Abstract
The histone code is among others established via differential acetylation catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs). To unambiguously determine the histone tail specificity of HDAC-containing complexes, we have established an in vitro system consisting of nucleosomal templates reconstituted with hyperacetylated histones or recombinant histones followed by acetylation with native SAGA or NuA4. Selective targeting of the mammalian Sin3/HDAC and N-CoR/SMRT corepressor complexes by using specific chimeric repressors created a near physiological setting to assess their histone tail specificity. Recruitment of the Sin3/HDAC complex to nucleosomal templates preacetylated with SAGA or NuA4 resulted in deacetylation of histones H3 and H4, whereas recruitment of N-CoR/SMRT resulted in deacetylation of histone H3 only. These results provide solid evidence that HDAC-containing complexes display distinct, intrinsic histone tail specificities and hence may function differently to regulate chromatin structure and transcription.
In a eukaryotic cell, the nucleosome forms the fundamental repetitive unit of chromatin. DNA packed in nucleosomes is repressive to processes that require access of proteins to DNA, such as transcription initiation (10). Thus, in order for the transcription machinery to gain access to DNA, chromatin has to be actively remodeled. Similarly, transcriptionally active chromatin has to be converted back into a repressive configuration when needed. This is largely achieved by ATP-dependent chromatin remodeling and posttranslational modifications of core histones. ATP-dependent chromatin-remodeling enzymes use ATP hydrolysis to catalyze the mobilization of nucleosomes on DNA, thereby changing the accessibility of the DNA to regulatory factors (4, 27). Posttranslational modifications of core histones include acetylation, methylation, phosphorylation, and ubiquitination (10). During the last decade, a large number of proteins and protein complexes from different organisms have been identified and characterized that catalyze these modifications and thus contribute, either negatively or positively, to the accessibility of DNA. It has been postulated that modified histones serve as a binding scaffold for regulatory proteins and that distinct combinations of modifications ultimately determine the functional status of the chromatin. This hypothesis is known as the histone code theory (19). To gain further insight into how this dynamic code is established, it is of critical importance to unravel the specificities of chromatin-modifying enzymes with regard to the histone tails.
Of the currently known histone tail modifications, lysine acetylation has been studied most extensively. Acetylation of histones is a reversible and highly dynamic process catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs). A large number of in vivo studies have shown that HATs and HDACs can be recruited to target genes by activators and repressors. In particular, chromatin immunoprecipitation (ChIP) has provided insight into the putative roles of HAT and HDAC complexes. These studies suggest that targeting of HATs to promoter regions renders hyperacetylated and transcriptionally active chromatin, whereas targeting of HDACs results in hypoacetylated and transcriptionally silent chromatin (7). Gene disruption studies in yeast have revealed that HATs and HDACs may also serve a more global role in affecting the acetylation status throughout the genome (21). Although these experiments are very informative, it has been difficult to attribute an observed effect to a particular protein or protein complex, probably due to cellular compensatory effects and functional redundancy in vivo. Furthermore, it is difficult to determine whether a particular protein directly or indirectly causes an effect. Therefore, in vitro reconstituted nucleosomal templates and purified native HATs (9, 30) and HDACs are required to gain insights into the specificity and eventually the mechanistics of these multiprotein chromatin regulators.
Although considerable progress has been made in deciphering the specificity of different HAT complexes by using specific targeting to in vitro reconstituted templates (20, 35), such systems have not been described for HDACs. Several HDAC-containing complexes have been described thus far in mammalian cells. The Sin3/HDAC and Mi-2/NuRD complexes both appear to contain two HDACs, HDAC1 and -2, whereas the N-CoR/SMRT complex appears to contain only one deacetylase, HDAC3 (13, 23, 37, 38, 40-42, 44). These HDAC-containing complexes are recruited by transcription factors to regulate gene expression. The Sin3/HDAC complex can be recruited by the Mad1 protein (2). Mad1 is a repressor that is part of the Myc/Mad/Max network of transcription factors that plays an important role in keeping the balance between cell proliferation and differentiation (12, 26). The N-CoR/SMRT complex is recruited by unliganded nuclear hormone receptors (6, 16, 23). In the absence of ligand, the receptor recruits the N-CoR/SMRT complex to repress transcription. In the presence of ligand, a conformational change in the ligand binding domain of the receptor causes the corepressors to dissociate and to be substituted for by coactivators, leading to activation of gene expression (11).
The above-described roles for the Sin3/HDAC and N-CoR/SMRT complexes in the functioning of several transcription factors in different biological processes underscore their importance in the regulation of gene expression and development. However, the histone tail specificity, and therefore their mechanistic role in establishing a histone code, remains largely unknown. We purified the Sin3/HDAC and N-CoR/SMRT complexes from HeLa cells and analyzed their biochemical properties in an in vitro reconstituted chromatin system. Upon specific targeting to chromatin by using chimeric repressor molecules, the Sin3/HDAC and N-CoR/SMRT complexes displayed different histone tail specificities. These findings provide evidence that distinct HDAC-containing corepressor complexes may play different roles in the regulation of chromatin structure and transcription.
