Abstract
Nuclear pore complexes (NPCs) traverse the nuclear envelope (NE), providing a channel through which nucleocytoplasmic transport occurs. Nup358/RanBP2, Nup214/CAN, and Nup88 are components of the cytoplasmic face of the NPC. Here we show that Nup88 localizes midway between Nup358 and Nup214 and physically interacts with them. RNA interference of either Nup88 or Nup214 in human cells caused a strong reduction of Nup358 at the NE. Nup88 and Nup214 showed an interdependence at the NPC and were not affected by the absence of Nup358. These data indicate that Nup88 and Nup214 mediate the attachment of Nup358 to the NPC. We show that localization of the export receptor CRM1 at the cytoplasmic face of the NE is Nup358 dependent and represents its empty state. Also, removal of Nup358 causes a distinct reduction in nuclear export signal-dependent nuclear export. We propose that Nup358 provides both a platform for rapid disassembly of CRM1 export complexes and a binding site for empty CRM1 recycling into the nucleus.
The nucleus is the defining feature of a eukaryotic cell and is surrounded by a double membrane known as the nuclear envelope (NE), which prevents free diffusion of macromolecules between the nucleus and the cytoplasm. Nuclear pore complexes (NPCs) are protein channels residing in the NE, through which the active and highly specific transport of RNA and protein between the nucleus and the cytoplasm occurs, a process known as nucleocytoplasmic transport (24, 54, 65). The NPC is a modular and complex structure, displaying eightfold rotational symmetry (10, 50). It is composed of a series of concentric rings at the plane of the NE, with 80- to 100-nm filaments extending into the nucleus, distally connected to form a basket structure, and ∼50-nm filaments extending into the cytoplasm (22, 30, 50, 53). Approximately 30 proteins, termed nucleoporins, constitute the vertebrate NPC and contribute to many of its functions (10). Many nucleoporins form subcomplexes, and they collectively afford the structural integrity of the NPC and its assembly and disassembly during mitosis in higher eukaryotes, as well as playing a functional role in nucleocytoplasmic transport (60, 65).
Immunoelectron microscopy (EM) studies using a variety of techniques and antibodies have revealed ultrastructural localizations of nucleoporins within the NPC (for example, see reference 63). These localizations can be used to explain how certain substructures of the NPC contribute to specific functions. Definitive localization of nucleoporins has been important for developing models to explain selective translocation through the NPC (8, 51, 54). For example, members of a subset of nucleoporins containing FG repeats are thought to generate a hydrophobic barrier at the NPC, permeable only to transport-competent macromolecules, which suggests that they are localized at accessible regions of the NPCs, lining the translocation route (51, 54).
Three vertebrate nucleoporins are reported to localize exclusively to the cytoplasmic face of the NPC, Nup214/CAN, Nup88, and Nup358/RanBP2. Nup214 has been localized close to the midplane of the NE, possibly as a component of the cytoplasmic ring (34, 64), and interacts with Nup88 to form a stable subcomplex (6, 20, 41). The mechanism for targeting this Nup88-Nup214 subcomplex to the NPC during nuclear assembly apparently requires both proteins, since depletion of Nup214 from mouse embryos caused mislocalization of Nup88 from the NPC, and the Nup214 interaction domain of Nup88 expressed in BHK cells mislocalized Nup214 to the cytoplasm (6, 18, 20). Nup88 is present at an estimated 32 copies/NPC, compared to only 8 copies of Nup214 (10), and the ultrastructural localization of Nup88 at the NPC is currently unknown. Nup358/RanBP2 is localized to the cytoplasmic filaments of the NPC (64, 67, 69). Transmission electron microscopy (TEM) analysis of purified Nup358/RanBP2 revealed a ∼36-nm filamentous structure, and depletion of Nup358 from Xenopus egg extracts caused assembly of NPCs lacking detectable cytoplasmic filaments, indicating Nup358 as a major, and possibly the only, nucleoporin constituent of these filaments (12, 64). No nucleoporin binding partners have been found for Nup358; therefore, the molecular association of the cytoplasmic filaments with the NPC is unknown. Nup88 and also Nup214 represent possible candidates, although in vitro-assembled Nup214-depleted NPCs did have cytoplasmic filaments (64).
Soluble transport receptors are carriers that mediate the active transport of macromolecules through the NPC (23, 60). Biochemical studies have shown that for groups of transport substrates there is a specific transport receptor that utilizes a subset of nucleoporins to translocate the NPC (46). Many nucleoporins have been shown to bind certain transport receptors in vitro, providing primary indication of their roles in specific transport pathways. However, the precise roles of these proteins in vivo largely remain to be determined, and in vertebrates only a few nucleoporins have been shown to play dominant roles in specific transport pathways using model systems, such as in vitro nuclear assembly of Xenopus egg extracts, the use of antibodies, or overexpression in cultured cells and knockout mice (for example, see references 5, 58, and 63). Nup214 has been shown to interact with several import receptors in vitro (45, 68); however, depletion of Nup214 from Xenopus egg extracts resulted in assembly of synthetic nuclei still capable of nuclear protein import (64), illustrating that biochemical evidence is not necessarily indicative of an important functional role. With the advent of new techniques, including RNA interference (RNAi), more direct functional roles of nucleoporins in specific nucleocytoplasmic transport pathways in vertebrates can be investigated, as has been demonstrated for the role of the Nup107 nucleoporin subcomplex in NPC assembly (26, 61).
Proteins to be exported from the nucleus, including transcription factors and certain shuttling proteins, carry a short and hydrophobic nuclear export signal (NES), which was originally discovered in human immunodeficiency virus type 1 (HIV-1) REV and PKI (16, 66). CRM1 is the transport receptor that recognizes NES-containing substrates (1, 19, 21, 48, 56), and it belongs to a group of export receptors or exportins that bind their substrates with RanGTP in the nucleus (3, 19, 31, 37, 38, 57). Like other nuclear transport receptors, CRM1 is thought to interact directly with specific nucleoporins at the NPC to mediate transport. Immunoprecipitation studies predict that the most stable interaction of CRM1 at the NPC is with Nup214, and this complex is more stable in the presence of RanGTP and NES substrate (4, 33). Nup358 has also been identified in a complex with CRM1, mediated by the zinc finger domains of Nup358 (55), which suggests a role in NES protein export; however, more direct data supporting an important role in vivo are lacking.
In this study we investigate the organization of the three asymmetrically localized cytoplasmic nucleoporins identified in vertebrates, Nup214, Nup88, and Nup358, in vivo to determine the mechanism of assembly of the cytoplasmic filaments. We have localized Nup88 to a position proximal to Nup358 on the cytoplasmic filaments and Nup214 near the cytoplasmic ring, and we show a novel interaction between Nup88 and Nup358. We show that both Nup88 and Nup214 play a combined role in anchoring Nup358 to the NPC and display an interdependence for their own stability and NPC localization. We also show a functional role for Nup358 in CRM1-mediated protein export, which elaborates and extends earlier biochemical data.
