endogenous p40 gene, the results can reveal whether IL-10 makes the locus completely refractory to events associated with gene activation or, alternatively, targets a later step in the activation pathway. The results described here demonstrate that, in the presence of IL-10, recruitment of RNA polymerase II to the p40 promoter is efficiently blocked. However, IL-10 does not render the chromatin associated with the p40 locus resistant to decondensation or nucleosome remodeling and does not inhibit the activation or in vivo binding of transcription factors that were studied. These results suggest that IL-10 signaling inhibits one or more relatively late events that are required for RNA polymerase II recruitment to the p40 promoter.
MATERIALS AND METHOD
Cells
Macrophages were isolated from the peritoneal cavity of thioglycolate-treated mice as described earlier (54) and were allowed to adhere to culture plates for 24 to 36 h in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% low-endotoxin fetal bovine serum (Omega Scientific) and penicillin/streptomycin (Omega Scientific). Nonadherent cells were removed by washing with phosphate-buffered saline (PBS). Following the addition of fresh complete medium, cells were left untreated or were activated for 4 h with either 100 ng or 10 μg of LPS (Sigma)/ml plus 10 U of recombinant murine IFN-γ (PharMingen)/ml, with or without 10 ng of IL-10 (PharMingen)/ml.
Bone marrow-derived macrophages were prepared from C57BL/6 mice. Cells (n = 3.5 × 106) were plated in Dulbecco's modified Eagle's medium with 20% fetal bovine serum, 30% L929-conditioned medium, 1% l-glutamine, 0.5% sodium pyruvate, 0.1% β-mercaptoethanol, and penicillin/streptomycin. The cells were cultured for 7 days with fresh medium added as necessary and were then stimulated under the same conditions employed for peritoneal macrophages.
ELISA
Peritoneal macrophages (n = 2 × 107) in 10 ml of culture medium were left inactivated or were activated for 4 h. Combined concentrations of IL-12 p40 homodimer and p35/p40 (p70) heterodimer were determined by sandwich enzyme-linked immunosorbent assay (ELISA) by using 100 μl of the supernatants and 10-fold serial dilutions as described elsewhere (5, 54).
Northern blot
Total RNA was extracted from peritoneal macrophages by using TRI-reagent (MRC). Northern blot analysis was performed as described earlier (2) by using 50 μg of total RNA. Briefly, after agarose/formaldehyde gel electrophoresis, the RNA samples were transferred to Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) and were cross-linked by UV irradiation. Radiolabeled probes were prepared from p40 and β-actin cDNAs by using the Prime-it II random primer labeling kit (Stratagene). The blots were hybridized at 42°C overnight, followed by extensive washes and autoradiography.
RT-PCR
RNA isolated by using TRI-reagent was treated with RNase-free DNase I and was purified by phenol:chloroform extraction, followed by ethanol precipitation. Reverse transcription (RT) was performed with p40 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers as previously described (5). cDNAs derived from p40 primary transcripts and GAPDH mRNA were amplified by PCR with the primers and under the reaction conditions described previously (5). cDNA derived from p40 mRNA was amplified by using the following primers: 5′-TTATGTTGTAGAGGTGGACTGG-3′ (exon 3) and 5′-TTTCTTTGCACCAGCCATGAGC-3′ (exon 4). PCR conditions for this amplification were as follows: 94°C for 180 s; 19 cycles of 94°C for 60 s, 64°C for 60 s, and 72°C for 60 s; and final elongation at 72°C for 10 min. PCR products were resolved on a 1% agarose gel, followed by transfer to nylon membrane and cross-linking by UV irradiation. Oligonucleotides derived from p40 exon 4 (5′-ATTTGGTGCTTCACACTTCAGG-3′) and GAPDH cDNA (5′-GAAGACACCAGTAGACTCCACGAC-3′) were radiolabeled with [γ-32P]ATP and T4 DNA kinase. Probes were incubated with the membrane at 42°C by using ULTRAhyb (Ambion). The membranes were washed and were subjected to autoradiography.
Restriction enzyme accessibility and DNase I genomic footprinting
The ligation-mediated PCR (LM-PCR)-based restriction enzyme accessibility assay and DNase I genomic footprinting assay were performed as described earlier (54). For the Southern blot-based restriction enzyme assay, cell nuclei (n = 2 × 107) were suspended in New England Biolabs Buffer 2 (50 μl). SpeI was added, and reaction materials (in a total volume of 50 μl) were incubated at 37°C for 15 min. Genomic DNA was prepared by using the DNeasy Tissue Kit (Qiagen). Purified DNA was then digested in vitro with various restriction enzymes. Restriction enzymes PstI and EcoRI were used to monitor nucleosome remodeling at the p40 promoter; KpnI and SphI were used to monitor remodeling at the DNase I hypersensitivity region. The digested fragments were examined by Southern blotting. Probes were generated by PCR and were 32P labeled by using the Prime-it II kit (Stratagene). Primers for generating the promoter probe were as follows: 5′-CTCTCCTCTTCCCTGTCGCTAACTC-3′ and 5′-CCTTTCTATCAAATACACATCTGTCC-3′. Primers for generating the enhancer probe were as follows: 5′-GGCTAGGTGTACATGTATGTGCATATATC-3′ and 5′-GAAAGAAATGAATGAGTTCCCACC-3′.
Gel shift and Western blot assays
Nuclear extracts and gel shift probes were prepared as described elsewhere (36). Gel shift probes for NF-κB and C/EBP contained p40 promoter sequences from −142 to −107 and from −110 to −69, respectively. Antibodies from Santa Cruz Biotechnology were p50 (D-17), p65 (A), and c-Rel (C). A second c-Rel antibody (no. 1266NR) was a gift from Nancy Rice. Gel shift assays were performed as described earlier (5, 42). A Western blot assay was performed as described earlier (36).
ChIP assay
Peritoneal macrophages were fixed at room temperature by adding formaldehyde to the culture medium to a final concentration of 1%. After 10 min, cross-linking was stopped by addition of 1 M glycine to a final concentration of 0.125 M, and the incubation was continued at room temperature for 5 min. Cells were washed on plates with ice-cold PBS three times and were then collected. Cells were lysed for 10 min on ice in cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 8.0), 85 mM KCl, 0.5% NP-40, and protease inhibitors]. Nuclei were suspended in nuclear lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate, and protease inhibitors) and were incubated on ice for 10 min. Chromatin was sheared into 500- to 1,000-bp fragments by sonication and was then precleared with protein A-Sepharose beads. The purified chromatin was diluted with ChIP dilution buffer (0.01% sodium dodecyl sulfate, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl, and protease inhibitors) and was immunoprecipitated overnight at 4°C by using antibodies directed against C/EBPβ (SC-150×, 10 μg; Santa Cruz), RNA polymerase II (SC-899, 2 μg; Santa Cruz), or glutathione S-transferase (GST) (5 μl). Immune complexes were collected with protein A-Sepharose beads and were washed and eluted. The protein-DNA cross-links were then reversed. Following DNA purification, the presence of selected DNA sequences was assessed by PCR. Primer pairs for monitoring C/EBPβ binding to the p40 promoter were described elsewhere (5). Primer pairs for monitoring the C/EBPβ binding to the DNase I hypersensitive region were as follows: fragment A, 5′-ATGTCTCTCACATTGGTCATCTGC-3′ and 5′-AACTTTTTCTTTCTGTGTGACATAATTTATG-3′; and fragment B, 5′-AACTGTTACGGTCTTAGGCATGGTCTGG-3′ and 5′-TAGCCATGGGCAGGTGATTTAAAC-3′. PCR conditions were described elsewhere (5).