MATERIALS AND METHOD
Purification of the Sin3/HDAC and N-CoR/SMRT complexes
HeLa cell nuclear extract was prepared as described previously (8). Approximately 5 g of protein was loaded onto a P11 phosphocellulose column (Whatman) as described previously (36). The majority of the N-CoR protein was found in the 300 mM KCl fraction, whereas the majority of the Sin3a protein was found in the 500 mM KCl fraction. To further purify the Sin3/HDAC complex, proteins in the 500 mM fraction were precipitated with 50% ammonium sulfate, redissolved, dialyzed against buffer C (750 mM ammonium sulfate, 10% [vol/vol] glycerol, 50 mM potassium phosphate [pH 7.9], 0.2 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]), and then loaded onto a phenyl-Sepharose column (Pharmacia). Proteins binding to the column were eluted stepwise with buffer C containing 500 or 0 mM ammonium sulfate. The 0 M ammonium sulfate fraction was dialyzed against buffer D (100 mM KCl, 10% [vol/vol] glycerol, 20 mM Tris [pH 8.0], 0.2 mM EDTA, 2 mM DTT, 1 mM PMSF), loaded onto a DEAE-Sepharose column (Pharmacia), and eluted with buffer D containing 350 mM KCl. Finally, the DEAE eluate was loaded onto a Superose 6 column (Pharmacia) equilibrated in buffer E (100 mM KCl, 20% [vol/vol] glycerol, 20 mM Tris [pH 8.0], 0.2 mM EDTA, 2 mM DTT, 1 mM PMSF, 0.05% NP-40). The 300 mM KCl fraction of the phosphocellulose column containing N-CoR was dialyzed against buffer E, loaded onto a DEAE-Sepharose column, and eluted with buffer E containing 350 mM KCl. This eluate was loaded onto a Superose 6 column as described above for the Sin3/HDAC complex. Superose 6 fractions containing either the Sin3A or N-CoR protein were pooled, concentrated with a Centricon spin column, and stored at −80°C. A DNA affinity pull-down approach was used to recruit the Sin3/HDAC and N-CoR/SMRT complexes to immoblized templates. An oligonucleotide containing a LexA binding site (28) was multimerized, after which a Klenow fragment (Invitrogen) fill-in with bio-ATP (Gibco) was performed. Next, multimerized oligonucleotides were bound to streptavidin-conjugated Dynabeads (Dynal) and incubated with LexA-Mad, LexA-TR(DE), or the LexA DNA-binding domain (DBD) alone at 4°C for 20 min. The beads were washed twice with buffer F (100 mM KCl, 5% glycerol, 10 mM HEPES [pH 7.9], 2 mM MgCl2, 5 mM DTT, 1 mM PMSF) containing 0.25% Triton X-100 and incubated with the Superose 6 fractions containing the Sin3/HDAC and N-CoR/SMRT complexes, respectively, supplemented with 0.25% NP-40, 2 mM MgCl2, and 0.1-μg/μl dIdC for 3 h at 4°C. Beads were extensively washed with buffer F containing 175 mM NaCl and 0.5% Triton X-100, after which bound proteins were eluted with 1 M NaCl. These eluates were analyzed by Western blotting, silver staining and nanoscale liquid chromatography-tandem mass spectrometry (nLC-MS/MS) for the Sin3/HDAC complex.
Protein identification by LC-MS/MS
The purified SIN3/HDAC complex (50 μl in 1 M NaCl, 50 mM Tris [pH 8.0]) was reduced by 10 mM DTT for 30 min at room temperature and alkylated in 55 mM iodoacetamide for 30 min. The sample was diluted 10 times to a final concentration of 100 mM NaCl, 2 M urea, and 100 mM Tris (pH 8.0) and digested with 1 μg of LysC at 37°C for 4 h and 2 μg of modified trypsin at 37°C for 16 h. The digested sample was acidified with formic acid and purified by Poros R3 (Applied Biosystems) beads.
Peptide mixtures were separated by nano reversed-phase LC and subjected on-line to MS and MS/MS by using a QSTAR pulsar quadrupole time-of-flight tandem mass spectrometer (ABI/Sciex MDS). Nano columns of 5 cm with a 75-μm inside diameter and 8-μm opening size packed with 3-μm C18 beads were applied. Peptides were separated by applying an acetonitrile gradient for 2 h with a flow rate of 200 μl/min. Peptide selection and fragmentation was set by the Analyst software for cycles of 7.5 containing precursor selection in the mass range of 400 to 1,200 during 1.5 s and four MS/MS experiments in the mass range of 350 to 1,600 for 1.5 s each. Signals were enhanced around 730.
Data analysis
Lists of peaks containing the precursor masses and the corresponding MS/MS fragments were generated from the original data file by an Analyst script file. The peak lists were searched against the human National Center for Biotechnology Information database by using the Mascot algorithm, with an accuracy of 0.2 Da. Proteins identified by at least one first-ranked peptide were verified by manual inspection of the MS/MS spectra.
Purification of yeast HATs
Native HAT complexes from Saccharomyces cerevisiae were purified as described previously (9). After separation of the four major S. cerevisiae HAT complexes on a MonoQ column (Pharmacia), the NuA4 and SAGA complexes were further purified on a Superose 6 column. In vitro HAT assays of HeLa core histones were performed to determine the activity of the purified HAT complexes.
Expression and purification of recombinant proteins
LexA-Mad amino acids 5 to 24 and LexA-TR(DE) proteins were expressed as six-His-tagged fusion proteins in the pET28a vector (Novagen). Plasmids were transformed into BL21 DE3 LysS bacteria. Cells were grown at 30°C, and protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. The expressed recombinant proteins were purified from the soluble fraction with Ni2+-nitrilotriacetic acid agarose beads (Qiagen). Proteins were further purified over a MonoQ column (Pharmacia). LexA-Mad was found in the flowthrough of the column, whereas the LexA-TR protein eluted at approximately 300 mM NaCl.
Histone preparations and nucleosome reconstitution
Xenopus core histones were expressed and purified from Escherichia coli essentially as described previously (25). Core histones were reconstituted into octamers, purified on a Superose 12 gel filtration column (Pharmacia), and subsequently used for nucleosomal reconstitutions. Hyperacetylated core histones from HeLa cells as well as chicken oligonucleosomes were purified as described previously (24, 39). Plasmid L8G5E4T15S was obtained by cloning eight LexA sites in the PstI and HindIII sites of vector G5E4T (14). A Lychetinus variegatus 5S positioning element from vector PCL115 (18) was cloned in the XbaI and BamHI sites of this plasmid. The plasmid was linearized with BamHI, filled-in with Klenow polymerase by using bio-ATP (Gibco), and subsequently cleaved with HindIII. The resulting 520-bp fragment was purified and reconstituted with recombinant Xenopus octamers or hyperacetylated HeLa core histones as described previously (33).
