Abstract
The Snf1/AMP-activated kinases are involved in a wide range of stress responses in eukaryotic cells. We discovered a novel role for the Snf1 kinase in the cellular response to genotoxic stress in yeast. snf1 mutants are hypersensitive to hydroxyurea (HU), methyl-methane sulfonate, and cadmium, but they are not sensitive to several other genotoxic agents. HU inhibits ribonucleotide reductase (RNR), and deletion of SNF1 also increased the growth defects of an rnr4 ribonucleotide reductase mutant. The snf1 mutant has a functional checkpoint response to HU insofar as cells arrest division normally and derepress the transcription of RNR genes. The sensitivity of snf1 to HU or to RNR4 deletion may be due to posttranscriptional defects in RNR function or to defects in the repair of, and recovery from, stalled replication forks. The Mig3 repressor was identified as one target of Snf1 in this pathway. Genetic and biochemical analyses suggest that a weak kinase activity is sufficient to confer resistance to HU, whereas a high level of kinase activity is required for optimal growth on carbon sources other than glucose. Quantitative regulation of Snf1 kinase activity may contribute to the specificity of the effector responses that it controls.
The Snf1/AMP-activated protein kinase family is highly conserved in eukaryotes and is implicated in a variety of cellular stress responses (21, 31, 47). Interest in this kinase family has been greatly stimulated by emerging evidence of its involvement in the pathogenesis or treatment of several human diseases, including type 2 diabetes, obesity, heart disease, and cancer (31). The regulation of the Snf1/AMP-activated kinases is elaborate. The catalytic alpha subunit (encoded by the SNF1 gene in budding yeast) is only weakly active. Full activity requires phosphorylation of a threonine residue in the T loop of the kinase by distinct activating kinases (23, 41, 52) and the binding of beta (encoded by the SIP1, SIP2, and GAL83 genes in yeast) and gamma (encoded by the SNF4 gene in yeast) subunits. The beta subunits target the protein kinase to specific substrates and intracellular locations (43, 48, 56), and the binding of the gamma subunit stabilizes an active conformation of the kinase (29, 30, 34). In mammalian cells, the activation of these kinases in response to ATP depletion triggers events that tend to conserve ATP levels and modify gene expression (22, 35). In yeast cells, Snf1 is activated when cells are starved for glucose, and this activation leads to the inhibition of acetyl-coenzyme A-carboxylase and the expression of a large set of genes required for gluconeogenesis, for respiration, and for the metabolism of alternative carbon sources (4, 21). One of the best-studied targets of Snf1 is the Mig1 (multicopy inhibitor of GAL gene expression) transcriptional repressor (49). Mig1 is inactivated by phosphorylation on multiple sites within minutes when cells are glucose depleted. This phosphorylation is dependent on Snf1 and leads to the nuclear exclusion of Mig1 and the derepression of genes required for the metabolism of carbon sources other than glucose (10). Mig2 is another transcriptional repressor that is similar in sequence to Mig1. Although Mig2 is partially redundant with Mig1 in repressing transcription, its inactivation when cells are glucose depleted does not require Snf1 (39).
In addition to its roles in the cellular response to glucose depletion, the Snf1 kinase has also been implicated in the starvation response to other nutrients (55), in meiotic regulation (24, 46), in the regulation of filamentous and invasive growth (9, 33, 45), in resistance to high concentrations of sodium or lithium ions (1), in the regulation of glycogen accumulation and autophagy (58), and in yeast aging (2, 36). In this work, we show that Snf1 is also involved in the cellular response to DNA replication stress, a specific type of genotoxic stress.
Genotoxic stress refers to all situations in which the duplication or the integrity of the genome is compromised. Cells have developed extensive responses to this type of stress, including inhibition of cell cycle progression; transcriptional and posttranscriptional regulation of DNA replication, repair, and recombination pathways; and apoptosis (62). The exact nature of the responses depends on cell type, cell cycle phase, and the specific constitution and extent of the genotoxic stress. Conditions that lead to inhibition or blockage of replication fork progression have been previously called DNA replication stress (44). Hydroxyurea (HU) specifically blocks DNA synthesis by inhibiting ribonucleotide reductase (RNR) (12). RNR inhibition leads to decreases in deoxynucleoside triphosphate (dNTP) levels (32), and the subsequent stalling of the replication forks triggers a DNA checkpoint pathway composed of the Mec1 kinase (homologous to human ATR), the Rad53 kinase (homologous to human Chk2), and a third downstream kinase called Dun1 (also similar to Chk2) (17). Activation of the DNA checkpoint kinases stabilizes stalled replication forks, inhibits late-origin firing, blocks mitosis, and leads to derepression of the RNR genes (17). RNR is a heterotetrameric enzyme composed of two large subunits and two small subunits (12). The large subunits contain the active and allosteric sites of the enzyme. The small subunit contains an oxodiferric center that is required for forming a catalytically essential tyrosyl radical. HU inhibits RNR function by quenching the tyrosyl radical found in the small subunit. The yeast genome contains two genes (RNR1 and RNR3) encoding large subunits and two genes (RNR2 and RNR4) encoding small subunits (13, 57). RNR1, RNR2, and RNR4 are transcribed at the beginning of S phase and in response to DNA damage. RNR3 is significantly expressed only in response to DNA damage, but even under these conditions, it appears to make only a minor contribution to RNR activity, and rnr3Δ mutants have no obvious phenotypes (11, 13). In this paper, we describe a novel role for the Snf1 kinase in a cellular response to genotoxic stress in concert with the DNA checkpoint kinases.
MATERIALS AND METHOD
Strains and media
The yeast strains used in this work are listed in Table 1. Unless indicated otherwise, all strains were in the MCY/S288C background from Marian Carlson. Deletion and epitope-tagged strains were made by using PCR-amplified cassettes (38), except for the MEC1::HIS3 disruption, which was derived by transforming with an EcoRI mec1::HIS3 fragment from Sp309 obtained from Stéphane Marcand, and GFP-TUB1, which was derived by transforming with StuI-digested pGFP-TUB1 (51). SNF1-HA and snf1-T210A-HA plasmids were generous gifts of Martin Schmidt (41). Standard genetic methods were followed, and yeast cultures were grown in yeast extract, Bacto peptone, 2% dextrose (yeast-peptone-dextrose [YPD]), YP plus 2% galactose, or synthetic complete medium. The multicopy pFL44-MIG3 plasmid was obtained by AgeI-SalI digestion of a pFL44 plasmid containing a BamHI genomic region that included MIG3, GAL83, and CHO1.
TABLE 1.