MATERIALS AND METHOD
Antibodies
To generate specific antibodies against Xenopus Nup88 (anti-XNup88), a C-terminal fragment spanning amino acids 312 to 741 of XNup88B was expressed as an N-terminal His6-tagged fusion protein in pRSET A (Qiagen) in BL21(DE3) CodonPlusRIL (Stratagene), isolated from inclusion bodies, and dialyzed against phosphate-buffered saline (PBS)-8.7% glycerol to generate soluble protein. Antibodies were affinity purified and used at 3 μg/ml. To generate antibodies against human Nup214 (anti-hNup214), a cross-linked C-terminal peptide (amino acids 2076 to 2090) was synthesized. Antibodies were raised in rabbits, affinity purified against the antigen cross-linked to an Ultralink iodoacetyl gel (Pierce), and used for Western blotting at 2.5 μg/ml. Anti-hNup358/RanBP2 antiserum, anti-hNup358V, and anti-hNup358F were generously provided by V. Cordes (Karolinska Institute, Stockholm, Sweden) and by A. Gast and F. Melchior (Max Planck Institute for Biochemistry, Munich, Germany), respectively. Anti-hCRM1 (20), anti-hNup88 (BD Transduction Laboratories), anti-CAN9977 (17), monoclonal antibody (MAb) 414 (Eurogentec/Babco), anti-DNA 2C10 antibody, a gift from Yoshiuki Kanai and Tetsuo Kubota (University of Tokyo, Tokyo, Japan), and anti-hNup98 (32) were previously described.
EM immunolocalization
Nup88 immunolocalization on isolated Xenopus oocyte NEs using TEM and in field emission scanning EM (FESEM) was carried out as previously described (63). Briefly, antibodies were diluted 1:100 with PBS and incubated with isolated Xenopus oocyte NEs. The primary antibody was labeled with a 1:20 dilution of 10-nm-gold-conjugated anti-rabbit secondary antibodies (Amersham), and the samples were prepared for analysis by FESEM or TEM. Negative controls minus primary antibody were performed and revealed the secondary antibodies to be specific (data not shown). For FESEM analysis, isolated NEs were visualized at magnification ×100,000 to ×300,000, and the positions of gold-labeled antibodies were measured in relation to the center of the NPC. For TEM quantification, the distance of the gold particles from the midplane of the NE was determined. All measurements were calculated using AnalySIS software (SIS, Munster, Germany).
Xenopus egg extracts and immunoprecipitation
Fractionated Xenopus egg extracts were prepared as previously described (63). For dephosphorylation of Nup88, 400 U of Lambda protein phosphatase (New England Biolabs) was added to 50 μl of Xenopus egg extract according to the manufacturer's instructions and incubated at 30°C for 15 min. For immunoprecipitation, extracts were diluted 1:4 in binding buffer (200 mM NaCl, 20 mM HEPES-KOH [pH 7.9], 1 mM β-mercaptoethanol, Complete protease inhibitor cocktail [Roche], 8.7% glycerol) and incubated with 4 μg of antibody for 1 h at 4°C. Ten microliters of bed-volume protein A-Sepharose beads (Pharmacia Biotech) were added and incubated for 1 h at 4°C. Beads were collected by centrifugation, washed three times in binding buffer and once in binding buffer supplemented with 500 mM NaCl, and eluted in 2% sodium dodecyl sulfate (SDS) or 0.2% and 2% SDS. Samples were mixed with SDS protein sample buffer, boiled for 5 min, and analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting.
Cell culture
HeLa cells (ATCC CCL-2) and MCF-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (GibcoBRL) and antibiotics at 37°C and 5% CO2 in a humidified incubator. Xenopus A6 cells were cultured in L-15 (LEIBOVITZ) medium with Glutamax-1 W/L-amino acids (Gibco) supplemented with antibiotics at 20°C in a humidified incubator.
RNAi of nucleoporins
The oligonucleotides used for silencing of Nup88 (TGCTTTGTTGAACACATCC), Nup214 (TTGCCCAAGGAACGCTCGA), and Nup358 (CGAGGTCAATGGCAAACTA) were purchased from Sigma (United Kingdom) and cloned into the pSUPER vector as previously described (9). Empty pSUPER vector was used as a control. MCF-7 cells or low-passage HeLa cells were transfected at an estimated efficiency of 50 to 95% with either 3 to 4 μg of pSUPER plasmid, using electroporation as described previously (2), or 2 μg of pSUPER plasmid in 6-cm-diameter dishes, using Fugene-6 (Roche), according to the manufacturer's instructions. Forty-eight, seventy-two, and ninety-six hours posttransfection, cells were either fixed for immunofluorescence or lysed directly in boiling SDS sample buffer, and knockdown efficiency was analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting. The nuclear export assay using pRev-GFP was performed as described previously (28), except that 240 ng of plasmid DNA was electroporated along with 4 μg of pSUPER plasmid and expression was allowed to proceed up to 72 h.
Immunofluorescence microscopy and image analysis
Cells were fixed for 15 min in fresh 3.7% formaldehyde and permeabilized with either 0.001% digitonin for 10 min (15 min for Xenopus cells) at room temperature or 0.2% Triton X-100 for 10 min at room temperature. For immunofluorescence, cells were blocked in blocking buffer (1% skimmed milk in PBS) for 15 min at room temperature and incubated in primary antibody diluted in blocking buffer for 3 h at room temperature. Cells were washed three times in blocking buffer and incubated in fluorescently conjugated secondary antibody (Molecular Probes). For double immunolabelings, cells were incubated simultaneously in the two antibodies, except for Nup88 and Nup214 double immunolabeling, where cells were incubated for 2 h in anti-hNup88 (0.6 μg/ml) followed by addition of anti-CAN9977 for 1 h. Cells were washed once in blocking buffer and once in PBS and then mounted in Vectashield (Vector Laboratories). Images were recorded with a Leica TCS SP2 confocal microscope. For quantification of immunofluorescence at the NE, confocal images were analyzed using Image J software. Nuclear rim intensity measurements at four points per cell were averaged and subtracted from the intranuclear intensity and intercellular background. To exclude possible experimental error due to staining variability between samples, non-knocked-down cells in the same imaged field were used as internal controls.
Nucleotide sequence accession numbers
The Xenopus laevis Nup88A and Nup88B mRNA sequences are available under EMBL accession no. AJ617672 and AJ617673, respectively.