Real-time quantitative PCR
Eluted DNA samples from ChIP experiments were analyzed by real-time quantitative PCR in triplicate by using iQ SYBR Green Supermix (Bio-Rad) and the iCycler Sequence Detection System (Bio-Rad). The starting quantity (SQ) of the initial DNA template input for each ChIP sample was calculated from primer-specific standard curves by using the iCycler Data Analysis Software. The enrichment (n-fold) of C/EBPβ or RNA polymerase II was determined as {[IL-12 (mean of SQ)]/[TdT (mean of SQ)]}LPS/IFN-γ ± IL-10/{[IL-12 (mean of SQ)]/[TdT (mean of SQ)]}inactivated. The primer sets for real-time PCR were as follows: IL-12 p40, 5′-TAGTATCTCTGCCTCCTTCC-3′ and 5′-GAACTTTCTGATGGAAACCC-3′; and TdT, 5′-ACCAAGACTGACAACCCACGTT-3′ and 5′-GTGGCAGTCAGAGGCATCTTT-3′.
DNase I hypersensitivity assay
After one ice-cold PBS wash on culture dishes, peritoneal macrophages were collected and lysed for 5 min on ice in NP-40 lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, 0.15 mM spermine, and 0.5 mM spermidine). Nuclei were then washed in buffer A (100 mM NaCl, 50 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 0.15 mM spermine, and 0.5 mM spermidine) plus 1 mM CaCl2. After centrifugation, nuclear pellets were resuspended in buffer A plus 1 mM CaCl2. Then 50-μl aliquots of nuclei (2.0 × 107 to 2.5 × 107 cells) were mixed with increasing amounts of DNase I (0, 0.2, 0.4, 0.8, and 1.2 μg), followed by incubation at 37°C for 2 min. The reactions were terminated by addition of Stop solution (2 μl of 0.5 M EDTA, 20 μl of 20-mg/ml protease K solution, and 150 μl of 1× PBS). Genomic DNA was extracted by using DNeasy Tissue Kit (Qiagen), according to the manufacturer's protocol. Purified DNA (20 μg) was then digested with AseI (New England Biolabs). DNA samples were purified by phenol:chloroform extraction followed by ethanol precipitation and were then separated on a 0.7% agarose gel. Southern blots were performed by using a standard procedure (2). The radiolabeled probe was generated by PCR with the Prime-it II kit (Stratagene). The primer sequences were as follows: 5′-CTCAATTCGTAAAGTGAGCAGGATTGC-3′ and 5′-TAGCTCAGGCTAGCCTCAACTCTTC-3′.
RESULT
IL-10 inhibits IL-12 p40 gene transcription in peritoneal macrophages
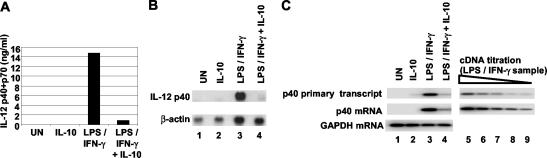
The goal of this study was to gain insight into the mechanism by which IL-10 inhibits IL-12 p40 expression in murine macrophages. Most experiments focused on events occurring at the endogenous p40 locus. Furthermore, primary peritoneal macrophages were used for most experiments because of the potential for increased physiological relevance and because the extent of inhibition was consistently greater than in macrophage cell lines (data not shown). In peritoneal macrophages stimulated for 4 h with LPS plus IFN-γ, addition of IL-10 at the time of stimulation routinely inhibited the accumulation of p40 protein by 90 to 95%, as determined by ELISA. The results obtained in a representative experiment are shown in Fig. 1A. The effect of IL-10 on steady-state p40 mRNA levels was determined by Northern blotting. p40 mRNA was greatly enhanced in peritoneal macrophages stimulated with LPS plus IFN-γ for 4 h, but this enhancement was strongly suppressed in the presence of IL-10 (Fig. 1B).
FIG. 1.
IL-10 inhibits IL-12 p40 gene transcription in peritoneal macrophages. Peritoneal macrophages were left untreated (UN) or were treated for 4 h with IL-10, LPS plus IFN-γ, or IL-10 and LPS plus IFN-γ. (A) IL-12 p40/p70 concentrations in culture supernatants were measured by ELISA. (B) p40 mRNA was detected by Northern blot by using 50 μg of total RNA. As a control, β-actin mRNA was monitored on the same blot. (C) Primary p40 transcripts, p40 mRNA, and GAPDH mRNA were analyzed by semiquantitative RT-PCR (lanes 1 to 4). To determine the extent of inhibition by IL-10, RT-PCR was used to analyze a twofold serial dilution of first-strand cDNA prepared from cells treated with LPS plus IFN-γ (lanes 5 to 9). The first sample in this dilution series (lane 5) contained half of the cDNA used for the reaction shown in lane 3.
Previously, a nuclear run-on assay was used to show that IL-10 inhibits p40 expression at the level of transcription initiation in human peripheral blood mononuclear cells (1). To determine whether IL-10 also acts at the level of transcription initiation in murine peritoneal macrophages, primary p40 transcripts and mature p40 mRNA were monitored by semiquantitative RT-PCR. Primary transcripts were monitored by using a primer that hybridizes to intron 3 and a reverse primer that hybridizes to exon 4. Because introns are often rapidly excised and degraded, an analysis of primary transcripts provides an approximate measure of transcription initiation frequency, similar to the information provided by nuclear run-on assays (30, 39). Interestingly, p40 primary transcripts and the mature mRNA were reduced by similar magnitudes when cells were activated in the presence of IL-10 (Fig. 1C, lanes 3 and 4). The approximate linearity of the PCR was demonstrated by analysis of twofold serial dilutions of an input cDNA sample prepared from cells stimulated with LPS plus IFN-γ (lanes 5 to 9). Consistent with the ELISA results (Fig. 1A), the abundance of primary transcripts was reduced by approximately 16-fold (Fig. 1C, top panel, compare lanes 4 and 8). For each of the experiments described below, the extent to which IL-10 inhibited p40 expression was monitored by ELISA. p40 protein concentrations were reduced by greater than 90% in each experiment (data not shown).
IL-10 disrupts RNA polymerase II recruitment
To determine whether IL-10 inhibits transcription by acting before or after the recruitment of RNA polymerase II, a ChIP assay was used to monitor polymerase association with the endogenous p40 promoter in peritoneal macrophages. Sheared genomic DNA precipitated with antibodies directed against RNA polymerase II was analyzed by PCR with two primer pairs. One primer pair amplified the p40 promoter, and the second pair amplified an intron of the lymphocyte-specific TdT gene, which is not expressed in macrophages. In the DNA sample prepared from macrophages stimulated with LPS plus IFN-γ, the p40 promoter product was substantially enriched relative to the TdT product (Fig. 2A, lane 2). Interestingly, in the sample prepared from macrophages treated simultaneously with IL-10 and LPS plus IFN-γ, the p40 product was not enriched (Fig. 2A, lane 3). Quantitative real-time PCR analysis of the DNA samples revealed that the IL-12 p40 promoter was 1.6-fold enriched in the stimulated cells and confirmed that IL-10 prevented this enrichment (Fig. 2B). These results provide strong support for a model in which IL-10 inhibits p40 expression by blocking the recruitment of RNA polymerase II to the promoter. The results also confirm that IL-10 blocks transcription initiation at the p40 gene.