S. cerevisiae strains used in this study
| Strain (reference) | Genotype |
|---|---|
| BY4741 (20) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BY4742 (20) | MATαhis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| CDY55 | W303 MIG3-13myc::KanMX6 |
| CDY61 | MATα his3-Δ200 leu2-3,112 ura3-52 MIG3-13myc::HIS3MX6 |
| CDY75 | MATα his3 leu2 lys2 ura3 snf1Δ10 MIG3-13myc::HIS3MX6 |
| CDY79 | MATα his3Δ200 leu2-3,112 ura3-52 mig3Δ::URA3 |
| CDY80 | MATα his3 leu2 lys2 ura3 snf1Δ10 mig3Δ::URA3 |
| CDY131 | MATα his3Δ200 leu2-3,112 ura3-52 TUB1-GFP::URA3 |
| CDY132 | MATα his3 leu2 lys2 ura3 snf1Δ10 TUB1-GFP::URA3 |
| CDY149 | MATα his3Δ200 leu2-3,112 ura3-52 sml1Δ::KanMX6 |
| CDY150 | MATα his3 leu2 lys2 ura3 snf1Δ10 sml1Δ::KanMX6 |
| CDY163 | MATα his3Δ200 leu2-3,112 ura3-52 dun1::HIS3MX6 |
| CDY195 | MATα his3Δ200 leu2-3,112 ura3-52 sml1Δ::KanMX6 mec1Δ::HIS3 |
| CDY196 | MATα his3 leu2 lys2 ura3 snf1Δ10 sml1Δ::KanMX6 mec1Δ::HIS3 |
| CDY201 | MATahis3-Δ200 leu2-3,112 ura3-52 MIG3-13myc::HIS3MX6 |
| CDY205 | Obtained from CDY195 × CDY201; sml1Δ::Kan mec1Δ::HIS3 MIG3-13myc::HIS3MX6 |
| CDY212 | Obtained from CDY196 × CDY201; sml1Δ::Kan mec1Δ::HIS3 snf1Δ10 MIG3-13myc::HIS3MX6 |
| CMY1202 | MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 ade2-101 lys2-801 ASF1-13myc::kanMX6 |
| CMY1274 | MATα his3Δ200 leu2-3,112 ura3-52 MIG3-3HA::HIS3MX6 |
| MCY2634 (56) | MATα his3Δ200 leu2-3,112 ura3-52 LYS+snf4Δ2 |
| MCY2649 (56) | MATα his3Δ200 leu2-3,112 ura3-52 |
| MCY2693 (56) | MATα his3Δ200 leu2-3,112 ura3-52 snf1-K84R |
| MCY2916 (56a) | MATα his3 leu2 lys2 ura3 snf1Δ10 |
| MCY4040 (56) | MATα his3Δ200 leu2-3,112 trp1Δ ura3-52 lys2 sip1Δ::KanMX6 sip2Δ3::LEU2 gal83Δ::TRP1 |
| W303-1a | MATacan1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
Protein extraction and immunoblotting
We found that it was important to rapidly denature proteins to ensure that the Snf1 kinase was not activated by glucose starvation during cell harvesting and protein extraction. Mutant or wild-type yeast cells were harvested by centrifugation (3,000 × g, 4°C, 1 min), and 20% trichloroacetic acid (TCA) was added to the cell pellet. Cells were then broken with acid-washed glass beads. The supernatant was retained, and the glass beads were washed in 5% TCA. The supernatant and wash were then combined, and insoluble proteins were pelleted by centrifugation. The pellet was resuspended in sample buffer (pH 8.8) and boiled for 5 min. For Western blotting, 20 to 40 μg of proteins was separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes by electroblotting. Membranes were incubated with 9E10 anti-Myc or 12CA5 anti-HA polyclonal antibodies and then with anti-mouse immunoglobulin G antibody coupled to horseradish peroxidase; finally, membranes were subjected to chemiluminescent detection. For the determination of Mig3-Myc protein stability, CDY55 cells were harvested at 20, 40, 60, or 120 min after glucose-to-galactose transfer and/or the addition of cycloheximide. Protein extracts were prepared by boiling in 8 M urea for 4 min and then breaking with glass beads. After addition of 2% SDS plus 0.1 M Tris (pH 6.8), extracts were vortexed, boiled again (2 min), and centrifuged (13,000 × g, 4°C, 10 min). Twenty micrograms of total protein was analyzed by SDS-PAGE, and Mig3-Myc quantities were estimated by immunoblotting as described above.
Dephosphorylation of protein extracts
Fifteen micrograms of whole-cell extract (prepared as described above) from CDY61 grown in the presence or absence of 200 mM HU was dephosphorylated with 10 U of calf intestinal phosphatase (CIP) for 30 min at 37°C in a 20-μl total volume of reaction buffer (50 mM Tris-HCl [pH 8.0], 0.1 mM ZnCl2, 1 mM MgCl2). The Mig3-Myc phosphorylation state was then analyzed by SDS-PAGE and immunoblotting as described above. Phosphatase inhibitors (5 mM β-glycerophosphate, 1.5 mM p-NO2-phenylphosphate, 1 mM sodium orthovanadate) were added to some reactions to ensure that modifications in Mig3-Myc mobility were due to CIP activity.
GST-Mig3 in vitro kinase assay
The MIG3 and YAF9 coding sequences (from the ATG to the last amino acid before the stop codon) were cloned into the Gateway pDEST15-GST (glutathione S-transferase) plasmid by using the procedures recommended by the manufacturer (Invitrogen). GST-Mig3 and GST-Yaf9 were purified from 500 ml of Escherichia coli BL21(λDE3)-pLysS cells containing pDEST15-GST-MIG3 (CDp29) or pDEST15-GST-YAF9 grown in Luria-Bertani medium and induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Cells were harvested, resuspended in phosphate-buffered saline (PBS) plus a complete protease inhibitor cocktail (Roche Biochemicals), and freeze-thawed, which was followed by the addition of Triton X-100 to a concentration of 0.1%, incubation with 375 U of benzonase for 15 min at 37°C to digest nucleic acids, and ultracentrifugation at 100,000 × g at 4°C for 1 h. GST-Mig3 and GST-Yaf9 were purified from the S100 by batch incubation for 3 h at 4°C with 1 ml of glutathione-Sepharose beads (Amersham Pharmacia) previously washed with PBS plus 0.1% Triton X-100 and 10% glycerol (PBS-TG) buffer. Beads were then washed once with PBS-TG, with PBS-TG plus 0.5 M NaCl, and again with PBS-TG. Bound proteins were eluted with PBS-TG plus 20 mM glutathione (pH 8.0).
Snf1-HA kinase was immunoprecipitated from yeast extracts prepared from cells broken in an Eaton press. First, 40 μl of Pan mouse immunoglobulin G (Dynal) magnetic beads were incubated with 2 μl of 12CA5 anti-HA ascites fluid. Beads were washed with PBS plus 0.1% bovine serum albumin and then with immunoprecipitation buffer (50 mM Tris [pH 7.4], 100 mM NaCl, 10 mM EDTA, 15% glycerol) plus 0.1% Triton X-100. Beads were then incubated (40°C, 3 h) with 40 μl (about 400 μg) of whole-cell extract from a snf1Δ (MCY2916) strain transformed with plasmids expressing Snf1 (5) or Snf1-3HA (41). Beads were washed with immunoprecipitation buffer and then equilibrated in kinase buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 10 μM ATP). The kinase assay was performed on the beads with 40 μl (approximately 1 μg) of purified GST-Mig3 or GST-Yaf9, 5 μl of 10× kinase buffer, and 1 μl of [γ-32P]ATP (10 μCi/μl) for 30 min at 30°C with occasional mixing. Loading buffer was added, and the incorporation of [32P]phosphate was analyzed with a PhosphorImager following SDS-4 to 12% PAGE and Coomassie blue staining. GST-Snf1 (37) was purified from E. coli BL21(λDE3)/pGEX-Snf1 cells as described above for GST-Mig3 and GST-Yaf9, and its kinase activity was assayed as was done for the Snf1-HA immunoprecipitates. Total calf thymus histones and purified calf thymus histones H3 and H4 for the kinase assays were purchased from Roche Applied Science.
Multicopy suppressors of snf1 HU sensitivity
MCY2916 cells were transformed with a yeast genomic DNA library in the multicopy vector pFL44. About 5,000 transformants were screened for growth on YPD containing 200 mM HU. The suppressing activity was checked twice under the same conditions. The SNF1, MSN4, and GIS4 genes were isolated from this screen.
Quantitative RT-PCR
RNA was prepared from cells with an RNeasy extraction kit (Qiagen). For reverse transcription-PCR (RT-PCR) analysis, reverse transcription with random hexanucleotide primers was performed with 1 μg of total RNA, 1 mM dNTPs, and 1 μl RNasin for 1.5 h at 42°C, which was followed by a 5-min heat inactivation at 95°C. For each gene, real-time quantitative PCR amplification was performed with an ABI Prism 7000 according to the manufacturer's instructions.