RESULT
Nup88 is a phosphorylated protein encoded by two genes in X. laevis and is localized adjacent to Nup358/RanBP2 and Nup214/CAN on the cytoplasmic face of the NPC
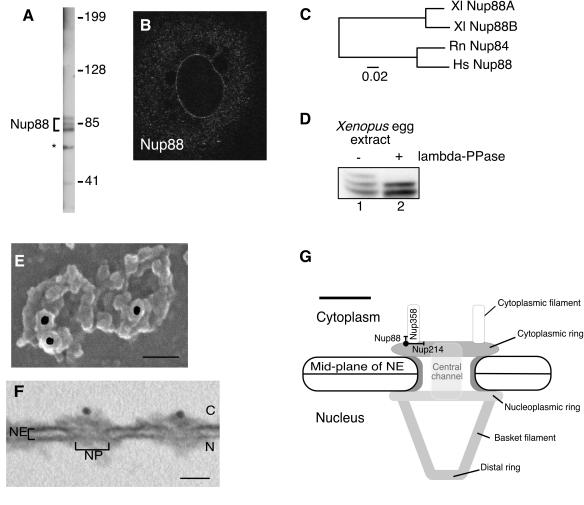
Three vertebrate nucleoporins are known to localize mainly to the cytoplasmic face of the NPC: Nup214, Nup358, and Nup88. Previous studies have determined the ultrastructural localization of Nup214 and Nup358 at the NPC (34, 49, 64, 67, 69). In order to investigate the relative organization of these three nucleoporins, we localized Nup88 on isolated Xenopus oocyte NEs using immuno-gold EM. The sequence of Xenopus Nup88 was extracted from expressed sequence tag (EST) databases using evolutionary conservation to the human and rat Nup88 homologues, and a polyclonal antibody was raised against a purified recombinant C-terminal fragment of Xenopus Nup88 comprising amino acids 312 to 741. After affinity purification, the antibody recognized a pattern of three bands at the approximate molecular weight of Nup88 on a Western blot of Xenopus egg extracts (Fig. 1A). A fourth band was also observed, which was subsequently found to be nonspecific, since it was absent with use of the XNup88 antibody raised in a different animal (not shown). To determine the specificity of the anti-XNup88 antibody in cells, Xenopus A6 cells were fixed, permeabilized, and immunostained with anti-XNup88. A punctate staining of the NE was observed (Fig. 1B), characteristic of nucleoporins, which overlapped with MAb 414 (11), which recognizes the FG repeat-containing nucleoporins Nup358, Nup214, Nup153, and p62 (data not shown).
FIG. 1.
Xenopus Nup88 is encoded by two genes and is phosphorylated, and it localizes adjacent to Nup214/CAN and Nup358/RanBP2. (A) Western blot of Xenopus egg extracts probed with anti-XNup88. The asterisk represents a nonspecific cross-reacting band. (B) Immunofluorescence of Xenopus A6 cells probed with anti-XNup88 to show specific staining of the NE. (C) A phylogram showing divergence of the two genes encoding Nup88 in X. laevis and their conservation with respect to human Nup88 and rat Nup84. (D) Western blot of Xenopus egg extracts before (−) and after (+) treatment with lambda protein phosphatase (lambda-PPase) probed with anti-XNup88. (E) Representative scanning EM of an isolated Xenopus oocyte NE labeled with anti-XNup88, which was secondarily labeled with 10-nm colloidal gold. Bar = 50 nm. (F) Representative TEM of a 70-nm cross-section through an isolated Xenopus oocyte NE labeled with anti-XNup88 and secondarily labeled with 10-nm colloidal gold. N, nucleus; C, cytoplasm. Bar = 50 nm. (G) Summary diagram of the NPC displaying the mean localization of Nup88. Bar = 50 nm. Error bars represent standard deviations of the mean.
X. laevis is a partially tetraploid organism, having duplicated its genome ∼30 million years ago. To determine if the Xenopus Nup88 protein is encoded by two divergent genes, which would partly explain the multiple banding pattern we observe, a more detailed analysis of the Xenopus EST database was undertaken. Two distinct mRNA species were found, encoding protein products homologous to human and rat Nup88 but only 91% identical to each other, too low an identity to be explained by intraspecies variation alone. In addition, the 3′ and 5′ UTR sequences were more divergent than the coding region, which further suggests that Nup88 is encoded by two separate genes in Xenopus. The two genes are designated XlNup88A and XlNup88B and a phylogram shows their evolutionary conservation with respect to human Nup88 and rat Nup84 (Fig. 1C). XlNup88A encodes a predicted protein product of 726 amino acids, compared to 728 amino acids for that of XlNup88B, and the predicted charge of the two proteins was strikingly different, −17.8 for XlNup88A and −9.6 for XlNup88B, both of which could contribute to a difference in electrophoretic mobility.
Many nucleoporins are phosphorylated during mitosis, coinciding with NPC disassembly (15), and phosphorylation also affects electrophoretic mobility of proteins. To further investigate the multiple banding pattern of Xenopus Nup88, Xenopus egg extracts were incubated with a nonspecific protein phosphatase from lambda (λ-PPase) before analysis by gel electrophoresis and Western blot. The three-banded pattern of Nup88 was reduced to two bands, presumably representing the unphosphorylated forms of the two Nup88 proteins (Fig. 1D). These data suggest that Xenopus Nup88 is a phosphorylated nucleoporin and is encoded by two highly homologous but independent genes.
The ultrastructural localization of Nup88 at the NPC was determined by labeling isolated Xenopus oocyte NEs with the anti-XNup88 antibody followed by 10-nm-gold-conjugated secondary antibody. The labeled envelopes were processed for visualization by FESEM to image the surface of the NE and transmission EM (TEM) of 70-nm cross-sections through the NE. Representative micrographs from FESEM and TEM are shown in Fig. 1E and F. Using FESEM, the localization of the gold particles along a radial axis was determined. The mean distance from the center of the NPC was 39 nm ± 17.4 nm (n = 87). Using TEM, the gold particles were measured distally from the midplane of the NE. The mean distance was 30.4 nm ± 7.6 nm (n = 22). The localization data are summarized in Fig. 1G. From these labeling data we can conclude that Nup88 localizes, at least in part, to a position close to both Nup358 and Nup214.