Micrococcal nuclease digestion of nucleosomal array
Approximately 750 ng of DNA reconstituted into nucleosomes was digested with 10 mU of micrococcal nuclease (Worthington Biochemicals) for 0, 20, 40, 60, and 180 s at 37°C in buffer F containing 50 mM KCl and 3 mM CaCl2. Reactions were stopped by adding 10 mM EGTA, after which DNA was phenol-chloroform extracted, precipitated, and loaded onto a 1.5% agarose gel.
Immobilized template pull-down assays
The biotinylated nucleosomal templates were coupled to paramagnetic streptavidin-conjugated Dynabeads (Dynal) as described previously (14). Approximately 200 ng of template reconstituted with recombinant Xenopus histone octamers bound to beads was acetylated with NuA4 or SAGA for 1 h at 30°C in 50 μl of buffer F containing 50 mM KCl. Beads were then washed extensively with buffer F containing 300 mM KCl and 0.5% Triton X-100 to remove the HATs from the templates. Next, templates were incubated with LexA-Mad, LexA-TR, or LexA DBD in 50 μl of buffer F containing 75 mM KCl, 0.1% NP-40, and complete protease inhibitor cocktail (Roche) for 20 min at 37°C. After being washed with buffer F, 5 μl of each of the Superose 6 fractions containing the N-CoR/SMRT or Sin3/HDAC complex was added in 50 μl of buffer F containing 75 mM KCl and 0.1% NP-40. Furthermore, when indicated, a 50- to 100-fold molar excess of competitor chicken oligonucleosomes and 1 μM trichostatin (TSA) were added. Finally, the beads were washed twice with buffer F containing 300 mM KCl and 0.25% NP-40, after which, Western blotting analysis was performed. Blots were probed with antibodies against HDAC2 (Santa Cruz), HDAC3 (Santa Cruz), Sin3a (Santa Cruz), acetyl H3 Lys 9,14 (Upstate Biotechnology, Inc.), and acetyl H4 tetra (Upstate Biotechnology, Inc.).
RESULT
Purification of the Sin3/HDAC and N-CoR/SMRT complexes
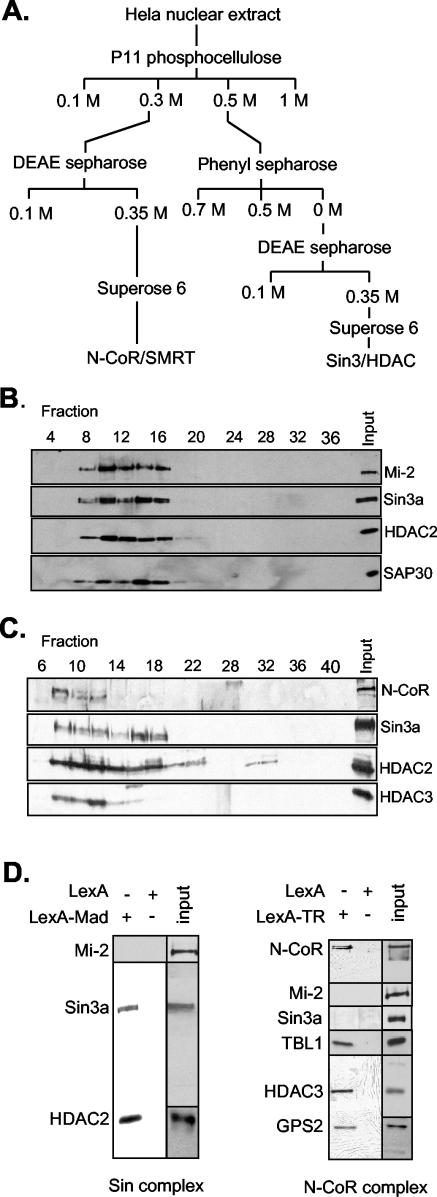
We set out to purify the Sin3/HDAC and N-CoR/SMRT complexes from HeLa cell nuclear extracts by conventional column chromatography (Fig. 1A). As shown in Fig. 1B, Sin3A, HDAC2, and SAP30 (all known to be part of the Sin3/HDAC complex) cofractionated on a Superose 6 gel filtration column. Intriguingly, Mi-2, a 240-kDa protein component of the Mi-2/NuRD corepressor complex, cofractionated with the Sin3/HDAC complex. HDAC1 and -2 and RbAp48 and -46 are shared subunits between these complexes. Besides Mi-2, we found the Swi/Snf complex proteins BRM and BAF170 in the fractions that also contained Sin3/HDAC complex, although these proteins did not exactly cofractionate with the Sin3/HDAC and Mi-2/NuRD complexes (data not shown). Thus, the Superose 6 fractions are highly enriched for the Sin3/HDAC complex, but they also contain other complexes that are involved in chromatin metabolism.
FIG.1.