FIG. 2.
IL-10 inhibits RNA polymerase II recruitment to the p40 promoter. (A) Binding of RNA polymerase II to the endogenous p40 promoter was measured by ChIP. Formaldehyde-cross-linked chromatin samples prepared from untreated peritoneal macrophages (lane 1) or peritoneal macrophages treated for 4 h with LPS plus IFN-γ (lane 2), or IL-10 and LPS plus IFN-γ (lane 3) were immunoprecipitated with RNA polymerase II antibodies (top panel) or control GST antibodies (middle panel). The presence of p40 promoter and TdT intronic DNAs was determined by semiquantitative PCR. Input samples are shown in the bottom panel. (B) Quantitative real-time PCR was performed with the same precipitated DNA samples analyzed in panel A. The bar graph shows the enrichment (n-fold) of RNA polymerase II at the p40 promoter in cells stimulated in the absence and presence of IL-10, relative to the amount observed in unstimulated (UN) cells. Mean values and standard errors of the mean from triplicate PCR experiments are shown.
Effect of IL-10 on C/EBPβ binding to the endogenous p40 promoter
To gain insight into the mechanism by which IL-10 inhibits RNA polymerase II recruitment, we first monitored the effect of IL-10 on a central regulator of p40 transcription, C/EBPβ. Two isoforms of C/EBPβ are expressed in peritoneal macrophages (5): LAP, which contains transactivation domains and the basic leucine zipper DNA-binding and dimerization domain, and LIP, which lacks the transactivation domains. We recently showed that the concentrations of both LAP and LIP increase in peritoneal macrophages stimulated with LPS plus IFN-γ (5). However, the results suggested that the increased concentrations were unimportant for p40 induction (5). Western blot assays revealed that IL-10 did not prevent the upregulation of LAP and LIP in peritoneal macrophages stimulated with LPS plus IFN-γ for 4 h (Fig. 3A). In fact, slight enhancements in LAP and LIP concentrations were consistently observed in the presence of IL-10 (Fig. 3A, lanes 3 and 4; data not shown). Similarly, in gel shift experiments performed with nuclear extracts from unstimulated and stimulated peritoneal macrophages, the DNA-binding activities of LAP homodimers and LAP/LIP heterodimers were slightly enhanced when IL-10 was added simultaneously with the LPS plus IFN-γ (Fig. 3B). Previous studies showed that these complexes, which form on a radiolabeled probe containing the C/EBP site from the p40 promoter, interact with C/EBPβ antibodies (5).
FIG. 3.
Effect of IL-10 on C/EBPβ binding to the endogenous p40 promoter. (A) C/EBPβ isoforms LAP and LIP were visualized by Western blotting by using nuclear extracts from untreated (UN) peritoneal macrophages (lane 1) or peritoneal macrophages treated for 4 h with IL-10 (lane 2), LPS plus IFN-γ (lane 3), or IL-10 and LPS plus IFN-γ (lane 4). A nonspecific protein recognized by the antibody (*) was used for the purpose of normalization. (B) The DNA-binding activities of LAP-LAP homodimers and LAP-LIP heterodimers were monitored by gel shift with a 32P-labeled probe containing the C/EBP site from the p40 promoter. The relevant complexes were identified on the basis of antibody supershift experiments and other criteria described previously (5). (C) DNase I genomic footprinting assays were performed with peritoneal macrophages that were left unstimulated or were stimulated for 4 h as described above. LM-PCR was performed by using primers specific to the p40 promoter region. The locations of the Rel, C/EBP, and AP-1 sites within the p40 promoter are indicated. Additional sites that exhibit hypersensitivity to DNase I cleavage in stimulated cells are also indicated (*). (D) Binding of C/EBPβ to the endogenous p40 promoter was monitored by ChIP and semiquantitative PCR by using C/EBPβ and GST antibodies and chromatin samples as described in the legend to Fig. 2. (E) Quantitative real-time PCR was performed with the precipitated DNA samples analyzed for panel D. The bar graph shows the enrichment (n-fold) of C/EBPβ at the p40 promoter in cells stimulated in the absence and presence of IL-10, relative to the amount observed in unstimulated cells. Mean values and standard errors of the mean from triplicate PCR experiments are shown. Similar results were obtained in two additional independent experiments by using peritoneal macrophages isolated from different mice.
To determine whether IL-10 affects the association of C/EBPβ with the endogenous p40 promoter, DNase I genomic footprinting assays were first performed. As previously reported (54), the C/EBP site and adjacent AP-1 site were strongly protected following stimulation for 4 h with LPS plus IFN-γ (Fig. 3C, lane 2). Other characteristic changes in the DNase I digestion pattern were apparent, which may be due to remodeling of the nucleosome encompassing this region (reference 54 and see below). Importantly, IL-10 only slightly diminished protection of the C/EBP and AP-1 sites and had no significant effect on the overall pattern of DNase I digestion (Fig. 3C, lanes 2 and 4). The minimal effect on occupancy of the AP-1 site is consistent with published evidence that IL-10 does not inhibit AP-1 activity (38, 52). We have also found that IL-10 does not inhibit AP-1 binding in gel shift assays performed with extracts from peritoneal macrophages stimulated in the absence and presence of IL-10 (data not shown).
To monitor more specifically the association of C/EBPβ with the endogenous p40 promoter, a ChIP assay was employed that used C/EBPβ antibodies. In cells stimulated with LPS plus IFN-γ, association of C/EBPβ with the p40 promoter was demonstrated by the increased abundance of the p40 promoter PCR product relative to the TdT product (Fig. 3D, lanes 1 and 2). The p40/TdT ratio was moderately reduced in cells that were treated simultaneously with IL-10 (Fig. 3D, lane 3). However, substantial enrichment of the p40 promoter product relative to the input sample was still apparent (Fig. 3D, lane 3, compare top and bottom panels). Quantitative real-time PCR analysis revealed that the IL-12 p40 promoter fragment was enriched by 3.8-fold in the stimulated cells and that IL-10 reduced this enrichment by 40% (Fig. 3E). Although the reduced association of C/EBPβ revealed by genomic footprinting and ChIP may contribute to the inhibition of p40 transcription by IL-10, the modest magnitudes of the reductions suggest that C/EBPβ inhibition is not a primary cause of the more dramatic inhibition of transcription and RNA polymerase II recruitment. Instead, the severe defect in transcription is likely to be caused by another mechanism, which may, as a secondary effect, result in the destabilization of C/EBPβ binding. This hypothesis is consistent with published evidence that eukaryotic regulatory proteins bind promoters cooperatively with components of the basal transcription machinery (15, 28, 43).
Effect of IL-10 on IL-12 p40 expression and C/EBPβ binding in bone marrow-derived macrophages
To determine whether the results obtained with peritoneal macrophages are relevant to the mechanism by which IL-10 inhibits IL-12 p40 expression in other macrophage populations, bone marrow-derived macrophages were analyzed. ELISA results revealed that IL-10 inhibited the production of IL-12 p40 protein following LPS-plus-IFN-γ stimulation by approximately eightfold (Fig. 4A), which was slightly less inhibition than typically observed with peritoneal macrophages. Strikingly, ChIP experiments analyzing the association of C/EBPβ with the p40 promoter following stimulation of the bone marrow-derived macrophages in the presence and absence of IL-10 yielded results that were virtually identical to those obtained with peritoneal macrophages (compare Fig. 4B and C with 3D and E). In the quantitative experiments (Fig. 4C and 3E), IL-10 reduced C/EBPβ association by 33% in the bone marrow-derived cells, in comparison to the 40% reduction observed in the peritoneal macrophages. Although we have not performed a complete analysis of IL-10 inhibition in the bone marrow-derived macrophages, these results suggest that the inhibition mechanism may be similar to the mechanism employed by peritoneal macrophages.