Nup88 is in a complex with both Nup358/RanBP2 and Nup214/CAN
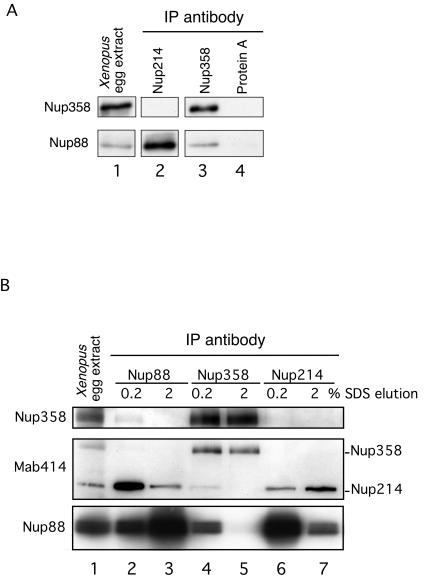
The immunolocalization studies position Nup88, Nup214, and Nup358 in close proximity at the cytoplasmic face of the NPC; however, only Nup214 and Nup88 have been linked biochemically, and nucleoporin binding partners for Nup358 have so far not been identified. To investigate interaction partners of Nup358, we immunoprecipitated Nup358 or Nup214 from fractionated Xenopus egg extracts, isolated the bound protein complexes using protein A-Sepharose after extensive washing, and analyzed the coimmunoprecipitating proteins by gel electrophoresis and Western blotting. Nup88 was found to specifically coimmunoprecipitate with Nup214 (Fig. 2A, lane 2), as has already been shown (6, 17, 18). Interestingly, Nup358 was also able to coimmunoprecipitate Nup88 from Xenopus egg extracts (Fig. 2A, lane 3). To more thoroughly investigate interactions between these three nucleoporins, we labeled immunoprecipitates from Nup88, Nup214, and Nup358 with MAb 414 to detect Nup214 and Nup358, a Nup358-specific antibody, Nup358V, and Nup88. These immunoprecipitates were isolated as above but eluted in two steps with 0.2% and 2% SDS. We were unable to detect Nup358 coimmunoprecipitating with Nup214 (Fig. 2A and B, lanes 6 and 7), even using a Nup358-specific antibody (Fig. 2B, lanes 6 and 7, upper panel). As shown in lane 2, Nup358 was found to be coimmunoprecipitated with Nup88. We could also detect low levels of Nup214 coimmunoprecipitating with Nup358 (Fig. 2B, lane 4). This suggests that Nup88 coprecipitates with Nup358 in part as a complex with Nup214. These data confirm that both Nup214 and Nup358 are interacting partners of Nup88, consistent with their ultrastructural localization.
FIG. 2.
(A) Nup88 is coimmunoprecipitated with both Nup214/CAN and Nup358/RanBP2. Antibodies to Nup214, Nup358, or protein A-Sepharose were incubated with Xenopus egg extract, and coimmunoprecipitating proteins analyzed by labeling a Western blot with anti-Nup358 or anti-XNup88. (B) Antibodies to Nup88 (lanes 2 and 3), Nup358 (lanes 4 and 5), and Nup214 (lanes 6 and 7) were used to coimmunoprecipitate protein complexes from Xenopus egg extracts. Proteins were isolated using protein A-Sepharose beads, and coimmunoprecipitating proteins were eluted using 0.2% (lanes 2, 4, and 6) followed by 2% (lanes 3, 5, and 7) SDS and analyzed by labeling a Western blot with anti-Nup358 (upper panel), MAb 414 (middle panel), and anti-Nup88 (lower panel).
RNAi of Nup88 and Nup214/CAN causes a reduction of Nup358/RanBP2 at the NE
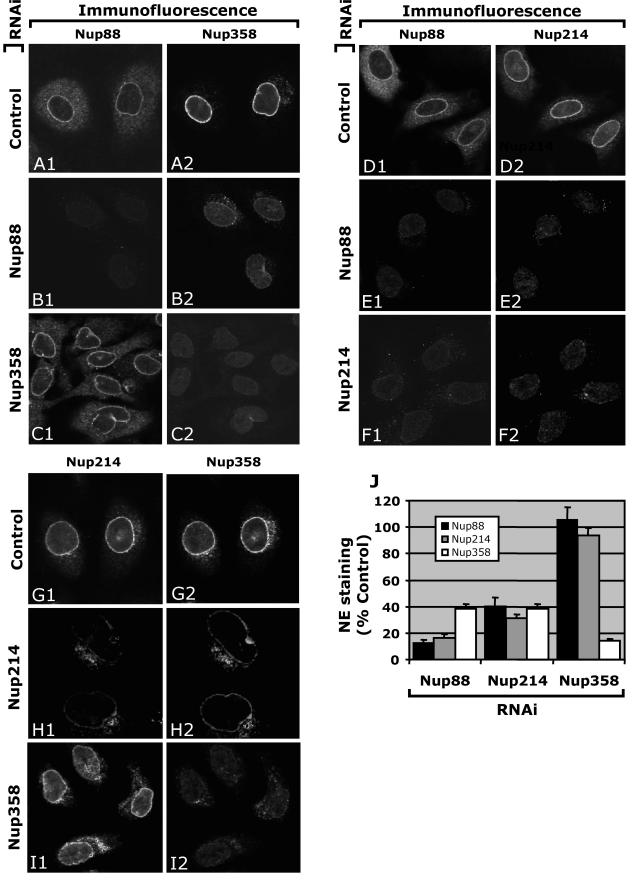
Based on immunoelectron microscopy, Nup358 is the most distal cytoplasmic nucleoporin from the midplane of the NE, apparently located above Nup88 and Nup214. The immunoprecipitation studies predict Nup88 and/or Nup214 as the sites of interaction through which Nup358 may dock to the NPC. To study the organization of Nup88, Nup358, and Nup214, we utilized a technique using small interfering RNAs (13) expressed by the pSUPER vector (9) to reduce endogenous expression of each nucleoporin and analyze the localization of the others using immunofluorescence in human cells. Oligonucleotides containing 19 bases from the mRNA sequences of Nup358, Nup214, and Nup88 were cloned into the pSUPER expression vector as described in Materials and Methods. The pSUPER expression vectors were transfected into HeLa or MCF-7 cells by either electroporation or lipofection. Empty pSUPER vector was transfected as a negative control in all experiments. After 72 h, double immunolabelings were performed, and NE staining intensities of the nucleoporins were quantified in cells which showed clear reduction of the nucleoporin targeted by RNA interference (RNAi) and were compared to the levels in control cells. As shown in Fig. 3, antibodies against Nup88, Nup214, and Nup358 decorated the NE in the punctate manner characteristic of nucleoporins. Cytoplasmic pools of these nucleoporins were also visible at different intensities. Nup88 antibodies showed more dispersed labeling and higher levels of labeling in the cytoplasm than Nup214 or Nup358, whose cytoplasmic staining was more discrete and concentrated in cytoplasmic bodies (Fig. 3A, D, and G). NE staining of each of the three nucleoporins was significantly reduced after transfection with their respective RNAi expression plasmids (Fig. 3B1 and E1 for Nup88 RNAi, F2 and H1 for Nup214, and C2 and I2 for Nup358), which was verified by gel electrophoresis and Western blotting (see Fig. 4). Interestingly, Nup358 NE staining was significantly reduced after RNAi of either Nup88 or Nup214 (Fig. 3B2 and H2, respectively, and J). Conversely, Nup358 RNAi had no effect on Nup88 or Nup214 staining (Fig. 3C1 and I1, respectively, and J); however, RNAi of either Nup88 or Nup214 provoked significant reduction of the other at the NE (Fig. 3E and F, respectively, and J). These data suggest that Nup88 and Nup214 are codependent to incorporate into the NPC, and docking of Nup358 to the NPC in vivo requires the presence of both Nup88 and Nup214.