Purification of the Sin3/HDAC and N-CoR/SMRT complexes and recruitment to DNA by using chimeric repressor molecules. (A) Conventional purification of the Sin3/HDAC and N-CoR/SMRT complexes. HeLa cell nuclear extract was fractionated as shown. The Sin3/HDAC and N-CoR/SMRT complexes were monitored throughout the purification by Western analysis with antibodies against HDAC2, mSin3a, SAP30, N-CoR, and HDAC3. (B) Elution profile of the Sin3/HDAC complex on a Superose 6 column analyzed by Western blotting against Sin3a, HDAC2, and SAP30. (C) As in panel B, using the N-CoR/SMRT fractions and antibodies against N-CoR, HDAC2, HDAC3, and Sin3a. (D) Recruitment of the Sin3/HDAC and N-CoR/SMRT complexes by the LexA-Mad and LexA-TR(DE) fusion proteins, respectively. Fractions 9 to 13 (Fig. 1B) and 7 to 11 (Fig. 1C) from the Superose 6 columns containing the Sin3/HDAC and N-CoR/SMRT complexes were used for the LexA-Mad and LexA-TR(DE) pull-down on paramagnetic streptavidin-conjugated Dynal Dynabeads containing multimerized LexA oligonucleotides. Bound proteins were eluted from DNA with 1 M NaCl, loaded on a sodium dodecyl sulfate-polyacrylamide (8%) gel, and analyzed by Western blotting with the indicated antibodies. Input fractions used for the pull-down are also shown.
Whereas the majority of the Sin3a protein was detected in the 0.5 M phosphocellulose (PC) fraction, most of the N-CoR protein was found in the 0.3 M PC fraction. The N-CoR/SMRT complex was subsequently further purified (Fig. 1A). Western blotting showed that N-CoR and HDAC3 cofractionate on a gel filtration column (Fig. 1C). Probing the fractions for components of other complexes revealed the presence of some HDAC2 and Sin3a, although their elution profile was clearly different from that of N-CoR/HDAC3.
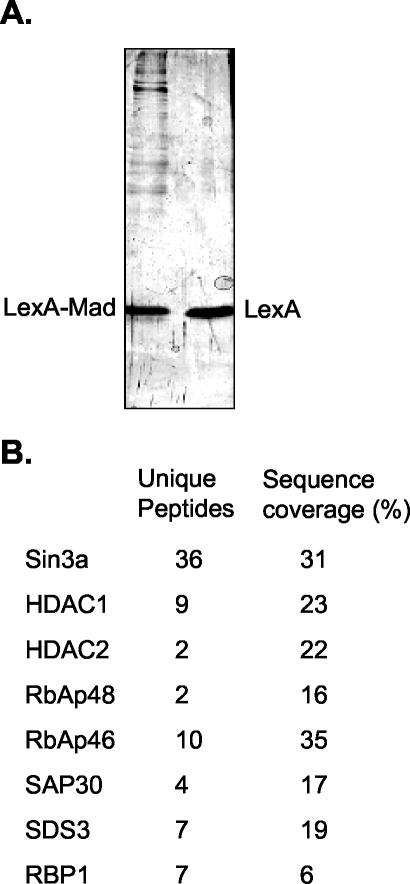
These results show that we obtained fractions highly enriched for the Sin3/HDAC and N-CoR/SMRT complexes, although these fractions are not homogeneous in composition. To obtain pure complex preparations, we performed affinity purification by using chimeric repressor molecules. A protein containing the LexA DBD fused to amino acids 5 to 24 of the Mad repressor (LexA-Mad) was used to target the Sin3/HDAC complex to multimerized LexA DNA binding sites coupled to magnetic beads. Both Sin3a and HDAC2 were specifically recruited to the LexA-Mad beads, whereas Mi-2 was not (Fig. 1D). In this experiment, dIdC was included to reduce binding of proteins that have nonspecific DNA binding affinity. Analysis of the recruited proteins by silver stain showed a number of proteins that were specifically retained on the LexA-Mad beads, but not on LexA beads (Fig. 2A). These proteins were analyzed by nLC-MS/MS. Besides matches to Sin3a, we obtained peptides matching HDAC1 and -2, RbAp48 and -46, SAP30, SDS3, and RBP1 (Fig. 2B). Surprisingly, we did not detect peptides matching SAP18 or Sin3b, two of the reported subunits of the core Sin3/HDAC complex. Moreover, we did not obtain peptides from Mi-2 or MTA2 or components of the N-CoR/SMRT complex. Similarly, Swi/Snf complex proteins, which have recently been reported to interact with the Sin3/HDAC complex, were absent, although these proteins were present in the input material. Thus, the LexA-Mad protein appears to specifically recruit a Sin3/HDAC core complex to DNA.
FIG. 2.
Analysis of the affinity-purified Sin3/HDAC complex by silver stain and MS. (A) Recruitment of the Sin3/HDAC complex by the LexA-Mad fusion protein. Fractions 9 to 13 (Fig. 1B) from the Superose 6 column containing the Sin3/HDAC complex were used for the LexA-Mad pull-down on paramagnetic streptavidin-conjugated Dynal Dynabeads containing multimerized LexA oligonucleotides. Bound proteins were eluted from DNA with 1 M NaCl, loaded onto an SDS-polyacrylamide (8%) gel, and analyzed by silver staining. (B) A pull-down similar to that described for panel A was performed. After elution of the recruited proteins with 1 M NaCl, they were analyzed by nLC-MS/MS.
A chimeric protein containing the LexA DBD fused to the DE domain of the thyroid hormone receptor was used to target the N-CoR/SMRT complex. The DE domain of the thyroid hormone receptor is known to interact with the N-CoR protein in the absence of hormone ligand. Western blot analysis revealed that N-CoR, HDAC3, TBL1, and GPS2, a recently identified subunit of the N-CoR complex, were specifically recruited by the LexA-TR(DE) fusion protein, but not by the LexA DBD alone (Fig. 1D). Furthermore, Western blot analysis with antibodies specific for Sin3a, HDAC2, and Mi-2 showed that these were not recruited by the LexA-TR(DE) fusion protein, confirming the specificity of the chimeric repressor for the N-CoR/SMRT complex.