FIG. 4.
Effect of IL-10 on IL-12 p40 expression and C/EBPβ binding in bone marrow-derived macrophages. Murine bone marrow-derived macrophages were left untreated (UN) or were treated for 4 h with IL-10, LPS plus IFN-γ, or IL-10 and LPS plus IFN-γ. (A) IL-12 p40/p70 concentrations in culture supernatants were measured by ELISA. (B) Binding of C/EBPβ to the endogenous p40 promoter was monitored by ChIP and semiquantitative PCR by using C/EBPβ antibodies and chromatin samples prepared from bone marrow-derived macrophages. (C) Quantitative real-time PCR was performed with the precipitated DNA samples analyzed for panel B. The bar graph shows the enrichment (n-fold) of C/EBPβ at the p40 promoter in cells stimulated in the absence and presence of IL-10, relative to the amount observed in unstimulated cells. Mean values and standard errors of the mean from triplicate PCR experiments are shown.
Effect of IL-10 on NF-κB DNA-binding activity
As described above, another critical control element in the p40 promoter binds NF-κB complexes (35, 36). The NF-κB family member c-Rel is required for p40 induction in peritoneal macrophages stimulated with LPS plus IFN-γ (42), and more recent studies suggest that the c-Rel requirement is due to selective binding of c-Rel to the p40 promoter (Shomyseh Sanjabi, Kevin Williams, and Smale, unpublished results). Unfortunately, we have been unable to detect direct binding of c-Rel to the p40 promoter in peritoneal macrophages by using genomic footprinting (Fig. 3C) or ChIP assays (data not shown). In the genomic footprinting experiments, cleavage at one position near the center of the NF-κB recognition sequence was actually enhanced in nuclei from cells stimulated with LPS plus IFN-γ, with or without IL-10 (Fig. 3C, lanes 2 and 4). The reason for this change in DNase I digestion pattern in the vicinity of the NF-κB site is not known, but a favored hypothesis is that it is the result of nucleosome remodeling rather than of NF-κB binding. Our inability to detect clear protection of an NF-κB site that is important for p40 promoter function in transient- and stable-transfection assays suggests that NF-κB binding may be relatively transient, as has been demonstrated for binding of the glucocorticoid receptor to the mouse mammary tumor virus long terminal repeat (33).
Although we have been unable to monitor the binding of NF-κB to the endogenous p40 promoter, gel shift experiments were performed with the goal of determining whether the activation of c-Rel-containing complexes might specifically be inhibited by IL-10. As previously described (42), stimulation of peritoneal macrophages for 4 h with LPS plus IFN-γ resulted in the induction of two protein-DNA complexes when a probe containing the NF-κB site from the p40 promoter was used (Fig. 5A, lanes 1 to 3). Neither complex was noticeably affected when IL-10 was added with the LPS-plus-IFN-γ stimulus (Fig. 5A, lane 8). Previous studies demonstrated that the lower complex contains p50 homodimers, whereas the upper complex contains a mixture of p65/p50 and c-Rel/p50 heterodimers (42). Consistent with these assignments, p50 antibodies supershifted both complexes and p65 antibodies eliminated a substantial fraction of the upper complex, leaving behind a diffuse band that is characteristic of the p50/c-Rel heterodimer (Fig. 5A, lanes 4 and 5; see reference 42). In the presence of a c-Rel antibody that disrupts protein-DNA interactions by c-Rel, little effect was apparent (Fig. 5A, lane 6). However, simultaneous addition of the p65 and c-Rel antibodies led to a complete loss of the upper complex (Fig. 5A, lane 7). Importantly, activation of the cells in the presence of IL-10 had little effect on any of the complexes detected when these antibodies were added (Fig. 5A, lanes 9 to 12). To determine more conclusively whether IL-10 inhibits the induction of the p50/c-Rel complex, an antibody was used that selectively supershifts c-Rel-containing complexes. Supershifted complexes of comparable abundance were detected with extracts from cells stimulated in the absence or presence of IL-10 (Fig. 5B, lanes 6 and 8), confirming that IL-10 does not prevent induction of the p50/c-Rel complex.
FIG. 5.
Effect of IL-10 on NF-κB DNA-binding activity. (A) Gel shift and supershift assays were performed with a 32P-labeled probe containing the NF-κB site from the p40 promoter and nuclear extracts from untreated peritoneal macrophages (lane 1) or peritoneal macrophages treated for 4 h with IL-10 (lane 2), LPS plus IFN-γ (lanes 3 to 7), or IL-10 and LPS plus IFN-γ (lanes 8 to 12). Specific antibodies (Ab) and locations of protein-DNA complexes are indicated. (B) The effect of IL-10 on the induction of c-Rel-containing protein-DNA complexes was determined by using a c-Rel antibody that supershifts c-Rel complexes with a high degree of specificity.
Effect of IL-10 on nucleosome remodeling at the IL-12 p40 promoter
Induction of the p40 gene is accompanied by the remodeling of a single nucleosome at the p40 promoter, called nucleosome 1 (54). To determine whether IL-10 inhibits this remodeling event, a restriction enzyme accessibility assay was used. With this assay, inducible remodeling at the p40 promoter is revealed by an increased cleavage efficiency when a restriction enzyme is added to nuclei from peritoneal macrophages stimulated with LPS plus IFN-γ, in comparison to the cleavage efficiency in nuclei from unstimulated cells. The restriction endonuclease SpeI was used for these experiments because it cleaves at two sites within nucleosome 1 (Fig. 6A) (54).
FIG. 6.
Effect of IL-10 on nucleosome remodeling at the IL-12 p40 promoter. (A) The locations of nucleosome 1 and of relevant restriction enzyme recognition sites are shown on a diagram of the p40 promoter region. Gene specific primer A used for LM-PCR and the 3′ probe used for the Southern blot assay are also indicated. The p40 transcription start site is depicted by a bent arrow. (B) Remodeling at nucleosome 1 was monitored by using a restriction enzyme accessibility/LM-PCR assay and nuclei from untreated peritoneal macrophages (lane 1) or peritoneal macrophages treated for 4 h with LPS plus IFN-γ (lane 2), IL-10 (lane 3), or IL-10 and LPS plus IFN-γ (lane 4). Digestion with SpeI was performed in isolated nuclei to monitor accessibility within nucleosome 1. Genomic DNA was then purified and cleaved to completion with AatII for the purpose of normalization. After quantification of the SpeI and AatII cleavage products by phosphorimager analysis, the total SpeI cleavage signal was divided by the total SpeI and AatII cleavage signals, yielding the percentage of DNA cleaved by SpeI (bottom). The accuracy of these values is limited by the poor linearity of the LM-PCR assay (see text). (C) Remodeling at nucleosome 1 was monitored by using a restriction enzyme accessibility/Southern blot assay and nuclei from three independent samples of untreated peritoneal macrophages (lanes 1 to 3) or peritoneal macrophages treated for 4 h with LPS plus IFN-γ (lane 4 to 6) or IL-10 and LPS plus IFN-γ (lane 7 to 9). Purified SpeI-cleaved DNA was digested to completion with PstI and EcoRI. The digested DNA fragments were visualized by Southern blotting with the 32P-labeled 3′ probe. The PstI-EcoRI fragment corresponds to the in vitro cleavage product. The SpeI-EcoRI fragments reveal the efficiency of SpeI cleavage in the isolated nuclei. An unrelated hybridization product is also apparent (*). (D) The results shown in panel C were quantified by phosphorimager analysis. The percentage of genomic DNA cleaved by SpeI was calculated as described in the legend to panel B. The bar graph shows the mean values and standard error of the mean from the three independent samples shown in panel C.