RESULT
Mig3 is phosphorylated and degraded in the absence of glucose
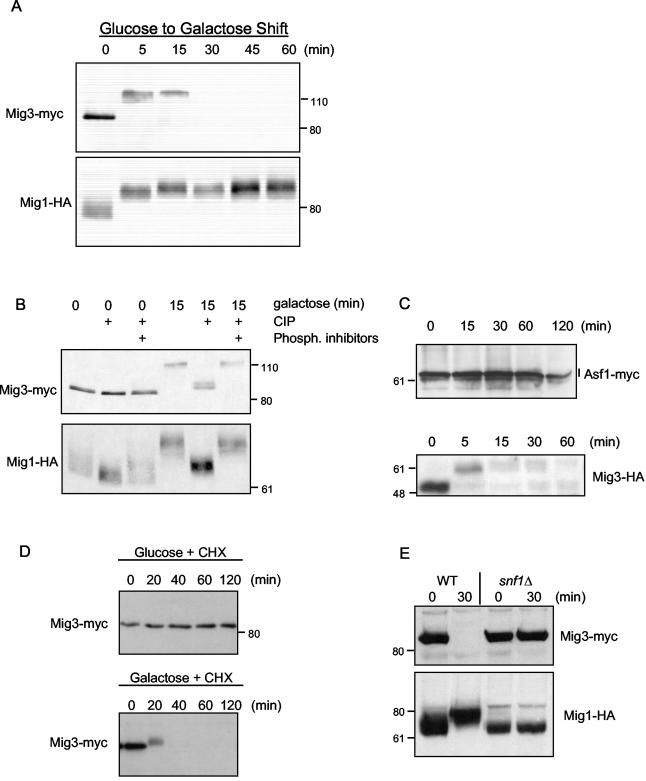
A genetic screen for multicopy plasmid suppressors of the dominant toxicity associated with the expression of a Rad53-green fluorescent protein (GFP) fusion protein was previously described (40). The expression of this fusion protein is toxic because it activates the DNA damage checkpoints in the absence of DNA damage. YER028c was one such dosage suppressor that codes for a putative transcriptional repressor with sequence similarity to the Mig1 and Mig2 repressors (39). We named this gene MIG3 because of its similarity to MIG1 and MIG2 and because it is a multicopy inhibitor of growth of the snf1 mutant in the presence of a genotoxic stress as described below. Mig1 and Mig2 are required for the repression of a large number of genes when yeast cells are grown in the presence of glucose (49). Mig3 is similar to Mig1 and Mig2 in terms of binding the same DNA sequence in vitro and of being able to recruit the Tup1-Ssn6 repressor complex. However, many glucose-repressed genes were derepressed in a mig1Δ mig2Δ double mutant, indicating that Mig3 could not repress these genes when Mig1 and Mig2 were absent (39). It was not clear why Mig3 was unable to functionally replace Mig1 and Mig2. We found that, similar to Mig1, Mig3 tagged with a 13Myc epitope (Mig3-Myc) was rapidly and extensively modified when yeast cells were transferred to galactose, as we observed with a much slower electrophoretic mobility in SDS-polyacrylamide gels (Fig. 1A). Treatment with CIP showed that, just as for Mig1, most of this modification was due to phosphorylation (Fig. 1B). However, a residual mobility shift was apparent even after phosphatase treatment, suggesting that Mig3 received additional modifications other than phosphorylation when cells were shifted from glucose to galactose. It was pointed out that the Myc epitope (GEQKLISEEDLN) contains a sequence that is similar to an Snf1/AMPK consensus phosphorylation site (ØxBxxSxxxØ, where B is a basic residue, Ø is a hydrophobic residue, and x is any amino acid) (50) and that Snf1 might be phosphorylating the 13Myc epitope tag rather than Mig3 itself. To test this possibility, we first determined whether an Asf1-13Myc fusion protein was phosphorylated when Snf1 is activated. Asf1 is a nuclear histone chaperone that has not been implicated in glucose derepression pathways. We found no evidence for induced phosphorylation of Asf1-13Myc when yeast cells were shifted from glucose to galactose (Fig. 1C). Thus, the 13Myc epitope is not necessarily a promiscuous substrate of Snf1 in vivo. We further tagged Mig3 with a triple HA epitope (Mig3-HA) and found that, similar to Mig3-Myc, it was also rapidly and highly modified when cells were shifted from glucose to galactose (Fig. 1C). Thus, Mig3 itself is highly phosphorylated when Snf1 is activated in vivo.
FIG. 1.
Mig3 is phosphorylated and degraded after shifting cells from glucose to galactose in an Snf1-dependent manner. (A) Mig3-Myc cells (CDY61) with a centromeric plasmid expressing Mig1-HA (48) were cultivated in a synthetic complete medium containing 2% glucose and lacking uracil. Cells were then washed once and resuspended in a synthetic complete medium containing 2% galactose and lacking uracil for the periods of time indicated in the figure. Proteins were extracted from TCA-precipitated cells, and Mig3-Myc and Mig1-HA levels as well as electrophoretic mobility were monitored by immunoblotting. (B) Protein extracts prepared from the 0- and 15-min galactose shifts described above were treated with or without 10 U of CIP. Phosphatase inhibitors were added to some reactions as a control showing that the mobility shifts were due to the action of the CIP. (C) Asf1-13Myc cells (CMY1202) and Mig3-HA cells (CMY1274) were shifted from glucose to galactose medium for the indicated periods of time, and proteins were extracted and analyzed by immunoblotting. (D) Mig3-Myc (CDY55) cells in YPD were split in two. One-half was treated with 50 μg of cycloheximide (CHX) per ml, whereas the other half was washed and resuspended in the same volume of YP plus 2% galactose plus 50 μg of CHX per ml; Mig3-Myc protein levels were evaluated by immunoblotting. (E) Mig3-Myc and Mig1-HA levels as well as electrophoretic mobility were monitored in wild-type (CDY61) and snf1Δ cells (CDY75) at 0 and 15 min after a glucose-to-galactose shift. The positions of molecular weight markers are shown next to the immunoblots.
Phosphorylation of Mig1 leads to its nuclear exclusion (10), but Mig3 phosphorylation was instead followed by its proteolysis (Fig. 1A, C, and D). Blocking protein synthesis with cycloheximide showed that Mig3 had a half-life of less than 20 min when cells were shifted to galactose compared to more than 120 min when cells were maintained in glucose (Fig. 1D). The hyperphosphorylation and proteolysis of Mig3 upon transfer of cells from glucose to galactose were blocked in a snf1Δ mutant (Fig. 1E). The Snf1-dependent proteolysis of Mig3 that occurred when cells were transferred to galactose suggests that Mig3 inactivation contributes to the extensive gene derepression that is observed when yeast cells are glucose depleted.