HDAC activity of the Sin3/HDAC and N-CoR/SMRT complexes
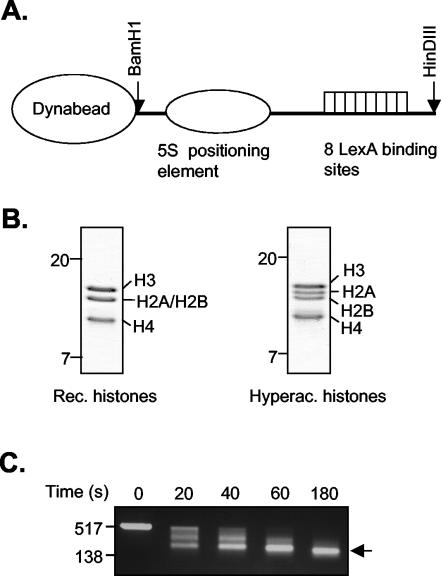
Having established that the LexA-Mad and LexA-TR(DE) fusion proteins can efficiently and specifically recruit the Sin3/HDAC and N-CoR/SMRT complexes to DNA, we extended our analyses to nucleosomal templates. To generate a nucleosomal template, DNA (Fig. 3A) was mixed with purified Xenopus octamers or hyperacetylated HeLa core histones (Fig. 3B) in a 1:1 molar ratio and reconstituted by a salt dilution protocol. Subsequently, the nucleosomal templates were bound to streptavidin-coupled Dynabeads. The reconstitutions were analyzed by agarose gel electrophoresis before binding them to the beads (data not shown). In addition, a partial micrococcal nuclease digestion of the template was performed (Fig. 3C).
FIG. 3.
Reconstitution and analysis of the nucleosomal template. (A) Schematic representation of the DNA template containing eight LexA binding sites and a 5S nucleosome positioning element. (B) Analysis of purified recombinant (Rec.) Xenopus octamers and hyperacetylated (Hyperac.) core histones purified from HeLa cells on SDS-polyacrylamide (15%) gel electrophoresis gel stained with Coomassie brilliant blue. (C) Partial micrococcal nuclease digestion. Nucleosomal templates were incubated with 10 mU micrococcal nuclease at 37°C for 0, 20, 40, 60, and 180 s. Reactions were stopped by adding 10 mM EGTA. DNA was phenol chloroform extracted, precipitated, and loaded onto a 1.5% agarose gel. DNA size markers are indicated on the left. An arrow indicates mononucleosomal DNA.
Next, we addressed whether the partially purified Sin3/HDAC and N-CoR/SMRT complexes could deacetylate nucleosomal templates reconstituted with hyperacetylated histones in vitro. Initially to confirm that the purified fractions contained HDAC activity, in vitro HDAC assays of 3H-labeled core histones were performed (data not shown). As shown in Fig. 4A, the Superose 6 fractions 9 to 13 containing the Sin3/HDAC complex displayed deacetylase activity. However the deacetylation reaction could not be driven to completion, even when increasing amounts of proteins were added (data not shown). This indicates that a subset of the acetyl groups could not be removed and/or that hyperacetylated histones contain modifications that inhibit or prevent histone deacetylation. Deacetylation of the templates, however, was complete when recombinant Xenopus histones were used that had been acetylated with purified S. cerevisiae SAGA complex (Fig. 4B). Similar results were obtained with Superose 6 fractions containing the N-CoR/SMRT complex (data not shown).
FIG. 4.
Deacetylase activity of the Sin3/HDAC complex. (A) Nucleosomal templates reconstituted with hyperacetylated (Hyperac.) histones were incubated with the Sin3/HDAC complex in the absence or presence of TSA. The amount of H3 deacetylation was determined by Western blotting with an antibody that recognizes diacetylated histone H3 Lys 9,14. (B) Nucleosomal templates reconstituted with recombinant (Rec.) Xenopus octamers were incubated with the S. cerevisiae SAGA complex, washed, and subsequently incubated with Sin3/HDAC complex in the presence or absence of TSA, after which the amount of H3 acetylation (Ac) was determined as described for panel A.
The Sin3/HDAC complex deacetylates histones H3 and H4 upon targeting to chromatin
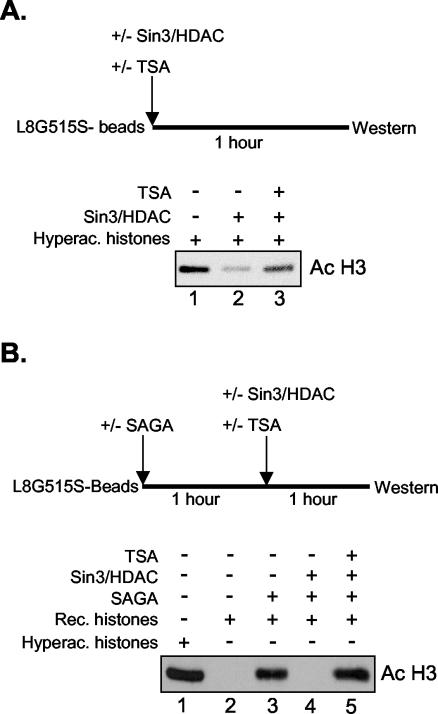
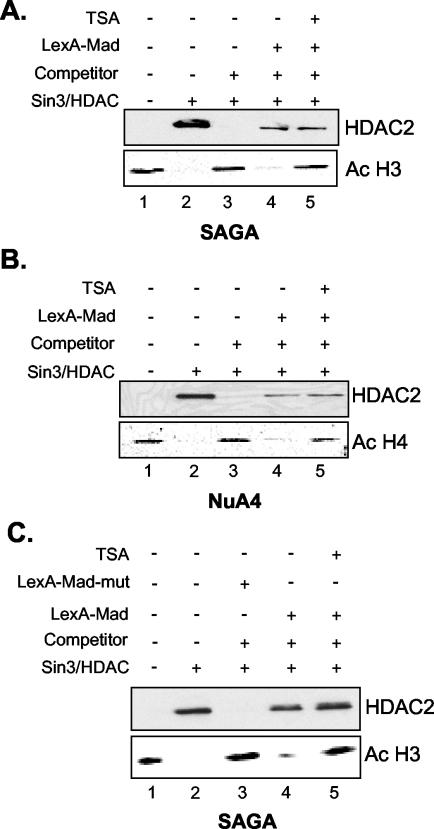
The experiments described above showed that the Superose 6 fractions have significant nucleosomal deacetylase activity. These fractions, however, also contain small amounts of the Mi-2/NuRD complex and possibly other HDAC-containing complexes. Therefore, we cannot unequivocally attribute the observed HDAC activity to the Sin3/HDAC and N-CoR/SMRT complexes. To circumvent this problem, we used an experimental setup in which the Sin3/HDAC and N-CoR/SMRT complexes are recruited to the immobilized nucleosomal template in a competitive setting. Highly enriched native S. cerevisiae HAT complexes (SAGA and NuA4) were used to acetylate the nucleosomes. After incubation for an hour, the beads were washed with high-salt buffer and detergent to strip the HAT complexes from the templates, as monitored by Western blotting (data not shown). The beads were then incubated with or without LexA-Mad, washed, and subsequently incubated