FIG.3.
Knockdown of Nup88 or Nup214/CAN causes a decrease in Nup358/RanBP2 at the NE. Immunofluorescence of HeLa cells after knockdown of Nup88 (B and E), Nup214 (F and H), and Nup358 (C and I) is shown. Cells were fluorescently double labeled with anti-hNup88 and anti-hNup358F (A, B, and C), anti-hNup88 and anti-CAN9977 (D, E, and F), or anti-CAN9977 and anti-Nup358V antibodies (G, H, and I). The A, D, and G panels show control levels 72 h after transfection with empty pSUPER vector. (J) Graphic representation of the results to show fluorescence levels of Nup88, Nup214, and Nup358 after knockdown of each individual nucleoporin as a percentage of the negative control. Nup358 analysis was performed using two different antibodies, leading to similar results.
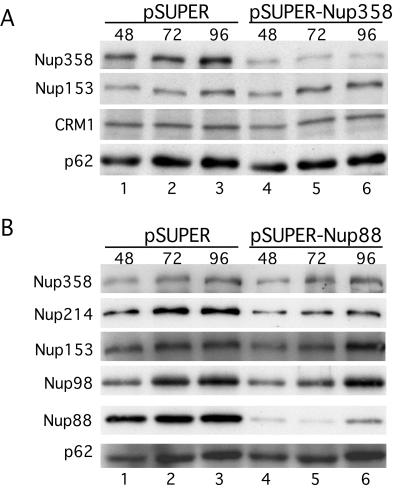
FIG. 4.
Efficient knockdown of Nup88 and Nup358/RanBP2 using RNAi and coreduction of Nup214/CAN on knockdown of Nup88. (A) Western blot of MCF-7 cells transfected by electroporation with pSUPER-Nup358 compared to the pSUPER negative control collected 48, 72, and 96 h posttransfection. The blot was probed with anti-Nup358V, MAb 414, and anti-CRM1. (B) Western blot of HeLa cells transfected by Fugene with pSUPER-Nup88 compared to the pSUPER negative control collected 48, 72, and 96 h posttransfection. The blot was probed with anti-Nup358V, anti-hNup88, anti-hNup214, anti-Nup98, and MAb 414.
Nup88 RNAi causes an associated decrease in the protein levels of Nup214/CAN but not Nup358/RanBP2
The specific reduction of Nup358 at the NE after RNAi of Nup88 or Nup214 could also be the result of a decrease in the stability of Nup358 and its subsequent degradation. To study the effect of nucleoporin RNAi on the protein levels of the remaining untargeted nucleoporins, 48, 72, and 96 h after transfection, cells were lysed directly in SDS sample buffer to minimize breakdown, and the proteins were analyzed by gel electrophoresis and Western blotting. Transfection of cells with either pSUPER-Nup358 (Fig. 4A) or pSUPER-Nup88 (Fig. 4B) results in a clear, specific knockdown of these proteins within 48 h which was stable until the last time point tested at 96 h, confirming the decrease observed by immunofluorescence analysis. Western blots were also labeled with other nucleoporin antibodies, including MAb 414. Nup358 RNAi showed no decrease in the levels of Nup214 or Nup88 at the NE as shown by immunofluorescence (Fig. 3), and there was no associated decrease in the protein levels of either Nup153 or p62 (Fig. 4A). Knockdown of Nup88, however, resulted in a significant decrease in the protein levels of Nup214 (Fig. 4B), indicating that Nup214 is less stable in the absence of Nup88. Importantly, there was no associated decrease in the protein levels of Nup358, indicating that it is the specific attachment of Nup358 to the NPC which is affected by Nup88 RNAi and not its protein level. On knockdown of Nup88, there was no detectable difference in protein levels of either Nup153 or p62, or indeed Nup98, which has also been shown to bind Nup88 in vitro (25).
RNAi of Nup358/RanBP2 mislocalizes CRM1
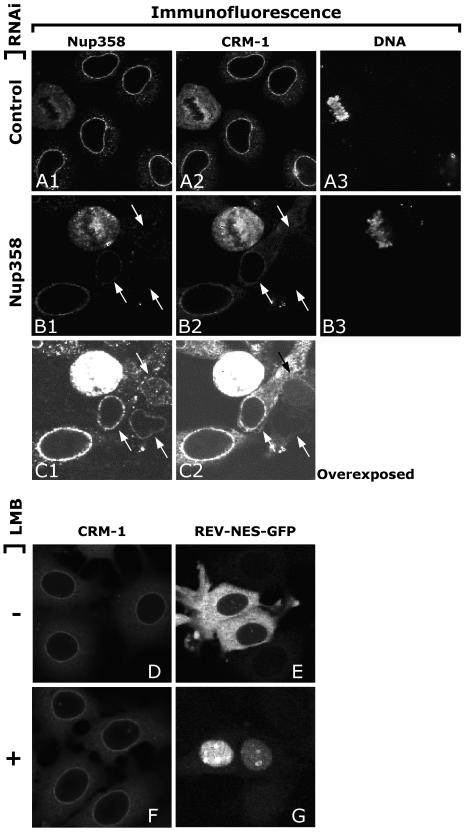
We have shown that Nup358 RNAi specifically reduced its own protein levels without affecting the protein levels of either Nup214, Nup88, or CRM1. Immunostaining of CRM1 in mammalian cells shows that this transport receptor is highly concentrated at the NE (1, 20, 35); however, its binding site(s) is currently unknown and was still present in Nup214-deficient mouse blastocysts (20). In order to investigate the localization of CRM1 at the NPC and a possible role for Nup358 in export, we immunolabeled HeLa cells transfected with either empty pSUPER or pSUPER-Nup358. Seventy-two hours after transfection, cells were fixed and digitonin permeabilized to visualize only the cytoplasmic side of the NE. Triple labeling of Nup358, CRM1, and DNA (to verify the digitonin treatment) was performed, and images were obtained using confocal microscopy. As shown in Fig. 5A, when the NE is intact and DNA antibodies are unable to enter the nucleus, CRM1 is visible at the cytoplasmic side of the NE. However, after Nup358 RNAi, CRM1 accumulation is lost (Fig. 5B2 and C2 [overexposed]), clearly indicating that CRM1 localization at the cytoplasmic side of the NE is dependent on Nup358.