Mig3 is implicated in the cellular response to genotoxic stress
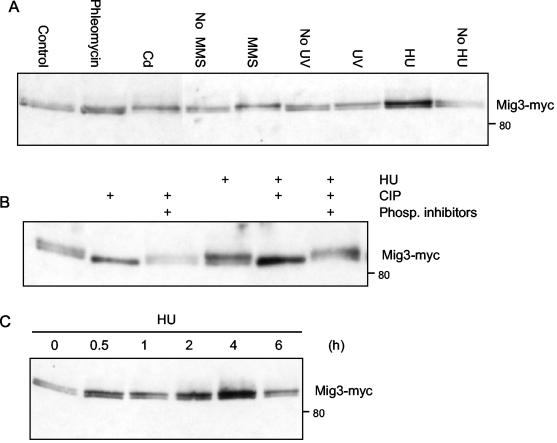
Since multicopy MIG3 could suppress the dominantly active Rad53-GFP, we tested it for a role in the cellular response to DNA damage. Although we found no DNA damage sensitivity associated with mig3Δ, mig2Δ mig3Δ, or mig1Δ mig2Δ mig3Δ mutants (data not shown), we did find that MIG3 mRNA levels increased when cells where subjected to genotoxic stress. RT-PCR experiments indicated that MIG3 mRNA was increased fivefold when cells were incubated with 200 mM HU for 2 h and increased twofold when cells were irradiated with 80 J of UV light per m2 (data not shown). Microarray analyses also indicated a three- to fourfold increase in MIG3 mRNA in cells incubated with 0.07 to 0.1% methyl methane sulfonate (MMS) for 1 h and a 4.6-fold increase in cells incubated with 0.2 mM 1,3bis(2-chloroethyl)-1-nitrosourea for 1 h (27, 42). Furthermore, a slight but reproducible shift in the electrophoretic migration of Mig3-Myc was observed when cells were treated with a range of genotoxic agents including MMS, HU, UV, phleomycin, and cadmium (Fig. 2A). A mobility shift was also seen for Mig3-HA after treating cells with HU or Cd (data not shown). In untreated cells, Mig3-Myc migrated as a closely spaced doublet of bands. The quantity of the slower-migrating form was increased after genotoxic stress. These bands were converted into a single, faster-migrating band when protein extracts were treated with CIP (Fig. 2B). These results showed that Mig3-Myc was phosphorylated in unstressed cells. After treatment with genotoxic agents, the amount of the slowly migrating, more highly phosphorylated form of Mig3-Myc was increased to various extents. The shift was most obvious with cadmium and MMS and was less so for HU, phleomycin, and UV treatment. An increase in phospho-Mig3 was visible after 30 min of treatment with 200 mM HU and increased slightly during 6 h of treatment (Fig. 2C). The half-life of the Mig3-Myc protein was unchanged after the limited phosphorylation induced by genotoxic stress when cells were grown in glucose (data not shown), a finding which contrasts with the decreased stability of the protein observed after its hyperphosphorylation when cells were transferred from glucose to galactose (Fig. 1A and C). The increase of MIG3 mRNA and the induced phosphorylation of the Mig3 protein in the presence of DNA damage suggested that it is part of a DNA damage response pathway.
FIG. 2.
Mig3 is phosphorylated during genotoxic stress. CDY61 cells expressing Mig3-Myc were grown in YPD (lanes labeled Control, No MMS, No UV, and No HU) or treated with 5 μg of phleomycin per ml for 30 min (Phleomycin), 200 μM cadmium for 1 h (Cd), 200 mM HU for 2 h, or 1% MMS for 30 min followed by neutralization with an excess of sodium thiosulfate (MMS). For UV irradiation (UV), cells were concentrated and spread on the surface of a YPD plate that was then irradiated with 80 J of 254-nm light per m2 in a Stratagene cross-linker. Cells were resuspended for an additional hour in YPD before TCA protein extraction and immunoblotting. (B) Protein extracts were prepared from cells grown in YPD or in YPD containing 200 mM HU for 2 h. The extracts were incubated with or without 10 U of CIP. Phosphatase inhibitors were added to some reactions as a control showing that the mobility shifts were due to the action of the CIP. (C) Time course of Mig3 phosphorylation after the addition of 200 mM HU to CDY61 cells growing exponentially in YPD. The position of an 80-kDa molecular mass marker is shown next to the immunoblots.
Mec1 checkpoint pathway and Snf1 kinase both contribute to DNA damage-induced phosphorylation of Mig3
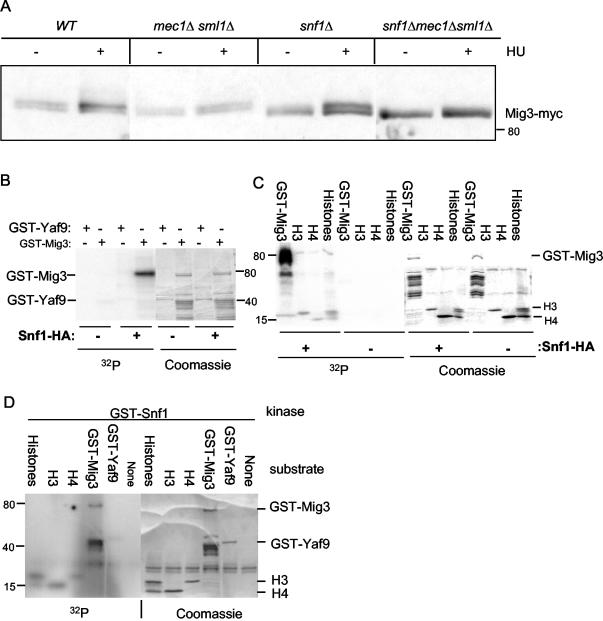
The principal DNA damage response pathway in budding yeast is activated by the Mec1 kinase. Mec1 has an essential function in activating ribonucleotide reductase by promoting the phosphorylation and inactivation of the Sml1 protein, an inhibitor of RNR (60, 61). A deletion of the MEC1 gene is viable in a sml1Δ mutant. Surprisingly, Mig3-Myc phosphorylation was still induced by HU in a mec1Δ sml1Δ mutant, although the degree of basal and induced phosphorylation appeared slighter than that in the wild type (Fig. 3A). Since Mig3 was phosphorylated in a Snf1-dependent fashion when cells were transferred from glucose to galactose (Fig. 1), we examined whether phosphorylation of Mig3 induced by HU in glucose-grown cells was also dependent on Snf1. As for the mec1Δ sml1Δ mutant, phosphorylation of Mig3-Myc was still induced after treating the snf1Δ mutant with HU, although the degree of basal and induced phosphorylation seemed slightly less than that in the wild type (Fig. 3A). We then examined Mig3-Myc phosphorylation in a snf1Δ mec1Δ sml1Δ mutant. In this double-kinase mutant, basal and HU-induced phosphorylation of Mig3-Myc were greatly inhibited (Fig. 3A). We conclude that Snf1 and the Mec1 checkpoint pathway contribute independently and in parallel to phosphorylation of Mig3.
FIG. 3.
Mig3 phosphorylation in vivo after genotoxic stress is dependent on both Mec1 and Snf1 kinases, and Mig3 is an in vitro substrate for the Snf1 kinase. (A) Immunoblot analysis of Mig3 in vivo phosphorylation before or after a 2-h treatment with 200 mM HU was done with the wild-type (WT) (CDY61), mec1Δ sml1Δ (CDY205), snf1Δ (CDY75), or snf1Δ mec1Δ sml1Δ (CDY212) strains expressing Mig3-Myc. Panels B through D show that Snf1 phosphorylates GST-Mig3 in vitro. (B and C) The Snf1-HA kinase was immunoprecipitated from protein extracts prepared from SNF1-HA yeast cells. A control immunoprecipitation was done on protein extracts from the wild-type untagged strain. The left-hand panels show PhosphorImager scans of SDS-polyacrylamide gels showing [32P]phosphate incorporation into GST-Mig3 and histones H3 and H4 but not into a control GST-Yaf9 fusion protein. The right-hand panels show the Coomassie blue-stained gels. (D) The GST-Snf1 kinase purified from E. coli weakly phosphorylates GST-Mig3 and the histones H3 and H4 in vitro but not the GST-Yaf9 protein. The left-hand panel shows a PhosphorImager scan of the kinase reaction products after SDS-PAGE, and the right-hand panel shows the Coomassie blue staining of the same gel.
We also tested whether Snf1 could phosphorylate Mig3 in vitro. Snf1-HA immunoprecipitated from yeast cells readily phosphorylated a GST-Mig3 fusion protein purified from E. coli but did not phosphorylate a GST-Yaf9 control protein (Fig. 3B). No phosphorylation was observed for anti-HA immunoprecipitates from an untagged control strain. Snf1 was shown to phosphorylate serine-10 of histone H3 (37), although this serine does not correspond to a typical consensus site for Snf1 phosphorylation. We compared the abilities of Snf1-HA immunoprecipitates to phosphorylate GST-Mig3 and calf thymus histones in vitro. Anti-HA immunoprecipitates from Snf1-HA cell extracts, but not those from control extracts, phosphorylated GST-Mig3 and histones H3 and H4 (Fig. 3C). Both histones H3 and H4 contain some potential Snf1/AMPK consensus phosphorylation sites that may contribute to their phosphorylation in vitro but whose physiological significance is unknown. Notably, however, GST-Mig3 was phosphorylated to a much greater extent than were the histones.