with the enriched Sin3/HDAC fractions, in the absence or presence of competitor oligonucleosomes. Finally as a control, a targeting experiment was performed in the presence of an HDAC inhibitor, TSA. In the absence of LexA-Mad and competitor oligonucleosomes, the Sin3/HDAC complex was associated with the nucleosomal template. This resulted in a complete deacetylation of histone H3 (Fig. 5A, lane 2). The association of the Sin3/HDAC complex was DNA sequence independent, because upon addition of a 50- to 100-fold molar excess of competitor oligonucleosomes, HDAC2 binding to the beads could not be detected, and deacetylation was not observed (compare lanes 2 and 3). However, the Sin3/HDAC complex was retained on the beads when specifically recruited by LexA-Mad to the nucleosomal templates in the presence of competitor oligonucleosomes, and deacetylation was observed (compare lanes 3 and 4). Finally, a control experiment in which TSA was added showed that HDAC activity could be inhibited (Fig. 5A, lane 5).
FIG. 5.
The Sin3/HDAC complex can deacetylate histones H3 and H4 upon targeting to a nucleosomal template. Nucleosomal templates reconstituted with recombinant Xenopus octamers were incubated with the S. cerevisiae SAGA complex (A and C) or NuA4 complex (B), washed, incubated with or without LexA-Mad (A and B) or a LexA-Mad mutant (C), and subsequently incubated with the Sin3/HDAC complex in the presence or absence of competitor oligonucleosomes, in the presence or absence of TSA. The amount of H4 acetylation (Ac) was determined by Western blotting with an antibody that recognizes tetra-acetylated H4.
Next, we wanted to address whether the Sin3/HDAC complex could deacetylate nucleosomal templates containing histone H4 molecules acetylated with highly enriched NuA4 complex. The NuA4 complex acetylates all four lysines in the N terminus of histone H4 (1). As for histone H3, the Sin3/HDAC complex deacetylated histone H4 (Fig. 5B). Again, in the presence of competitor oligonucleosomes, targeted recruitment by the LexA-Mad molecule was a prerequisite to achieve histone deacetylation. Thus, the Sin3/HDAC complex is able to deacetylate both histones H3 and H4 when targeted to nucleosomal templates preacetylated by the S. cerevisiae SAGA and NuA4 complexes, respectively.
To further illustrate the specificity of the LexA-Mad protein for the Sin3/HDAC complex, we purified a mutant LexA-Mad molecule containing two point mutations, L12P and A16P. This mutant was subsequently assayed for its ability to recruit the Sin3/HDAC complex to immobilized nucleosomal templates. As shown in Fig. 5C, HDAC2 recruitment and deacetylation of histone H3 could not be observed on nucleosomal templates incubated with the LexA-Mad mutant. In contrast, HDAC2 recruitment and almost complete deacetylation of histone H3 were observed on beads incubated with the wild-type LexA-Mad molecule (compare lanes 3 and 4 of Fig. 5C).
The N-CoR/SMRT complex only deacetylates histone H3 upon targeting to chromatin
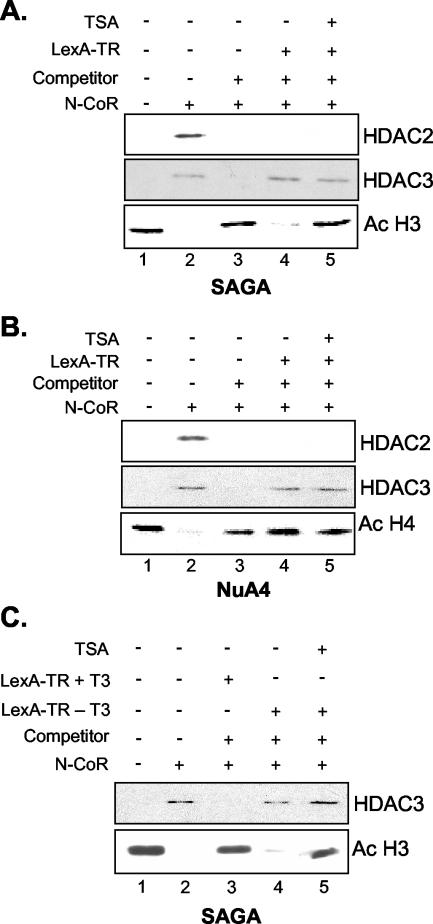
To assess whether the HDAC3-containing N-CoR/SMRT complex displays histone substrate specificity, we used the LexA-TR(DE) fusion protein to target the N-CoR/SMRT complex to acetylated nucleosomal templates. As shown in Fig. 6A and B, addition of the N-CoR/SMRT enriched fractions in the absence of competitor oligonucleosomes and the LexA-TR(DE) fusion protein resulted in deacetylation of both histones H3 and H4 (Fig. 6A and B, lane 2). Western blotting clearly showed that HDAC2 and HDAC3, and thus both the Sin3/HDAC and the N-CoR/SMRT complexes, associated with the nucleosomal templates in this nontargeted setting. Addition of competitor oligonucleosomes displaced HDAC2 and HDAC3 and abolished deacetylation of the immobilized template (Fig. 6A and B, lane 3). However, LexA-TR(DE) specifically recruited HDAC3 and thus the N-CoR/SMRT complex under these competitive binding conditions, whereas Sin3/HDAC recruitment was no longer observed, illustrating the specificity of the LexA-TR(DE) fusion protein for the N-CoR/SMRT complex. Strikingly, in this competitive setting, the N-CoR/SMRT complex deacetylated histone H3 but showed no detectable activity toward histone H4 (compare Fig. 6A and B, lane 4). This indicates that the N-CoR/SMRT complex specifically deacetylates histone H3 upon recruitment to nucleosomes.