FIG. 5.
CRM1 is mislocalized from the cytoplasmic side of the NE in Nup358 knocked-down cells but not when cargo substrate binding is inhibited. HeLa cells were fixed and permeabilized with 0.001% digitonin 72 h posttranfection with pSUPER (A) or pSUPER-Nup358 (B and C). Antibodies for Nup358, CRM1, and DNA were applied. (B and C) Nup358 knocked-down cells, indicated by arrows, show reduced CRM1 NE staining at the accessible side of the NE. Under these conditions, DNA antibodies can only access dividing cells (A3 and B3). CRM1 localization at the cytoplasmic side of the NE was not altered upon leptomycin B (LMB) treatment of MCF-7 cells (D and F). Treatment was sufficient to abolish CRM1-mediated export of a REV-GFP construct (E and G).
To investigate whether the Nup358-dependent localization of CRM1 at the nuclear periphery represented export complexes or empty CRM1, we treated MCF-7 cells with 100 nM leptomycin B for 3 h. Leptomycin B covalently binds to CRM1 and dissociates it from RanGTP and NES substrates (19, 36, 47). Indeed, leptomycin B efficiently blocked CRM1-dependent nuclear export as a Rev-green fluorescent protein (GFP)-NES substrate accumulated in the nucleus (Fig. 5G). Under these conditions, CRM1 was not reduced at the nuclear periphery (Fig. 5F). Leptomycin B treatment also did not change the reduction of CRM1 upon Nup358 depletion (not shown). These data indicate that the Nup358-dependent CRM1 localization at the nuclear periphery represents CRM1 in its empty state.
Nup358/RanBP2 plays a supportive role in CRM1-mediated NES protein export
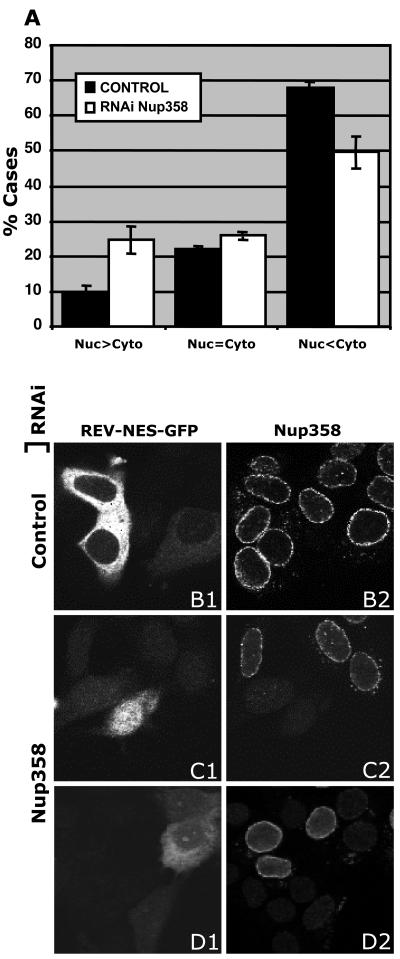
To determine whether loss of CRM1 from the NE after Nup358 RNAi is functionally significant for CRM1-mediated NES protein export, export assays were performed with MCF-7 cells by cotransfecting pSUPER with a GFP-linked export substrate that includes both the NLS and NES of the HIV-1 Rev protein (pRev-GFP [28]). Previous studies have shown that under normal conditions, Rev-GFP partially accumulates at the nucleoli, but on treatment of cells with actinomycin D, the protein is specifically exported and accumulates in the cytoplasm. Cells were cotransfected with pRev-GFP and either pSUPER empty vector or pSUPER-Nup358. Seventy-two hours after transfection, cells were treated with 5 μg of actinomycin D/ml for 3 h and then fixed and prepared for confocal microscopy. Protein export was quantified in three independent experiments by counting the number of cells (n > 100) with predominantly cytoplasmic accumulation of GFP, a predominantly nuclear accumulation, or an even distribution between the nucleus and the cytoplasm. Under control conditions, Rev-GFP was localized predominantly in the cytoplasm in 68% ± 2%, consistent with it being efficiently exported (Fig. 6A and B1). For 22% ± 1% of cells, Rev-GFP was dispersed evenly between the nucleus and the cytoplasm, and the remaining 10% ± 2% of cells showed nuclear accumulation. However, after Nup358 RNAi, the localization of Rev-GFP was significantly redistributed towards the nucleus (Fig. 6A, C1, and D1). Fifty percent ± five percent of cells now accumulated Rev-GFP in the cytoplasm, 26% ± 1% had an even distribution throughout the cell, and 25% ± 4% accumulated Rev-GFP predominantly in the nucleus (Fig. 6A). These results show that Nup358 plays a contributory role in CRM1-mediated export of NES cargoes.
FIG. 6.
Nup358/RanBP2 knocked-down cells are less efficient in CRM1-mediated export of Rev-GFP. (A) Rev-GFP-transfected MCF-7 cells were scored for predominant accumulation of GFP in the cytoplasm (Nuc<Cyto), the nucleus (Nuc>Cyto), or equal distribution (Nuc=Cyto) between the two compartments under control (black bars) or Nup358 RNAi (white bars) conditions. The mean distribution in three independent experiments is shown; error bars represent standard errors. Illustrative images of Nup358 show a strong (C1 and C2) and weak (D1 and D2) export defect. Empty pSUPER-transfected cells are shown in panels B1 and B2.
DISCUSSION
We have investigated the organization of three cytoplasmically orientated nucleoporins, Nup358, Nup88, and Nup214, at the NPC. Previous studies have localized Nup358 and Nup214 to specific NPC substructures (34, 49, 64, 67, 69). Here we immunolocalize Nup88 and study the role of each protein in assembling mature NPCs. We also study in detail the function of the cytoplasmic filaments in CRM1-mediated nuclear protein export.
Characterization of Xenopus Nup88
Nup88 was originally identified as an interacting partner for Nup214 (6, 20). Here we studied Nup88 initially in X. laevis and found it to be encoded by two divergent genes. The two genes, designated XlNup88A and XlNup88B, are 91% homologous, and gene A was found to be more abundant in the EST database, indicating that it is the more common form. We don't currently know which of the two bands identified by the antibody on Western blots of dephosphorylated Xenopus extracts represents which form of Nup88. It is also unclear at present whether the two Nup88 forms possess redundant functions. Xenopus Nup88 was found to be phosphorylated both in meiotic extracts and in extracts which have been induced to enter interphase from meiosis II (data not shown). It therefore appears that Nup88 phosphorylation is not exclusively mitotic or meiotic, as is a common feature for many nucleoporins (15, 39, 44, 62).