Snf1 immunoprecipitated from yeast is associated with activating β and γ subunits and may conceivably contain contaminating kinase activities that coprecipitate with Snf1. We used a GST-Snf1 protein purified from E. coli to further test its ability to phosphorylate Mig3 in vitro. The GST-Snf1 kinase catalytic subunit purified from E. coli has a weak but detectable protein kinase activity in the absence of the activating β and γ subunits (37). We found that GST-Snf1 could weakly phosphorylate GST-Mig3 and histones H3 and H4 but that it could not phosphorylate a GST-Yaf9 control protein (Fig. 3D). In keeping with its efficient phosphorylation by the Snf1 kinase complex, the Mig3 sequence contains several serines in a context that is favorable for phosphorylation by the Snf1/AMP-activated kinases (50). Mig3 was thus an excellent Snf1 substrate in vitro, which suggests that at least part of the Mig3 phosphorylation observed in vivo is due to direct phosphorylation by this kinase.
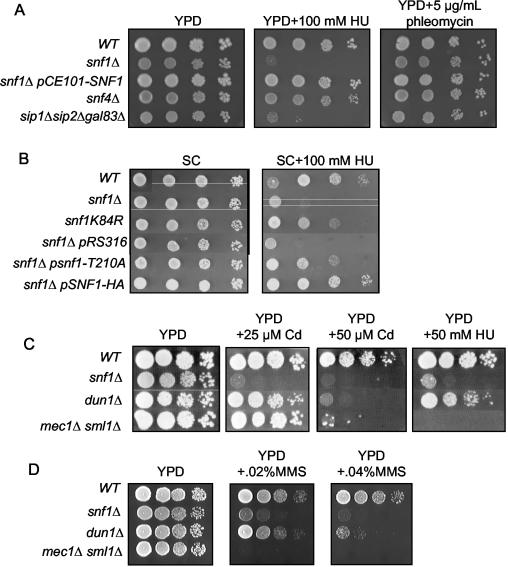
snf1 mutants are sensitive to HU, MMS, and cadmium
Since Snf1 contributed to Mig3 phosphorylation in response to DNA-damaging agents, we tested whether snf1 mutants were sensitive to genotoxic stress. snf1 mutants were sensitive to treatment with HU, MMS, and cadmium (Fig. 4) but not to UV or gamma irradiation or to treatment with hydrogen peroxide, camptothecin, or phleomycin (Fig. 4 and data not shown). HU is a specific inhibitor of RNR (12), MMS is a highly reactive alkylating agent that attacks both nucleic acids and proteins (28), and cadmium is known to bind the reactive thiols of proteins such as glutathione (16). Interestingly, treatment of yeast cells with low levels of MMS preferentially inhibits their progression through S phase (53), suggesting that snf1 sensitivity to these agents may involve DNA replication stress. Thiol binding by cadmium might directly or indirectly inhibit RNR activity and DNA replication. However, we did not observe a specific accumulation of wild-type or snf1 mutant cells in S phase when they were treated with a wide range of cadmium concentrations (data not shown). Furthermore, the mec1Δ sml1Δ mutant was much more sensitive than the snf1Δ mutant to HU and MMS treatment, whereas the opposite result was found for cadmium (Fig. 4C and D). These findings suggest that the cadmium sensitivity of the snf1 mutant cannot be accounted for solely by the genotoxic effects of cadmium. The mec1 mutant is highly sensitive to DNA replication stress because it is unable to stabilize blocked replication forks (54), whereas the dun1 mutant is thought to be moderately sensitive to such stress mainly because of defects in gene expression (18) and in the posttranscriptional activation of RNR by inactivation of the Sml1 repressor (61). Interestingly, although the snf1 mutant is less sensitive to HU and MMS than is the mec1 mutant, it is more sensitive than the dun1 mutant (Fig. 4C and D). We decided to focus our further study on the response of the snf1 mutant to HU, as it is the least pleiotropic of the three toxic molecules.
FIG. 4.
snf1Δ mutants are hypersensitive to HU, MMS, and cadmium. Wild-type (WT) (MCY2649), snf1Δ (MCY2916) transformed when indicated with pCE101-SNF1 expressing the WT Snf1 (5), pSNF1-HA expressing HA-tagged Snf1 (41), psnf1-T210A expressing the Snf1-T210A mutant (41), or the pRS316 empty vector, snf4Δ (MCY2634), sip1Δ sip2Δ gal83Δ (MCY4040), and snf1-K84R (MCY2693) strains were diluted to an optical density at 600 nm (OD600) of 0.2, and serial 10-fold dilutions were spotted on YPD plates (A) with HU or phleomycin at the indicated concentrations or on synthetic complete glucose medium (SC) plates (B) with (three upper lanes) or without (three lower lanes) uracil and containing HU at the indicated concentrations. (C and D) Serial 10-fold dilutions of the wild-type (MCY2649), snf1Δ (MCY2916), dun1Δ (CDY163), and mec1Δ sml1Δ (CDY196) strains were spotted on YPD plates containing the indicated concentrations of cadmium, HU, and MMS.
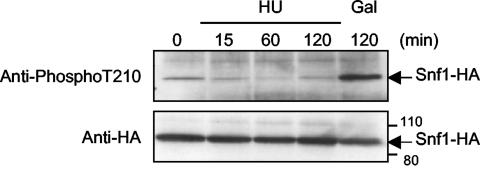
Members of the Snf1/AMP-activated kinase family require association with β and γ subunits for full kinase activity. Budding yeast contains three β-type proteins. The presence of any one of these β subunits conferred normal HU resistance to yeast cells (data not shown), but a triple-deletion mutant of all three β-subunit genes (i.e., a sip1Δ sip2Δ gal83Δ mutant) was as sensitive to HU, MMS, and cadmium as was the snf1Δ mutant (Fig. 4A and data not shown). Surprisingly, a mutant containing a deletion of the sole γ-subunit gene in yeast (snf4Δ) was much less sensitive to HU than was the snf1Δ mutant (Fig. 4A). Furthermore, the snf1-K84R and snf1-T210A mutants have greatly reduced kinase activity and nevertheless showed significant resistance to HU (Fig. 4B). K84R mutates a lysine implicated in ATP binding (5, 15), and T210A mutates a threonine residue in the T-loop of Snf1 that must be phosphorylated by upstream kinases to fully activate Snf1 (23, 41, 52). These results suggest that low-level Snf1 kinase activity suffices to confer significant HU resistance to yeast cells. Consistent with this notion, we found no evidence that Snf1 was activated by T210 phosphorylation when cells were treated with HU (Fig. 5), whereas such phosphorylation was readily apparent when cells were transferred from glucose to galactose as reported previously (41).
FIG. 5.