For the initial experiments, SpeI cleavage efficiency was monitored by LM-PCR. Genomic DNA was purified from SpeI-treated nuclei and was then cleaved to completion in vitro with a second restriction enzyme, AatII. The relative cleavage efficiencies at the SpeI and AatII sites were determined by standard LM-PCR by using gene-specific primers that hybridize in the vicinity of the p40 transcription start site (Fig. 6A). The in vitro AatII cleavage products are used for the purpose of normalization. The results of this analysis confirmed that stimulation of peritoneal macrophages with LPS plus IFN-γ results in a dramatic increase in cleavage at the two SpeI sites (Fig. 6B, lanes 1 and 2; see reference 54), consistent with the hypothesis that gene activation is accompanied by nucleosome remodeling. However, stimulation in the presence of IL-10 only slightly reduced the abundance of the SpeI cleavage products (Fig. 6B, lane 4). Overall, the approximate percentage of genomic DNA cleaved by SpeI in the absence and presence of IL-10 decreased from 90 to 61% (Fig. 6B).
One potential limitation of the LM-PCR assay is that the nonlinear PCR amplification step may mask true differences in cleavage efficiency. Because of this limitation, SpeI cleavage efficiencies were quantified by direct Southern blot analysis of the genomic DNA. For this analysis, purified DNA that had been cleaved in nuclei with limiting concentrations of SpeI was digested to completion in vitro with EcoRI and PstI, prior to gel electrophoresis and Southern blotting. When a radiolabeled probe derived from the p40 coding region was used, in vitro cleavage with EcoRI and PstI yielded a 1,280-bp fragment, whereas additional cleavages at the SpeI sites yielded fragments of 1,028 or 1,053 bp (Fig. 6A). The two SpeI cleavage products are indistinguishable on the Southern blot images.
Figure 6C shows the results obtained with three independent samples derived from unstimulated peritoneal macrophages (lanes 1 to 3), three samples derived from macrophages stimulated with LPS plus IFN-γ (lanes 4 to 6), and three samples derived from macrophages stimulated with LPS plus IFN-γ in the presence of IL-10 (lanes 7 to 9). On average, 0.8% of the genomic DNA was cleaved by SpeI in the unstimulated cells, and an average of 8.8% of the genomic DNA was cleaved following stimulation with LPS plus IFN-γ (Fig. 6D). In the presence of IL-10, an average of 3.2% of the genomic DNA was cleaved, representing a decrease of 70%. These results suggest that the inhibition of p40 transcription by IL-10 is accompanied by a threefold decrease in nucleosome remodeling at the p40 promoter. It is not clear from these data whether the decrease is due to stable remodeling in a smaller percentage of the cells within a macrophage population or to the existence of a remodeled state that is less stable in all of the cells. This threefold decrease appears comparable to the decrease in C/EBPβ binding, as monitored by genomic footprinting and ChIP (Fig. 3). In both cases, the decreases appear to be significant, but the magnitudes are modest in comparison to the much more dramatic decreases in transcription and RNA polymerase II recruitment. The results therefore suggest that reduced remodeling and C/EBPβ binding are not causes but are likelier to be effects of the block in RNA polymerase II recruitment.
Identification of a distal DNase I hypersensitivity site and the effect of IL-10
The above results demonstrate that the inhibition of p40 transcription by IL-10 is accompanied by a severe reduction in the recruitment of RNA polymerase II. However, the genomic footprinting, ChIP, and restriction enzyme accessibility assays revealed only modest reductions in transcription factor binding and nucleosome remodeling at the p40 promoter. One possibility is that IL-10 blocks the activity of a distant enhancer within the p40 locus and that inhibition of this enhancer is primarily responsible for inhibiting p40 transcription and polymerase recruitment. Because distant control regions have not previously been described in the murine or human p40 loci, a systematic search was initiated by using a DNase I hypersensitivity assay. Briefly, nuclei isolated from peritoneal macrophages were incubated with increasing amounts of DNase I. Genomic DNA was purified from each sample and was cleaved to completion in vitro with appropriate restriction enzymes. Southern blots were then performed to determine whether specific regions of the p40 locus could be identified that were hypersensitive to DNase I cleavage. DNase I hypersensitivity indicates the presence of a disrupted nucleosomal array, which often corresponds to a transcriptional control region (8).
This search led to the identification of one DNase I hypersensitive site that is detectable only in stimulated peritoneal macrophages. This hypersensitive site, HSS1, is located approximately 10 kb upstream of the p40 transcription start site (+1) and was detected with genomic DNA cleaved in vitro with AseI (Fig. 7A). In genomic DNA samples prepared from peritoneal macrophages stimulated with LPS plus IFN-γ but not in samples prepared from unstimulated macrophages, a DNase I-dependent 4-kb band cleaved at HSS1 was detected below the 5.6-kb AseI-AseI band (Fig. 7B, lanes 1 to 10). Importantly, the efficiency of DNase I cleavage at HSS1 was similar in cells stimulated in the presence and absence of IL-10 (Fig. 7B, lanes 6 to 15, and C).
FIG. 7.
Effect of IL-10 on an inducible DNase I hypersensitivity site within the IL-12 p40 locus. (A) The positions of the DNase I hypersensitive site (HSS1 site), the AseI recognition sites, and the Southern blot probe are shown. The transcription start site is depicted by a bent arrow (+1). The distance from the transcription start site to the first AseI site is approximately 5.9 kb, and the distance between the two AseI sites is approximately 5.6 kb. (B) Aliquots of nuclei isolated from untreated peritoneal macrophages (lanes 1 to 5) and peritoneal macrophages treated for 4 h with LPS plus IFN-γ (lanes 6 to 10) or IL-10 and LPS plus IFN-γ (lanes 11 to 15) were incubated with different amounts of DNase I (0, 0.2, 0.4, 0.8, and 1.2 μg). Purified DNAs were digested in vitro with AseI. Cleavage products were visualized by Southern blot. The AseI-AseI fragment and the fragment extending from HSS1 to the proximal AseI site are indicated. (C) The results in panel B were quantified by phosphorimager analysis and are depicted on the bar graph as the percentage of total genomic DNA cleaved at HSS1 versus the amount of DNase I added to the sample. The results shown are representative of two independent experiments.
To analyze in greater depth the effect of IL-10 on the changes in chromatin structure that result in DNase I hypersensitivity, the restriction enzyme accessibility-Southern blot assay was used. This assay provides a means of directly comparing changes in chromatin structure at this putative enhancer with those occurring at the p40 promoter. This direct comparison is possible because an SpeI site is located near HSS1 (Fig. 8A). Because this same enzyme was used to monitor restriction enzyme accessibility at the p40 promoter, the same SpeI-cleaved genomic DNA samples used for the experiment depicted in Fig. 6C can be used to monitor accessibility at the putative enhancer, after in vitro cleavage with appropriate restriction enzymes.
FIG. 8.