Assembly of the cytoplasmic filaments
Using a combination of approaches, we find that the association of Nup358, which is predicted to be a predominant component of the cytoplasmic filaments (12, 64), is dependent on interactions with and between both Nup214 and Nup88, providing evidence of a structural collaboration between these three nucleoporins to assemble mature NPCs. First, we have localized Xenopus Nup88 to a position in close proximity to both Nup214, a known interacting partner, and Nup358, placing it in a position to physically interact with the cytoplasmic filaments, possibly with the N-terminal leucine-rich domain of Nup358 that is suggested to be located at the more central position (64). The localization data were obtained using a combination of TEM and FESEM, since each of the techniques provides a different view of the NPC. Although TEM images potentially provide the ability to measure localization in both a distal and radial direction, only distal data were used because radial distances are influenced by the plane of each 70-nm section through the NPC. We show that Nup88 can be coimmunoprecipitated with both Nup358 and Nup214, indicating that the three proteins do indeed interact in vivo. It remains possible that the Nup88-Nup358 interaction is bridged by additional nucleoporins. The levels of Nup88 that were coimmunoprecipitated with Nup358 were significantly less than with Nup214; however, Nup214 is known to form a soluble and stable subcomplex with Nup88 in the cytoplasm (6, 20), and it is possible that the interaction of Nup358 with Nup88 occurs predominantly at the NPC. To show that the observed interactions and ultrastructural organization of these three proteins are relevant to the organization of the in vivo NPC, we showed, using an RNAi approach, that the association of Nup358 with the NPC was specifically dependent on the presence of both Nup214 and Nup88. This observation is not due to interference with NPC architecture per se, since Nup358 is able to assemble into NPCs after knockdown of Tpr, an unrelated nucleoporin on the nucleoplasmic face of the NPC (27). We also found that localization of Nup214 and Nup88 at the NPC occurred only in the presence of each other, indicating a codependence of NPC localization. This interdependence is consistent with the absence of Nup88 in Nup214 knockout blastocysts (59) and a mislocalization of Nup159, the closest yeast homologue of Nup214, in Nup82-Δ108 cells, which carry a mutation in the yeast homologue of Nup88 (29). The precise mechanism of this interdependence remains to be elucidated, although it may partially be explained by protein stability, since reduction of Nup88 caused a significant decrease in the protein levels of Nup214.
We have previously shown that the cytoplasmic filaments and Nup358 were still present in in vitro-synthesized NPCs that lack Nup214 (64), which appears inconsistent with the results shown here. However, in Xenopus egg extracts there is a large excess of stockpiled nucleoporins, and it is likely that after depletion of Nup214 there remained a significant and stable pool of Nup88 which was able to mediate assembly of the filaments, even in the absence of Nup214.
Nup358/RanBP2 and NES-mediated export
Using an in vivo export assay that was previously used to measure differences in nuclear export signal strength (28), we found a significant decrease in CRM1-mediated nuclear export when expression of Nup358 was substantially reduced, i.e., by 90% or more. In experiments where RNAi was less efficient, hardly any reduction in nuclear export could be observed. This may explain why RNAi of Nup88 did not affect NES-mediated export, since Nup358 levels, even though strongly reduced, were not as low.
Hydrolysis of GTP on Ran is not required for a single round of NES-mediated export (14, 52); however, continued export is predicted to be dependent on recycling of both Ran and CRM1. In addition, it appears that rapid export complex disassembly by RanGTP hydrolysis close to the cytoplasmic face of the NPC is required for overall export efficiency by enhancing the directionality of export (7). Biochemical experiments have indicated that CRM1-NES-RanGTP export complexes bind stably to Nup214 and are disassembled by the combined action of RanGAP and either RanBP1 or RanBP1-like domains of Nup358 (4, 33). A main candidate for providing these activities is Nup358, since it associates with RanGAP, contains RanBP1-like domains, and is located close to the cytoplasmic exit of the NPC (40, 42, 43, 67, 69). The presented results showing a decrease in export due to Nup358 depletion, therefore, provide direct in vivo support of these models. We do not currently have in vivo evidence for which is the terminal binding site of CRM1-containing export complexes. The strong in vitro interaction with Nup214 makes this a likely candidate. However, we do not observe accumulation of export complexes at the nuclear periphery when Nup358 is depleted, suggesting that interaction with Nup214 is at least not rate limiting. Conversely, we find a strong decrease of CRM1 at the nuclear periphery when Nup358 is absent. This Nup358-dependent nuclear rim localization does not represent export complexes because it is not affected by leptomycin B, which is known to dissociate CRM1 from RanGTP and cargo (19, 36). Our data therefore indicate that Nup358 both provides a platform for rapid disassembly of CRM1 export complexes and a binding site for empty CRM1 recycling into the nucleus.
Acknowledgments
We thank Terry Allen and Sandra Rutherford for help with EM, Reuven Agami for sharing RNAi technology, Beric Henserson and Josean Rodriguez for plasmid reagents and advice, Volker Cordes, Frauke Melchior, Yoshiuki Kanai, and Tetsuo Kubota (University of Tokyo, Japan) for antibody reagents, Lauran Oomen and Lenny Brocks for valuable assistance in confocal microscopy, Christos Samakovlis for discussions and communicating unpublished results, and Frauke Melchior, Sebastian Nijman, Judith Boer, Dieuwke Engelsma, and Jolita Hendriksen for critically reading the manuscript.
R.B. is supported by a grant from The Netherlands Science Foundation Earth and Life Sciences. H.P. is supported by a Marie Curie European Community Training and Mobility Fellowship.