Snf1 is activated by phosphorylation on threonine-210 after transfer from glucose to galactose medium but not after HU treatment. snf1Δ cells transformed with a centromeric plasmid expressing Snf1-HA (41) were grown to log phase in synthetic complete-glucose medium lacking uracil. HU was added to a concentration of 200 mM to one aliquot of cells, whereas a second aliquot of cells was harvested, washed, and resuspended in synthetic complete-galactose (Gal) medium lacking uracil. Cells were harvested at the indicated times after medium changes, Snf1-HA was immunoprecipitated from protein extracts, and Snf1 activation was monitored by immunoblotting using antibodies directed against phospho-T210 (41). The efficiency of Snf1-HA immunoprecipitation was monitored by immunoblotting using anti-HA antibodies (lower panel).
snf1 mutants do not show S-phase checkpoint defects
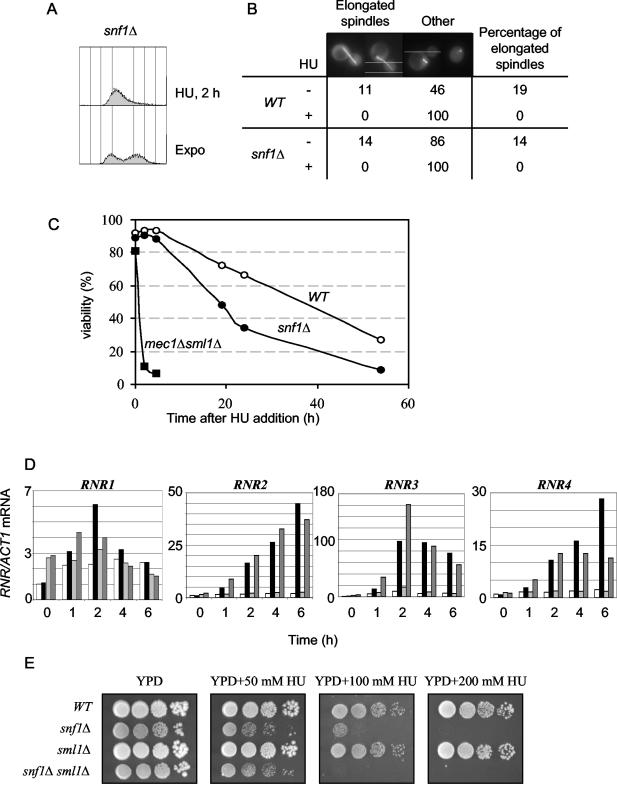
DNA synthesis was inhibited in the snf1 mutant treated with HU (Fig. 6A), and the mutant showed a normal checkpoint-mediated arrest of spindle elongation as determined by the visualization of GFP-Tub1 in these cells (Fig. 6B). These results showed that snf1 mutants arrested cell cycle progression correctly in the presence of HU. The snf1 mutant also lost viability very gradually in the presence of HU compared to the mec1Δ sml1Δ mutant (Fig. 6C), a finding which suggests that snf1 is not defective in stabilizing stalled replication forks. Activation of the DNA damage checkpoints also derepresses to various extents the four genes encoding RNR subunits in yeast and leads to the inactivation of Sml1, an inhibitor of Rnr1 (7, 25, 61). RT-PCR analyses showed that derepression of RNR1, RNR2, RNR3, and RNR4 occurred normally in the snf1 mutant (Fig. 6D). Furthermore, deletion of the SML1 gene did not suppress the HU sensitivity of the snf1 mutant (Fig. 6E), suggesting that Sml1 was inactivated normally in the snf1 mutant in the presence of HU.
FIG. 6.
The snf1 mutant does not show S-phase checkpoint defects during HU treatment. (A) Flow cytometry showed that the snf1Δ mutant accumulated with a 1 N DNA content after treating exponentially growing cells (Expo) with 200 mM HU for 2 h. (B) Visualization of mitotic spindles in wild-type (WT) (CDY131) and snf1Δ (CDY132) cells expressing GFP-Tub1 (51) before or after a 2-h treatment with 200 mM HU. As for the WT, the snf1Δ mutant showed no spindle elongation after HU treatment. (C) Exponentially growing WT (MCY2649), snf1Δ (MCY2916), or mec1Δ sml1Δ (CDY195) cells in YPD were treated with 200 mM HU, and cell viability was determined at the indicated time points. The viability of WT (MCY2649) and snf1Δ (MCY2916) cells cultivated in YPD without HU for 24 h was 96 and 89%, respectively. (D) The snf1 mutant derepresses RNR gene expression in the presence of HU. The relative levels of RNR1, RNR2, RNR3, and RNR4 to ACT1 mRNAs were determined by real-time RT-PCR analyses for the WT and snf1Δ mutant growing in YPD with and without HU for the indicated periods of time. White bar is WT in YPD, black bar is WT in YPD plus 200 mM HU, the light grey bar is snf1Δ in YPD, and the dark grey bar is snf1Δ in YPD plus 200 mM HU. (E) Serial 10-fold dilutions of WT (MCY2649), snf1Δ (MCY2916), sml1Δ (CDY149), and snf1Δ sml1Δ (CDY150) strains were spotted onto YPD or YPD plus HU at the indicated concentrations.
Mig3 is a biological target of Snf1 in the response of yeast cells to HU
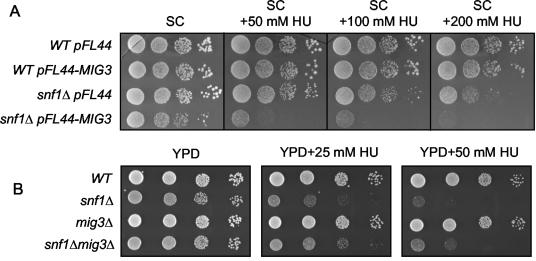
Overexpression of MIG3 in a wild-type strain had no obvious phenotype, but its overexpression in the snf1 mutant greatly sensitized it to treatment with HU (Fig. 7A). This result is consistent with a model in which Mig3 is a transcriptional repressor whose action is antagonized by Snf1. However, deleting the MIG3 gene in an snf1 mutant only slightly alleviated the HU sensitivity of snf1 (Fig. 7B). Thus, inactivation of Mig3 cannot be the sole function of Snf1 in conferring HU resistance.
FIG. 7.
Genetic analysis suggests that Mig3 is a target of the Snf1 kinase in the cellular response to HU. (A) Log-phase culture of wild-type (WT) (MCY2649) and snf1Δ (MCY2916) cells transformed with either the vector pFL44 or pFL44-MIG3 (CDp20) were diluted to an OD600 of 0.2, and serial 10-fold dilutions were spotted and grown for 5 days on synthetic complete plates without uracil and containing HU at the indicated concentrations. (B) Log-phase culture of WT (MCY2649), snf1Δ (MCY2916), mig3Δ (CDY79), and snf1Δ mig3Δ (CDY80) were diluted to an OD600 of 0.2, and serial 10-fold dilutions were spotted and grown for 2 days on YPD plates containing HU at the indicated concentrations.
Deletion of SNF1 enhances the growth defects and the HU sensitivity of a rnr4Δ mutant
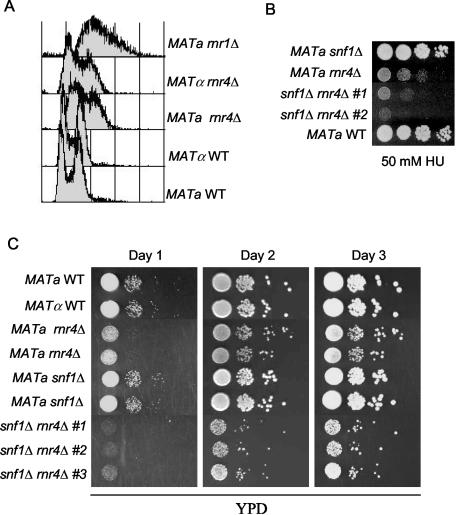
HU inhibits RNR and thereby leads to the reduction of dNTP pools and the stalling of replication forks. The HU hypersensitivity of the snf1 mutant might be due to defects at the level of HU detoxification or to defects in various aspects of the cellular response to RNR inhibition other than cell cycle arrest and RNR derepression. The yeast genome contains two genes (RNR1 and RNR3) encoding large subunits and two genes (RNR2 and RNR4) encoding small subunits (13, 57). However, only the Rnr2 subunit is competent for forming a tyrosyl radical, and it is essential for cell viability. The Rnr4 subunit is thought to facilitate the folding or the function of Rnr2 (6, 19). Deletion of RNR1 or RNR4 in the BY background (20) leads to a slow-growth phenotype with an accumulation of cells in S phase (Fig. 8A and C and unpublished data). We crossed a snf1Δ mutant to rnr1Δ and rnr4Δ mutants to test for genetic interactions. Deletion of SNF1 increased the growth defects and HU sensitivity of rnr4Δ mutants (Fig. 8B and C). This result suggests that the hypersensitivity of snf1Δ to HU cannot be explained solely by a defect in drug detoxification. In contrast to rnr4Δ, we found that snf1Δ rnr1Δ double mutants grew at the same rate and showed the same HU sensitivity as did rnr1Δ single mutants (data not shown). Thus, rnr1Δ is epistatic to snf1Δ, whereas snf1Δ has an additive effect on rnr4Δ phenotypes. These genetic interactions suggest that Snf1 may act at the level of Rnr1, although other explanations cannot be ruled out.