Nucleosome remodeling and C/EBPβ binding at HSS1. (A) The positions of HSS1, relevant restriction enzyme recognition sites, the Southern blot probe used for the restriction enzyme accessibility assay, and PCR primer pairs used for the ChIP assay (arrowheads labeled A and B) are indicated on a diagram of the p40 5′ flanking region extending from −5.9 kb to −11.5 kb relative to the transcription start site. (B) Nucleosome remodeling in the vicinity of HSS1 was monitored with a restriction enzyme accessibility-Southern blot assay. The same SpeI-cleaved genomic DNA samples used for the experiment in Fig. 6C were digested in vitro with KpnI and SphI. Cleavage products were visualized by Southern blot. The results were quantified by phosphorimager analysis. The percentage of total genomic DNA cleaved at the SpeI site near HSS1 is shown at the bottom of each lane. (C) C/EBPβ binding to the HSS1 region was monitored by ChIP with antibodies against C/EBPβ (lanes 1 to 3) or GST (lanes 4 to 6). PCR was used to analyze the DNA fragments present in the immunoprecipitates. Primer pair A amplifies a sequence within HSS1, whereas primer pair B amplifies a sequence located 2.4 kb downstream of HSS1.
The results of this analysis revealed efficient SpeI cleavage in macrophages stimulated with LPS plus IFN-γ but not in unstimulated macrophages (Fig. 8B, lanes 1 to 5). This result confirms that the SpeI site is within the region that undergoes changes in chromatin structure during macrophage stimulation. Interestingly, a comparison of the results in Fig. 8B with those in Fig. 6C revealed that SpeI cleaves much more efficiently within HSS1 than within the p40 promoter. That is, the same samples that yielded an average cleavage efficiency of 8.8% at the promoter yielded an average cleavage efficiency of approximately 50% at the distant enhancer. The reason for this difference is not known. Most importantly, the efficiency of SpeI cleavage at HSS1 in the presence of IL-10 was reduced by an average of only 19% (Fig. 8B, lanes 3 to 8). Since these same samples showed a 70% decrease in cleavage efficiency at the p40 promoter in the IL-10-treated samples, the results increase confidence that the 70% decrease is significant and does not reflect a global change in chromatin structure in IL-10-treated macrophages.
Because the results shown above establish that in vivo binding of C/EBPβ can readily be monitored by using a ChIP assay (see above), this assay was used to determine whether C/EBPβ might bind in the vicinity of HSS1. Sheared genomic DNA precipitated with C/EBPβ antibodies was analyzed by PCR by using a primer pair that amplifies a DNA sequence near HSS1 and a second primer pair that amplifies a sequence 2.4 kb downstream of HSS1 (Fig. 8A, primer pairs A and B). The results revealed that C/EBPβ associates with one or more elements in the vicinity of HSS1 (Fig. 8C). Association was observed in cells stimulated with LPS plus IFN-γ but not in unstimulated cells (Fig. 8C). C/EBPβ binding was not detected 2.4 kb downstream of HSS1 (Fig. 8C), which provides evidence that C/EBPβ interacts independently with HSS1 and with the promoter. Importantly, IL-10 had no effect on C/EBPβ binding to HSS1 (Fig. 8C).
DISCUSSION
IL-10 is a potent anti-inflammatory cytokine that inhibits the expression of a number of inducible genes in macrophages, including several proinflammatory cytokine genes (13). Despite the biological importance of IL-10, little is known about the mechanism by which it inhibits transcription. Previous efforts to elucidate the inhibition mechanism have focused on common activators of proinflammatory cytokine genes, with inconsistent findings (see the introduction). More recently, microarray studies have been used to identify proteins that may contribute to IL-10-mediated inhibition of transcription (7, 27). As an alternative approach, we examined the effect of IL-10 on molecular events occurring at the endogenous murine IL-12 p40 locus during activation by LPS plus IFN-γ. The results revealed that IL-10 is a potent inhibitor of RNA polymerase II recruitment to the p40 promoter, which confirms that IL-10 inhibits transcription initiation and demonstrates that it strongly inhibits at least one discrete event in the activation pathway. Surprisingly, despite the strong inhibition of RNA polymerase II recruitment, IL-10 had no effect or only a modest effect on other molecular events that were examined, including nucleosome remodeling at the p40 promoter, nucleosome remodeling at a putative p40 enhancer, and C/EBPβ binding to the endogenous promoter and enhancer. In peritoneal macrophages, IL-10 also had no effect on the activation of c-Rel-containing complexes that are known to be critical for p40 induction. These results demonstrate that IL-10 does not render the chromatin associated with the p40 locus resistant to activation. Instead, a number of events occur reasonably normally, suggesting that a specific event that acts at a relatively late stage of the activation pathway blocks RNA polymerase II recruitment and transcription.
Although the results of this study will help to direct future studies of the IL-10 inhibition mechanism, it is important to note that the results are consistent with several potential mechanisms. One possibility is that IL-10 induces the expression of a DNA-binding repressor protein, which binds directly to the promoter or enhancer. Such a protein could inhibit polymerase recruitment and could lead to a modest destabilization of the remodeled state at the promoter. The DNase I genomic footprinting studies failed to reveal an IL-10-dependent footprint at the p40 promoter. However, genomic footprinting studies generally reveal only a subset of the protein-DNA interactions at a control region, due to a variety of technical limitations of the assay (8).
A second possibility is that IL-10 inhibits the posttranslational modification of one of the transcription factors analyzed in this study, i.e., NF-κB, C/EBPβ, or AP-1. Although the DNA-binding activities of these factors were unaffected by IL-10, their abilities to stimulate transcription are known to depend on posttranslational modifications (20, 22, 37). A third possibility is that IL-10 blocks the induction of an as-yet-undiscovered DNA-binding protein that is critical for p40 gene activation in macrophages. The previous studies that led to the identification of NF-κB, C/EBPβ, and AP-1 as activators of p40 transcription relied on transient-transfection experiments, leaving open the strong possibility that other proteins required for activation of the endogenous murine p40 gene remain to be identified. It also remains possible, if not likely, that distant enhancers other than the putative enhancer identified in this study will be required for induction of the endogenous p40 gene. A final possibility is that IL-10 inhibits the activity of a critical non-DNA-binding coactivator or stimulates the activity of a corepressor of the endogenous p40 gene.
Because the results of this analysis are compatible with several mechanisms of inhibition, a central question that emerges is how to proceed to elucidate the relevant mechanism. Improved microarray screens may yield attractive candidates that can be pursued. In addition, further studies of the basic mechanism by which LPS plus IFN-γ stimulates p40 transcription may lead to the discovery of novel activators or coactivators, which may be targets of the IL-10 inhibition pathway. One valuable tool would be a functional assay employing p40 control regions that recapitulates the transcriptional inhibition by IL-10 observed at the endogenous p40 gene. Previous efforts to develop a functional assay have failed, as IL-10 inhibition was not faithfully recapitulated in transient- and stable-transfection assays that used p40 promoter-reporter plasmids (1; Zhou and Smale, unpublished). In addition, transformed macrophage lines do not support IL-10 inhibition at the same efficiency observed in peritoneal macrophages (Zhou and Smale, unpublished). Even if the mechanism responsible for the modest inhibition observed in macrophage cell lines could be elucidated, its relevance to the mechanism of inhibition in primary cells would remain uncertain. Thus, a functional assay in primary cells, involving either transient or stable transfection of p40 promoter-reporter plasmids, is likely to be required. Inclusion of the putative enhancer identified in this study and of other enhancers identified in the future may also be helpful. If a functional assay can be developed, promoter or enhancer mutant studies can be performed to determine whether IL-10 inhibition requires a specific repression element. If a systematic analysis fails to identify a dedicated repression element, the results would suggest that IL-10 inhibits transcription by targeting a critical activator or coactivator.