REFERENCE
- 1.Adachi, Y., and M. Yanagida. 1989. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kDa protein preferentially localized in the nucleus and its periphery. J. Cell Biol. 108:1195-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agami, R., and R. Bernards. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55-66. [DOI] [PubMed] [Google Scholar]
- 3.Arts, G. J., S. Kuersten, P. Romby, B. Ehresmann, and I. W. Mattaj. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17:7430-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastos, R., L. Ribas-de-Pouplana, M. Enarson, K. Bodoor, and B. Burke. 1997. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J. Cell Biol. 137:989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becskei, A., and I. W. Mattaj. 2003. The strategy for coupling the RanGTP gradient to nuclear protein export. Proc. Natl. Acad. Sci. USA 100:1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, L. I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84:7552-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delphin, C., T. Guan, F. Melchior, and L. Gerace. 1997. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell 8:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 14.Englmeier, L., J. C. Olivo, and I. W. Mattaj. 1999. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 9:30-41. [DOI] [PubMed] [Google Scholar]
- 15.Favreau, C., H. J. Worman, R. W. Wozniak, T. Frappier, and J. C. Courvalin. 1996. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry 35:8035-8044. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Lührmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod, M., J. Boer, S. van-Baal, M. Jaeglé, M. von-Lindern, K. G. Murti, D. Davis, J. Bonten, A. Buijs, and G. Grosveld. 1995. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 10:1739-1748. [PubMed] [Google Scholar]
- 18.Fornerod, M., J. Boer, S. van-Baal, H. Morreau, and G. Grosveld. 1996. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene 13:1801-1808. [PubMed] [Google Scholar]
- 19.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 20.Fornerod, M., J. van-Deursen, S. van-Baal, A. Reynolds, D. Davis, K. G. Murti, J. Fransen, and G. Grosveld. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg, M. W., and T. D. Allen. 1993. The nuclear pore complex: three-dimensional surface structure revealed by field emission, in-lens scanning electron microscopy, with underlying structure uncovered by proteolysis. J. Cell Sci. 106:261-274. [DOI] [PubMed] [Google Scholar]
- 23.Görlich, D. 1998. Transport into and out of the cell nucleus. EMBO J. 17:2721-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 25.Griffis, E. R., S. Xu, and M. A. Powers. 2003. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell 14:600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harel, A., A. V. Orjalo, T. Vincent, A. Lachish-Zalait, S. Vasu, S. Shah, E. Zimmerman, M. Elbaum, and D. J. Forbes. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11:853-864. [DOI] [PubMed] [Google Scholar]
- 27.Hase, M. E., and V. C. Cordes. 2003. Direct interaction with nup153 mediates binding of tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14:1923-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 29.Hurwitz, M. E., C. Strambio-de-Castillia, and G. Blobel. 1998. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc. Natl. Acad. Sci. USA 95:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarnik, M., and U. Aebi. 1991. Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 107:291-308. [DOI] [PubMed] [Google Scholar]
- 31.Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang, and E. K. O'Shea. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396:482-486. [DOI] [PubMed] [Google Scholar]
- 32.Kasper, L. H., P. K. Brindle, C. A. Schnabel, C. E. J. Pritchard, M. L. Cleary, and J. M. A. van Deursen. 1999. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19:764-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraemer, D., R. W. Wozniak, G. Blobel, and A. Radu. 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA 91:1519-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudo, N., S. Khochbin, K. Nishi, K. Kitano, M. Yanagida, M. Yoshida, and S. Horinouchi. 1997. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J. Biol. Chem. 272:29742-29751. [DOI] [PubMed] [Google Scholar]
- 36.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 37.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Görlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipowsky, G., F. R. Bischoff, P. Schwarzmaier, R. Kraft, S. Kostka, E. Hartmann, U. Kutay, and D. Görlich. 2000. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19:4362-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macaulay, C., E. Meier, and D. J. Forbes. 1995. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J. Biol. Chem. 270:254-262. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 41.Matsuoka, Y., M. Takagi, T. Ban, M. Miyazaki, T. Yamamoto, Y. Kondo, and Y. Yoneda. 1999. Identification and characterization of nuclear pore subcomplexes in mitotic extract of human somatic cells. Biochem. Biophys. Res. Commun. 254:417-423. [DOI] [PubMed] [Google Scholar]
- 42.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, M. W., M. R. Caracciolo, W. K. Berlin, and J. A. Hanover. 1999. Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367:51-60. [DOI] [PubMed] [Google Scholar]
- 45.Moroianu, J., G. Blobel, and A. Radu. 1995. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92:2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 47.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 49.Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichelt, R., A. Holzenburg, E. L. Buhle, Jr., M. Jarnik, A. Engel, and U. Aebi. 1990. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 110:883-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribbeck, K., U. Kutay, E. Paraskeva, and D. Görlich. 1999. The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr. Biol. 9:47-50. [DOI] [PubMed] [Google Scholar]
- 53.Ris, H., and M. Malecki. 1993. High-resolution field emission scanning electron microscope imaging of internal cell structures after Epon extraction from sections: a new approach to correlative ultrastructural and immunocytochemical studies. J. Struct. Biol. 111:148-157. [DOI] [PubMed] [Google Scholar]
- 54.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 55.Singh, B. B., H. H. Patel, R. Roepman, D. Schick, and P. A. Ferreira. 1999. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J. Biol. Chem. 274:37370-37378. [DOI] [PubMed] [Google Scholar]
- 56.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 57.Ullman, K. S., M. A. Powers, and D. J. Forbes. 1997. Nuclear export receptors: from importin to exportin. Cell 90:967-970. [DOI] [PubMed] [Google Scholar]
- 58.Ullman, K. S., S. Shah, M. A. Powers, and D. J. Forbes. 1999. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10:649-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van-Deursen, J., J. Boer, L. Kasper, and G. Grosveld. 1996. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 15:5574-5583. [PMC free article] [PubMed] [Google Scholar]
- 60.Vasu, S. K., and D. J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363-375. [DOI] [PubMed] [Google Scholar]
- 61.Walther, T. C., A. Alves, H. Pickersgill, I. Loiodice, M. Hetzer, V. Galy, B. B. Hulsmann, T. Kocher, M. Wilm, T. Allen, I. W. Mattaj, and V. Doye. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113:195-206. [DOI] [PubMed] [Google Scholar]
- 62.Walther, T. C., P. Askjaer, M. Gentzel, A. Habermann, G. Griffiths, M. Wilm, I. W. Mattaj, and M. Hetzer. 2003. RanGTP mediates nuclear pore complex assembly. Nature 424:689-694. [DOI] [PubMed] [Google Scholar]
- 63.Walther, T. C., M. Fornerod, H. Pickersgill, M. Goldberg, T. D. Allen, and I. W. Mattaj. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walther, T. C., H. S. Pickersgill, V. C. Cordes, M. W. Goldberg, T. D. Allen, I. W. Mattaj, and M. Fornerod. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 66.Wen, W., A. T. Harootunian, S. R. Adams, J. Feramisco, R. Y. Tsien, J. L. Meinkoth, and S. S. Taylor. 1994. Heat-stable inhibitors of cAMP-dependent protein kinase carry a nuclear export signal. J. Biol. Chem. 269:32214-32220. [PubMed] [Google Scholar]
- 67.Wu, J., M. J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209-14213. [DOI] [PubMed] [Google Scholar]
- 68.Yaseen, N. R., and G. Blobel. 1997. Cloning and characterization of human karyopherin beta3. Proc. Natl. Acad. Sci. USA 94:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, and U. Aebi. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]