FIG. 8.
Deletion of SNF1 increases the growth defects and HU sensitivity of the rnr4Δ mutant. (A) Flow cytometry shows that rnr1Δ and rnr4Δ mutants have higher levels of cells in S phase than do the wild-type (WT) controls. Log-phase cells were prepared for flow cytometry as described previously (26). (B) snf1Δ rnr4Δ double mutants are more sensitive to HU than is either single-mutant parent strain. Two independent double-mutant segregants of a meiotic cross are shown. Serial 10-fold dilutions of the indicated strains were spotted onto a YPD plate containing 50 mM HU and incubated at 30°C. Note that the snf1Δ mutant in the BY strain background shown here is less sensitive to HU inhibition than is the same mutant in the MCY strain background used elsewhere in this paper. (C) snf1Δ rnr4Δ double mutants grow more slowly than either single-mutant parent on YPD plates at 30°C incubated for the indicated periods of time at 30°C. Three different snfΔ rnr4Δ double mutants are shown. The WT MATa strain is BY4741, and the WT MATα strain is BY4742. The rnr1Δ, rnr4Δ, and snf1Δ mutant versions of these strains were obtained from the BY yeast gene deletion collection (20).
DISCUSSION
Snf1 kinase is required for resistance to HU, MMS, and cadmium
We found that snf1 mutants are hypersensitive to HU, MMS, and cadmium but not to other genotoxic stresses such as UV and gamma irradiation or treatment with hydrogen peroxide, camptothecin, and phleomycin. HU is a small-molecule inhibitor of RNR that functions by diffusing into the center of the R2 subunit and quenching the tyrosyl radical that is essential for the catalytic activity of the enzyme (12). RNR inhibition then leads to a reduction of dNTP pools and the stalling of replication forks (44). MMS and cadmium are expected to have a much more pleiotropic effect on cells than that of HU. MMS is an alkylating agent that reacts broadly with nucleophiles, including DNA bases and the reactive cysteines of many proteins. Cadmium toxicity is also associated with its binding to reactive cysteines (16). At low concentrations, MMS leads to a selective inhibition of S-phase progression (53). This effect has been suggested to be due to the alkylation of bases that would prevent conventional DNA polymerases from extending DNA chains. It is also possible that MMS directly or indirectly inhibits RNR activity by alkylating reactive cysteines of RNR and of cellular reducing agents such as glutathione, thioredoxin, and glutaredoxins that are required for RNR activity (12). However, dNTP levels clearly increased when yeast cells were treated with 0.01% MMS (8), thereby demonstrating that RNR remains active when yeast cells are treated with this MMS concentration. It remains possible that RNR is inhibited at higher MMS concentrations or that the snf1 mutant is sensitive to the presence of a large number of stalled replication forks. Phenotypic analyses showed that the snf1 mutant arrests cell division and derepresses the RNR genes normally in response to HU. Its HU hypersensitivity is presumably due to some other aspect of the cellular response to RNR inhibition. Since snf1Δ rnr4Δ double mutants grow more poorly than does either single mutant, it is likely that Snf1 is not required just for drug detoxification. Preliminary microarray analyses indicate that there are some gene expression defects in the snf1 mutant treated with HU (data not shown), but these defects do not explain in an obvious manner its sensitivity to HU. Further phenotypic characterization of snf1 is required to pinpoint the reasons for its sensitivity to RNR inhibition.
Cadmium reacts with and depletes cellular glutathione (16). It could thus inhibit RNR activity indirectly by depleting physiological reducing agents required for RNR activity, and it could also potentially bind directly to reactive cysteines of RNR and inhibit its activity. However, the fact that the snf1Δ mutant is much more sensitive to cadmium than is the mec1Δ mutant (whereas the opposite is true for HU) strongly suggests that the cadmium sensitivity of snf1Δ cannot be due to RNR inhibition. The basis of the cadmium sensitivity of snf1Δ thus remains to be determined.
Mig3 repressor is a novel target of Snf1 and the DNA checkpoint pathway
Overexpression of the Mig3 repressor protein sensitizes the snf1 mutant to HU, and conversely, the deletion of MIG3 slightly increases the HU resistance of snf1. Mig3 is also phosphorylated in a Snf1- and Mec1-dependent fashion when cells are subjected to a genotoxic stress. Mig3 is likely to be a direct target of Snf1 because it is an excellent substrate for Snf1 in vitro, but we do not know if Mec1-dependent phosphorylation is due to Mec1 itself or a downstream kinase such as Rad53 whose activity depends on Mec1. We have ruled out a role for the Dun1 kinase because Mig3 phosphorylation in response to HU was still observed in a dun1Δ snf1Δ double mutant (unpublished data). Finally, overexpression of MIG3 suppresses the toxicity of a constitutively active Rad53-GFP fusion protein (40). This suppression does not seem to occur by Mig3 binding the toxic fusion protein because we have not observed coimmunoprecipitation of Rad53 and Mig3 (unpublished data). These results suggest the following model. In agreement with previous work (39), we suggest that Mig3 is a transcriptional repressor of some unidentified target genes when cells are grown with glucose as the carbon source. In the presence of a genotoxic stress, Mig3 is phosphorylated and inhibited in a manner dependent on the Snf1 and Mec1 kinases. The resulting gene derepression would promote cell survival. The increase in MIG3 mRNA levels observed after genotoxic stress may facilitate a return to the basal repressed state after the repair of DNA damage, as has been proposed previously for the Crt1 repressor (25). The relevant target genes regulated by Mig3 remain to be identified. Further work is also required to identify additional downstream targets of Snf1 in this pathway. We sought such targets by isolating multicopy plasmid suppressors of the HU sensitivity of the snf1 mutant. MSN4 and GIS4 were identified as two dosage suppressors that partially alleviated the HU sensitivity of the snf1 mutant (unpublished data). Both of these genes were previously shown to be partial suppressors of the poor growth of snf1 mutants in the absence of glucose and were thus not considered to be specific downstream effectors of Snf1 in response to HU (3, 14).
Unusual aspects of Snf1 kinase function in cellular tolerance to HU
The Snf1 kinase is weakly active when cells are grown in the presence of glucose. Under conditions of glucose starvation, the kinase is rapidly activated by phosphorylation on Thr-210 by distinct upstream activating kinases (23, 41, 52, 59). Snf1 then phosphorylates key proteins acting in several pathways that enable yeast cells to grow in the absence of glucose. Snf1 is also required for growth in the presence of high concentrations of sodium or lithium ions (1). Activating phosphorylation of Snf1 was also observed when yeast cells were treated with high concentrations of sodium in the presence of glucose (41). However, we were unable to observe activating phosphorylation of Snf1 when cells were treated with HU. Furthermore, a snf4 mutant from which the activating γ subunit of the Snf1 kinase was deleted retained good resistance to HU even though it is unable to grow in a medium without glucose (5, 34). Likewise, the snf1-K84R and snf1-T210A kinase mutants have greatly reduced kinase activities and are unable to grow without glucose (5, 15, 41), yet they showed some resistance to HU. One explanation of these results would be to imagine that Snf1 kinase activity is not at all required for its function in the cellular tolerance to HU. However, although the snf1-K84R and snf1-T210A mutants had significant residual function, they nevertheless clearly did not work as well as the wild-type protein in conferring resistance to HU. It thus seems more likely to us that a low level of Snf1 kinase activity in association with one of the three beta subunits may be necessary and sufficient to render cells resistant to HU. Quantitative regulation of Snf1- or AMP-dependent kinase activity may contribute to the selectivity of the pathways that they control under different physiological conditions. Mig3 phosphorylation may be an example of such quantitative regulation. Snf1 modestly contributes to Mig3 phosphorylation in the presence of genotoxic stress when cells are grown in glucose, a condition in which the Snf1 kinase is weakly active. This phosphorylation does not induce Mig3 turnover, but genetic data suggest that it may inactivate Mig3 repression of genes involved in HU resistance. In contrast, when cells are shifted from glucose to galactose and when Snf1 is highly active, Mig3 is extensively phosphorylated and rapidly proteolyzed. This complete inactivation would ensure that Mig3 does not interfere with the derepression of genes when cells are starved for glucose.