Although some studies have suggested that IL-10 inhibits common activators of proinflammatory cytokine genes, such as NF-κB and AP-1, other studies, including this study, have failed to detect an effect on these factors (3, 10-12, 29, 38, 41, 44, 45, 47, 52). One possibility is that IL-10 inhibits transcription via different mechanisms in different species, in different macrophage populations, and in response to different stimuli. However, even in those cell populations in which IL-10 appears to diminish activation of NF-κB and AP-1, experiments are needed to rigorously test whether those proteins are the principal targets that are responsible for the downregulation of proinflammatory cytokine genes by IL-10. In this study, IL-10 resulted in a modest decrease in nucleosome remodeling and C/EBPβ binding at the p40 promoter. It is possible that these events are direct targets of the IL-10 signaling pathways and are critical events leading to the inactivation of p40 transcription. However, the modest magnitudes of the effects relative to the more dramatic inhibition of p40 transcription and RNA polymerase II recruitment lead us to favor a model in which these events are not the primary targets of IL-10 signaling. It remains possible that IL-10 signaling modestly, yet directly, inhibits a number of specific events involved in p40 activation that, when combined, result in a dramatic decrease in polymerase recruitment and transcription. However, until strong support for such a model is obtained, it seems most appropriate to focus on the possibility that IL-10 is a potent inhibitor of a specific step in the activation pathway that remains to be elucidated.
Acknowledgments
We thank Michelle Bradley, Bradley Cobb, and Vladimir Ramirez-Carrozzi for critical reading of the manuscript. We are also grateful to Valerie Schuman and Dana Russell for performing the thioglycolate injections.
A.A.N. was supported by the Ingram Research Endowment. S.T.S. is an Investigator with the Howard Hughes Medical Institute.
REFERENCE
- 1.Aste-Amezaga, M., X. Ma, A. Sartori, and G. Trinchieri. 1998. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J. Immunol. 160:5936-5944. [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Berlato, C., M. A. Cassatella, I. Kinjyo, L. Gatto, A. Yoshimura, and F. Bazzoni. 2002. Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide-induced macrophage activation. J. Immunol. 168:6404-6411. [DOI] [PubMed] [Google Scholar]
- 4.Bogdan, C., J. Paik, Y. Vodovotz, and C. Nathan. 1992. Contrasting mechanisms for suppression of macrophage cytokine release by transforming growth factor-beta and interleukin-10. J. Biol. Chem. 267:23301-23308. [PubMed] [Google Scholar]
- 5.Bradley, M. N., L. Zhou, and S. T. Smale. 2003. C/EBPβ regulation in lipopolysaccharide-stimulated macrophages. Mol. Cell. Biol. 23:4841-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, C. Y., C. A. Lagnado, M. A. Vadas, and G. J. Goodall. 1996. Differential regulation of the stability of cytokine mRNAs in lipopolysaccharide-activated blood monocytes in response to interleukin-10. J. Biol. Chem. 271:20108-20112. [DOI] [PubMed] [Google Scholar]
- 7.Cao, S., J. Liu, M. Chesi, P. L. Bergsagel, I. C. Ho, R. P. Donnelly, and X. Ma. 2002. Differential regulation of IL-12 and IL-10 gene expression in macrophages by the basic leucine zipper transcription factor c-Maf fibrosarcoma. J. Immunol. 169:5715-5725. [DOI] [PubMed] [Google Scholar]
- 8.Carey, M., and S. T. Smale. 2000. Transcriptional regulation in eukaryotes. Concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Cassatella, M. A. 1998. The neutrophil: one of the cellular targets of interleukin-10. Int. J. Clin. Lab. Res. 28:148-161. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, C. J., A. Hales, A. Hunt, and B. M. Foxwell. 1998. IL-10-mediated suppression of TNF-alpha production is independent of its ability to inhibit NF kappa B activity. Eur. J. Immunol. 28:1719-1726. [DOI] [PubMed] [Google Scholar]
- 11.Denys, A., I. A. Udalova, C. Smith, L. M. Williams, C. J. Ciesielski, J. Campbell, C. Andrews, D. Kwaitkowski, and B. M. J. Foxwell. 2002. Evidence for a dual mechanism for IL-10 suppression of TNF-α production that does not involve inhibition of p38 mitogen-activated protein kinase or NF-κB in primary human macrophages. J. Immunol. 168:4837-4845. [DOI] [PubMed] [Google Scholar]
- 12.Dokter, W. H., S. B. Koopmans, and E. Vellenga. 1996. Effects of IL-10 and IL-4 on LPS-induced transcription factors (AP-1, NF-IL6 and NF-kappa B) which are involved in IL-6 regulation. Leukemia 10:1308-1316. [PubMed] [Google Scholar]
- 13.Donnelly, R. P., H. Dickensheets, and D. S. Finbloom. 1999. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J. Interferon Cytokine Res. 19:563-573. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly, R. P., S. L. Freeman, and M. P. Hayes. 1995. Inhibition of IL-10 expression by IFN-gamma up-regulates transcription of TNF-alpha in human monocytes. J. Immunol. 155:1420-1427. [PubMed] [Google Scholar]
- 15.Ellwood, K., W. Huang, R. Johnson, and M. Carey. 1999. Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol. Cell. Biol. 19:2613-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foey, A. D., S. L. Parry, L. M. Williams, M. Feldmann, B. M. Foxwell, and F. M. Brennan. 1998. Regulation of monocyte IL-10 synthesis by endogenous IL-1 and TNF-alpha: role of the p38 and p42/44 mitogen-activated protein kinases. J. Immunol. 160:920-928. [PubMed] [Google Scholar]
- 17.Gately, M. K., L. M. Renzetti, J. Magram, A. S. Stern, L. Adorini, V. Gubler, and D. H. Presky. 1998. The interleukin-12/interleukin-12-receptor system: role in normal and pathological immune responses. Annu. Rev. Immunol. 16:495-521. [DOI] [PubMed] [Google Scholar]
- 18.Geng, Y., E. Gulbins, A. Altman, and M. Lotz. 1994. Monocyte deactivation by interleukin 10 via inhibition of tyrosine kinase activity and the Ras signaling pathway. Proc. Natl. Acad. Sci. USA. 91:8602-8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerard, C., C. Bruyns, A. Marchant, D. Abramowicz, P. Vandenabeele, A. Delvaux, W. Fiers, M. Goldman, and T. Velu. 1993. Interleukin 10 reduces the release of tumor necrosis factor and prevents lethality in experimental endotoxemia. J. Exp. Med. 177:547-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109:S81-S96. [DOI] [PubMed] [Google Scholar]
- 21.Howard, M., T. Muchamuel, S. Andrade, and S. Menon. 1993. Interleukin 10 protects mice from lethal endotoxemia. J. Exp. Med. 177:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karin, M. 1995. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270:16483-16486. [DOI] [PubMed] [Google Scholar]
- 23.Kishore, R., J. M. Tebo, M. Kolosov, and T. A. Hamilton. 1999. Cutting edge: clustered AU-rich elements are the target of IL-10-mediated mRNA destabilization in mouse macrophages. J. Immunol. 162:2457-2461. [PubMed] [Google Scholar]