FIG. 6.
The N-CoR/SMRT complex deacetylates histone H3 upon targeting to a nucleosomal template. Nucleosomal templates reconstituted with recombinant Xenopus octamers were incubated with the S. cerevisiae SAGA complex (A and C) or NuA4 complex (B), washed, incubated with or without LexA-TR(DE) (A and B) or LexA-TR(DE) in the presence of 5 μM T3 (C), and subsequently incubated with the N-CoR/SMRT complex in the presence or absence of competitor oligonucleosomes, in the presence or absence of TSA. Ac, acetylation.
The thyroid hormone receptor is thought to interact with the N-CoR/SMRT complex only in the absence of thyroid hormone. In the presence of thyroid hormone, a conformational switch in the DE domain of the receptor causes the corepressors to dissociate, which are then replaced by coactivators. In agreement with this, N-CoR/SMRT recruitment and histone H3 deacetylation were not observed when nucleosomal templates were incubated with the LexA-TR(DE) protein in the presence of thyroid hormone (compare lanes 3 and 4, Fig. 6C). This result shows the specificity of the unliganded LexA-TR(DE) molecule for the N-CoR/SMRT complex.
In conclusion, these experiments demonstrate that HDAC-containing complexes can be efficiently and specifically recruited to in vitro reconstituted nucleosomal templates by chimeric recombinant repressor molecules. As is observed for the HAT complexes, these HDAC complexes display differences in substrate specificities, which probably reflects different functions in vivo.
DISCUSSION
Purification of the Sin3/HDAC and N-CoR/SMRT complexes
In this study, we have used a combination of conventional column chromatography and a DNA affinity pull-down approach to purify the Sin3/HDAC and N-CoR/SMRT complexes from HeLa cell nuclear extracts. The purified N-CoR/SMRT complex was analyzed extensively by Western blotting, revealing that N-CoR, HDAC3, GPS2, and TBL1—all known to be part of the N-CoR/SMRT complex—were recruited by LexA-TR(DE). Sin3a and HDAC2 were not recruited by the LexA-TR(DE) fusion protein, despite the facts that these proteins have been described as N-CoR/SMRT-interacting proteins (15, 29) and were present in the fraction used for the pull-down. The Sin3/HDAC complex recruited on LexA-Mad beads was of sufficient purity to unequivocally identify the recruited proteins by nLC-MS/MS. Eight previously reported proteins were identified as core components of the Sin3/HDAC complex. Surprisingly, we did not obtain peptide hits for SAP18 and Sin3b, two of the reported subunits of the core Sin3/HDAC complex. Recently a novel SAP18-containing complex was reported, which does not contain Sin3a or HDACs, indicating that SAP18 might not be a component of the core Sin3/HDAC complex (31a). Moreover, Swi/Snf complex components that have been described as Sin3/HDAC- and N-CoR/SMRT-interacting proteins (22, 32, 34) could not be detected by nLC-MS/MS. Protein components of the human Swi/Snf complex, such as Brg1, BRM, BAF170, and BAF60, were detected in the Superose 6 fractions that were used for the LexA-Mad and LexA-TR(DE) pull-down. These proteins were partially coeluting with the Sin3/HDAC and N-CoR/SMRT complexes on the gel filtration column (data not shown). A possible explanation for these observations could be that the interactions between the Sin3/HDAC or N-CoR/SMRT complexes with Swi/Snf complex are lost during the stringent washing procedures that were used in our experiments. Intriguingly, when competitor dIdC was omitted and when the beads were washed with lower stringency, BAF170 and BRM were both retained on LexA-Mad beads, arguing in favor of a weak physical interaction between Sin3/HDAC and Swi/Snf (data not shown). Whatever the nature of the interaction, ATP-dependent nucleosome-remodeling activity was not required for efficient deacetylation of the immobilized templates, since all reactions described in this study were performed in the absence of ATP.
Histone tail specificity of HDAC-containing complexes
To date, the issue of histone tail specificity of different HDACs has mainly been addressed in vivo. However, potential indirect effects and compensatory mechanisms due to redundancy hamper a direct functional analysis of different HDACs. The in vitro approach described here allowed us to surmount these obstacles and enabled us to directly analyze the enzymatic properties of different HDAC-containing corepressor complexes. We have shown that the Sin3/HDAC and N-CoR/SMRT complexes can be selectively recruited to in vitro reconstituted nucleosomal templates by specific chimeric repressors in the presence of a large excess of competitor nucleosomes. This is in agreement with the reported specific recruitment of these corepressor complexes by several transcription factors, as determined in particular by ChIP experiments (3, 31). In addition, by using native SAGA and NuA4 HAT complexes to acetylate nucleosomal templates reconstituted with recombinant histones rather than using hyperacetylated nucleosomes, we have reconstituted near physiological conditions. Our analyses revealed that the Sin3/HDAC complex can deacetylate both histone H3 and H4 upon specific recruitment to nucleosomes, whereas the N-CoR/SMRT complex only deacetylates histone H3. Thus, different HDAC-containing complexes display distinct histone tail specificities, suggesting that they play different roles in the regulation of transcription.