Acknowledgments
We thank Marian Carlson for providing a large number of yeast strains and plasmids; Martin Schmidt for the generous gift of anti-Snf1-phospho-Thr-210 antibodies and plasmids; Shelley Berger, Stéphane Marcand, and Aaron Straight for plasmids; and Marie-Claude Marsolier, Cyril Buhler, and Lea Jessup for a critical review of the manuscript.
C.D. was supported by an Allocation Couplée de l'Ecole Normale Supérieure de Paris. This work was also supported by grants from the Association pour la Recherche sur le Cancer (ARC grant number 4470) and Electricité de France.
REFERENCE
- 1.Alepuz, P. M., K. W. Cunningham, and F. Estruch. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 26:91-98. [DOI] [PubMed] [Google Scholar]
- 2.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 3.Balciunas, D., and H. Ronne. 1999. Yeast genes GIS1-4: multicopy suppressors of the Gal phenotype of snf1 mig1 srb8/10/11 cells. Mol. Gen. Genet. 262:589-599. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 5.Celenza, J. L., and M. Carlson. 1989. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell. Biol. 9:5034-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chabes, A., V. Domkin, G. Larsson, A. Liu, A. Graslund, S. Wijmenga, and L. Thelander. 2000. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. USA 97:2474-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabes, A., V. Domkin, and L. Thelander. 1999. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem. 274:36679-36683. [DOI] [PubMed] [Google Scholar]
- 8.Chabes, A., B. Georgieva, V. Domkin, X. Zhao, R. Rothstein, and L. Thelander. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112:391-401. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8:1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domkin, V., L. Thelander, and A. Chabes. 2002. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 277:18574-18578. [DOI] [PubMed] [Google Scholar]
- 12.Eklund, H., U. Uhlin, M. Farnegardh, D. T. Logan, and P. Nordlund. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177-268. [DOI] [PubMed] [Google Scholar]
- 13.Elledge, S. J., and R. W. Davis. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4:740-751. [DOI] [PubMed] [Google Scholar]
- 14.Estruch, F., and M. Carlson. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauchon, M., G. Lagniel, J. C. Aude, L. Lombardia, P. Soularue, C. Petat, G. Marguerie, A. Sentenac, M. Werner, and J. Labarre. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9:713-723. [DOI] [PubMed] [Google Scholar]
- 17.Foiani, M., A. Pellicioli, M. Lopes, C. Lucca, M. Ferrari, G. Liberi, M. Muzi Falconi, and P. Plevani. 2000. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res. 451:187-196. [DOI] [PubMed] [Google Scholar]
- 18.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge, J., D. L. Perlstein, H. H. Nguyen, G. Bar, R. G. Griffin, and J. Stubbe. 2001. Why multiple small subunits (Y2 and Y4) for yeast ribonucleotide reductase? Toward understanding the role of Y4. Proc. Natl. Acad. Sci. USA 98:10067-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 21.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 22.Hardie, D. G., and S. A. Hawley. 2001. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23:1112-1119. [DOI] [PubMed] [Google Scholar]
- 23.Hong, S. P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honigberg, S. M., and R. H. Lee. 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 26.Hutter, K. J., and H. E. Eipel. 1979. Microbial determinations by flow cytometry. J. Gen. Microbiol. 113:369-375. [DOI] [PubMed] [Google Scholar]
- 27.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20:8157-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jelinsky, S. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105-3115. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp, B. E., D. Stapleton, D. J. Campbell, Z. P. Chen, S. Murthy, M. Walter, A. Gupta, J. J. Adams, F. Katsis, B. Van Denderen, I. G. Jennings, T. Iseli, B. J. Michell, and L. A. Witters. 2003. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 31:162-168. [DOI] [PubMed] [Google Scholar]
- 32.Koc, A., L. J. Wheeler, C. K. Mathews, and G. F. Merrill. 2004. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 279:223-230. [DOI] [PubMed] [Google Scholar]
- 33.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leech, A., N. Nath, R. R. McCartney, and M. C. Schmidt. 2003. Isolation of mutations in the catalytic domain of the Snf1 kinase that render its activity independent of the Snf4 subunit. Eukaryot. Cell 2:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leff, T. 2003. AMP-activated protein kinase regulates gene expression by direct phosphorylation of nuclear proteins. Biochem. Soc. Trans. 31:224-227. [DOI] [PubMed] [Google Scholar]
- 36.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J. Biol. Chem. 278:13390-13397. [DOI] [PubMed] [Google Scholar]
- 37.Lo, W. S., L. Duggan, N. C. Tolga, Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Lutfiyya, L. L., V. R. Iyer, J. DeRisi, M. J. DeVit, P. O. Brown, and M. Johnston. 1998. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150:1377-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsolier, M. C., P. Roussel, C. Leroy, and C. Mann. 2000. Involvement of the PP2C-like phosphatase Ptc2p in the DNA checkpoint pathways of Saccharomyces cerevisiae. Genetics 154:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 42.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nath, N., R. R. McCartney, and M. C. Schmidt. 2002. Purification and characterization of Snf1 kinase complexes containing a defined beta subunit composition. J. Biol. Chem. 277:50403-50408. [DOI] [PubMed] [Google Scholar]
- 44.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 45.Palecek, S. P., A. S. Parikh, J. H. Huh, and S. J. Kron. 2002. Depression of Saccharomyces cerevisiae invasive growth on non-glucose carbon sources requires the Snf1 kinase. Mol. Microbiol. 45:453-469. [DOI] [PubMed] [Google Scholar]
- 46.Purnapatre, K., S. Piccirillo, B. L. Schneider, and S. M. Honigberg. 2002. The CLN3/SWI6/CLN2 pathway and SNF1 act sequentially to regulate meiotic initiation in Saccharomyces cerevisiae. Genes Cells 7:675-691. [DOI] [PubMed] [Google Scholar]
- 47.Sanz, P. 2003. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 31:178-181. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt, M. C., and R. R. McCartney. 2000. Beta subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuller, H. J. 2003. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43:139-160. [DOI] [PubMed] [Google Scholar]
- 50.Scott, J. W., D. G. Norman, S. A. Hawley, L. Kontogiannis, and D. G. Hardie. 2002. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J. Mol. Biol. 317:309-323. [DOI] [PubMed] [Google Scholar]
- 51.Straight, A. F., W. F. Marshall, J. W. Sedat, and A. W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277:574-578. [DOI] [PubMed] [Google Scholar]
- 52.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. R. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 53.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 54.Tercero, J. A., M. P. Longhese, and J. F. Diffley. 2003. A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11:1323-1336. [DOI] [PubMed] [Google Scholar]
- 55.Thompson-Jaeger, S., J. Francois, J. P. Gaughran, and K. Tatchell. 1991. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics 129:697-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Vyas, V. K., S. Kuchin, and M. Carlson. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, P. J., A. Chabes, R. Casagrande, X. C. Tian, L. Thelander, and T. C. Huffaker. 1997. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol. Cell. Biol. 17:6114-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, Z., W. A. Wilson, M. A. Fujino, and P. J. Roach. 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 21:5742-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, X., A. Chabes, V. Domkin, L. Thelander, and R. Rothstein. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20:3544-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao, X., and R. Rothstein. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99:3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]