- 24.Kontoyiannis, D., A. Kotlyarov, E. Carballo, L. Alexopoulou, P. J. Blackshear, M. Gaestel, R. Davis, R. Flavell, and G. Kollias. 2001. Interleukin-10 targets p38 MAPK to modulate ARE-dependent TNF mRNA translation and limit intestinal pathology. EMBO J. 20:3760-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn, R., J. Lohler, D. Rennick, K. Rajewsky, and W. Muller. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263-274. [DOI] [PubMed] [Google Scholar]
- 26.Lang, R., A. L. Pauleau, E. Parganas, Y. Takahashi, J. Mages, J. N. Ihle, R. Rutschman, and P. J. Murray. 2003. SOCS3 regulates the plasticity of gp130 signaling. Nat. Immunol. 4:546-550. [DOI] [PubMed] [Google Scholar]
- 27.Lang, R., D. Patel, J. J. Morris, R. L. Rutschman, and P. J. Murray. 2002. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169:2253-2263. [DOI] [PubMed] [Google Scholar]
- 28.Lehman, A. M., K. B. Ellwood, B. E. Middleton, and M. Carey. 1998. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J. Biol. Chem. 273:932-939. [DOI] [PubMed] [Google Scholar]
- 29.Lentsch, A. B., T. P. Shanley, V. Sarma, and P. A. Ward. 1997. In vivo suppression of NF-kappa B and preservation of I kappa B alpha by interleukin-10 and interleukin-13. J. Clin. Investig. 100:2443-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipston, K. E., and R. Baserga. 1989. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl. Acad. Sci. USA 86:9774-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, X., J. M. Chow, G. Gri, G. Carra, F. Gerosa, S. F. Wolf, R. Dzialo, and G. Trinchieri. 1996. The interleukin 12 p40 gene promoter is primed by interferon gamma in monocytic cells. J. Exp. Med. 183:147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchant, A., C. Bruyns, P. Vandenabeele, M. Ducarme, C. Gerard, A. Delvaux, D. De Groote, D. Abramowicz, T. Velu, and M. Goldman. 1994. Interleukin-10 controls interferon-gamma and tumor necrosis factor production during experimental endotoxemia. Eur. J. Immunol. 24:1167-1171. [DOI] [PubMed] [Google Scholar]
- 33.McNally, J. G., W. G. Müller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 34.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, T. L., M. G. Cleveland, P. Kulesza, J. Magram, and K. M. Murphy. 1995. Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol. Cell. Biol. 15:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plevy, S. E., J. H. Gemberling, S. Hsu, A. J. Dorner, and S. T. Smale. 1997. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 17:4572-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramji, D. P., and P. Foka. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri, B., C. J. Fisher, C. F. Farver, A. Malur, J. Drazba, M. S. Kavuru, and M. J. Thomassen. 2000. Interleukin 10 (IL-10)-mediated inhibition of inflammatory cytokine production by human alveolar macrophages. Cytokine 12:1348-1355. [DOI] [PubMed] [Google Scholar]
- 39.Reddy, S. T., R. S. Gilbert, W. Xie, S. Luner, and H. R. Herschman. 1994. TGF-beta inhibits both endotoxin-induced prostaglandin synthesis and expression of the TIS10/prostaglandin synthase 2 gene in murine macrophages. J. Leukoc. Biol. 55:192-200. [DOI] [PubMed] [Google Scholar]
- 40.Rodig, S. J., M. A. Meraz, J. M. White, P. A. Lampe, J. K. Riley, C. D. Arthur, K. L. King, K. C. Sheehan, L. Yin, D. Pennica, E. M. Johnson, Jr., and R. D. Schreiber. 1998. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell 93:373-383. [DOI] [PubMed] [Google Scholar]
- 41.Romano, M. F., A. Lamberti, A. Petrella, R. Bisogni, P. F. Tassone, S. Formisano, S. Venuta, and M. C. Turco. 1996. IL-10 inhibits nuclear factor-kappa B/Rel nuclear activity in CD3-stimulated human peripheral T lymphocytes. J. Immunol. 156:2119-2123. [PubMed] [Google Scholar]
- 42.Sanjabi, S., A. Hoffmann, H. C. Liou, D. Baltimore, and S. T. Smale. 2000. Selective requirement for c-Rel during IL-12 p40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA 97:12705-12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawadogo, M., and R. G. Roeder. 1985. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell 43:165-175. [DOI] [PubMed] [Google Scholar]
- 44.Schottelius, A. J., M. W. Mayo, R. B. Sartor, and A. S. Jr. Baldwin. 1999. Interleukin-10 signaling blocks inhibitor of kappaB kinase activity and nuclear factor kappaB DNA binding. J. Biol. Chem. 274:31868-31874. [DOI] [PubMed] [Google Scholar]
- 45.Song, S., H. Ling-Hu, K. A. Roebuck, M. F. Rabbi, R. P. Donnelly, and A. Finnegan. 1997. Interleukin-10 inhibits interferon-gamma-induced intercellular adhesion molecule-1 gene transcription in human monocytes. Blood 89:4461-4469. [PubMed] [Google Scholar]
- 46.Takeda, K., B. E. Clausen, T. Kaisho, T. Tsujimura, N. Terada, I. Forster, and S. Akira. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10:39-49. [DOI] [PubMed] [Google Scholar]
- 47.Takeshita, S., J. R. Gage, T. Kishimoto, D. L. Vredevoe, and O. Martinez-Maza. 1996. Differential regulation of IL-6 gene transcription and expression by IL-4 and IL-10 in human monocytic cell lines. J. Immunol. 156:2591-2598. [PubMed] [Google Scholar]
- 48.Trinchieri, G. 1995. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 13:251-276. [DOI] [PubMed] [Google Scholar]
- 49.Trinchieri, G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 50.Trinchieri, G., and F. Gerosa. 1996. Immunoregulation by interleukin-12. J. Leukoc. Biol. 59:505-511. [DOI] [PubMed] [Google Scholar]
- 51.Wang, P., P. Wu, M. I. Siegel, R. W. Egan, and M. M. Billah. 1994. IL-10 inhibits transcription of cytokine genes in human peripheral blood mononuclear cells. J. Immunol. 153:811-816. [PubMed] [Google Scholar]
- 52.Wang, P., P. Wu, M. I. Siegel, R. W. Egan, and M. M. Billah. 1995. Interleukin (IL)-10 inhibits nuclear factor kappa B (NF kappa B) activation in human monocytes. IL-10 and IL-4 suppress cytokine synthesis by different mechanisms. J. Biol. Chem. 270:9558-9563. [DOI] [PubMed] [Google Scholar]
- 53.Weinmann, A. S., D. M. Mitchell, S. Sanjabi, M. N. Bradley, A. Hoffmann, H. C. Liou, and S. T. Smale. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2:9-10. [DOI] [PubMed] [Google Scholar]
- 54.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 55.Yasukawa, H., M. Ohishi, H. Mori, M. Murakami, T. Chinen, D. Aki, T. Hanada, K. Takeda, S. Akira, M. Hoshijima, T. Hirano, K. R. Chien, and A. Yoshimura. 2003. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 4:551-556. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, C., K. Gagnidze, J. H. Gemberling, and S. E. Plevy. 2001. Characterization of an activation protein-1-binding site in the murine interleukin-12 p40 promoter. Demonstration of novel functional elements by a reductionist approach. J. Biol. Chem. 276:18519-18528. [DOI] [PubMed] [Google Scholar]