Interestingly, the Sin3/HDAC complex contains two deacetylases, HDAC1 and HDAC2, whereas the N-CoR/SMRT complex contains only one deacetylase, HDAC3. It is tempting to speculate that a division of labor exists in the Sin3/HDAC complex and that one of the two HDACs deacetylates histone H3, whereas the other deacetylates histone H4. At present, this question cannot be addressed experimentally, since both HDAC1 and HDAC2 are present in one complex. Furthermore, recombinant HDAC1 and HDAC2 molecules are not active on nucleosomal substrates (43), and hence the division of labor hypothesis remains purely speculative at this point.
HDACs and the histone code
In our initial experiments, we made use of nucleosomal templates that contained hyperacetylated histones purified from HeLa cells. Surprisingly, the purified HDAC complexes were not able to fully deacetylate these templates. A possible explanation could be that a fraction of the purified bulk HeLa histones contain modifications such as phosphorylation or methylation that may inhibit HDAC activity. Recombinant nucleosomal templates uniformly and physiologically acetylated by native SAGA or NuA4 could, however, be efficiently deacetylated. These findings corroborate and extend the observed phenomenon of cross talk between different histone modifications.
The fact that the Sin3/HDAC complex can counteract the activities of both the SAGA and NuA4 complexes suggests that the Sin3/HDAC complex may impinge on processes other than regulation of transcription. Whereas the SAGA complex is predominantly involved in transcription-related processes, the NuA4 complex also plays a role in DNA repair and possibly other processes that involve chromatin modifications (5, 17). On the other hand, the apparent specificity of the N-CoR/SMRT complex for histone H3 acetylation suggests that this complex predominantly antagonizes transcription promoted by histone H3 K9 and K14 acetylation.
The work presented here clearly illustrates the specificity of the interaction between transcription factors and corepressors. Evidently, these transcription factors appear to recruit distinct corepressor complexes that display a general (“broad”) or rather restricted (“narrow”) histone tail specificity. The physiological implications of the recruitment of an HDAC complex with a broad or narrow tail specificity by the tumor supressor Mad and the unliganded thyroid hormone receptor, respectively, remain to be elucidated.
Acknowledgments
We thank Coen Campsteijn for purifying recombinant Xenopus histone octamers and Martijn van Aart for assistance. Furthermore, we thank M. Mann (CEBI, University of Southern Denmark) for the kind use of the QSTAR (and for the stay of E.L. in his laboratory). We are grateful to Danny Reinberg, Robert G. Roeder, and Jiemin Wong for providing antibodies.
This work was supported by The Netherlands Organization for Scientific Research (NWO) and in part by a grant from the National Institute of General Medical Sciences to J.L.W. M.J.C. is a postdoctoral fellow of the American Cancer Society (grant number PF-02-012-01-GMC). J.L.W. is a Howard Hughes Medical Institute Associate Investigator.
REFERENCE
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 3.Baek, S. H., K. A. Ohgi, D. W. Rose, E. H. Koo, C. K. Glass, and M. G. Rosenfeld. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110:55-67. [DOI] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 7.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberharter, A., S. John, P. A. Grant, R. T. Utley, and J. L. Workman. 1998. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15:315-321. [DOI] [PubMed] [Google Scholar]
- 10.Fischle, W., Y. Wang, and C. D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172-183. [DOI] [PubMed] [Google Scholar]
- 11.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 12.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 13.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 15.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 16.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 17.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 18.Jaskelioff, M., I. M. Gavin, C. L. Peterson, and C. Logie. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 21.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33ING1. Mol. Cell. Biol. 22:835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logie, C., and C. L. Peterson. 1999. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 304:726-741. [DOI] [PubMed] [Google Scholar]
- 25.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 26.Luscher, B. 2001. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene 277:1-14. [DOI] [PubMed] [Google Scholar]
- 27.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 28.Mohana-Borges, R., A. B. Pacheco, F. J. Sousa, D. Foguel, D. F. Almeida, and J. L. Silva. 2000. LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J. Biol. Chem. 275:4708-4712. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 30.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rietveld, L. E., E. Caldenhoven, and H. G. Stunnenberg. 2002. In vivo repression of an erythroid-specific gene by distinct corepressor complexes. EMBO J. 21:1389-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Schwerk, C., J. Prasad, K. Degenhardt, H. Erdjument-Bromage, E. White, P. Tempst, V. J. Kidd, J. L. Manley, J. M. Lahti, and D. Reinberg. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 23:2981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steger, D. J., and J. L. Workman. 1999. Transcriptional analysis of purified histone acetyltransferase complexes. Methods 19:410-416. [DOI] [PubMed] [Google Scholar]
- 34.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 35.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 36.Valcarcel, R., and H. G. Stunnenberg. 1996. Retinoid-dependent in vitro transcription. Methods Enzymol. 274:149-161. [DOI] [PubMed] [Google Scholar]
- 37.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843-846. [DOI] [PubMed] [Google Scholar]
- 38.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Workman, J. L., I. C. Taylor, R. E. Kingston, and R. G. Roeder. 1991. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 35:419-447. [DOI] [PubMed] [Google Scholar]
- 40.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021-1031. [DOI] [PubMed] [Google Scholar]