Abstract
Despite the recent advances in single molecule manipulation techniques, purely mechanical approaches can not detect subtle conformational changes in the biologically important regime of weak forces. We developed a hybrid scheme combining force and fluorescence which allowed us to examine the effect of sub-pN forces on the nanometer scale motion of the Holliday junction (HJ) at 100 Hz bandwidth. The HJ is an exquisitely sensitive force sensor whose force response is amplified with an increase in its arm lengths, demonstrating a lever-arm effect at the nanometer length scale. Mechanical interrogation of the HJ in three different directions helped elucidate the structures of the transient species populated during its conformational changes. This method of mapping two dimensional reaction landscapes at low forces is readily applicable to other nucleic acid systems and their interactions with proteins and enzymes.
Many biological processes are dependent on tension. In recent years, single molecule force measurements have shown directly that biochemical reactions can be influenced by applied force (1). Yet, purely mechanical tools can not detect small scale conformational changes unless strong enough and persistent force is applied. At weak forces, the flexible tether connecting the mechanical probe to the biological molecule is not stretched enough to transmit small movements. This is unfortunate because weak and transient forces are likely more prevalent in vivo but the experimental limitations confine single molecule mechanical studies to examining the effect of relatively large forces. We aimed to study the effect of weak external forces on the biomolecular conformational dynamics by combining single molecule fluorescence resonance energy transfer (smFRET) (2–4) with manipulation using optical trap (5). smFRET has high spatial resolution (≤ 5 Å) (6, 7)) and can be measured at arbitrarily low forces. Previous attempts to combine FRET and optical trap using the DNA hairpin as a model system (8, 9) did not reveal new information because the hairpin unzips at high forces (~ 15 pico Newton (pN)), a regime that had been extensively investigated using force-based techniques (10, 11). Here, we report an approach to detect nanometer-scale motion at sub-pN forces. We used the approach to gain insight into the reaction landscape of the Holliday junction (HJ) by gently stretching it along different directions.
The HJ is a four-stranded DNA structure that forms as an intermediate during recombination (12). To understand the mechanisms of cellular enzymes that function with the HJ, a detailed description of the static and dynamic structural properties of the HJ itself is needed. In the absence of added ions the HJ adopts an open structure, where the four helical arms point toward the corners of a square (13, 14) (Fig. 1A). In the presence of physiological concentrations of magnesium ions, the HJ becomes more compact by pairwise coaxial stacking of helical arms into a right-handed antiparallel stacked-X structure(13–15). There are two ways of forming this stacked structure that depend upon the choice of helical stacking partners (isoI and isoII) (Fig. 1B). For these studies, we have chosen a sequence with nearly equal population of stacking conformers isoI and isoII (16) (17) (Fig. S1). smFRET studies showed that a HJ continually switches between the two stacking conformations (18). At present, there is no structural information on the transient species populated during these conformational changes.
Fig. 1.
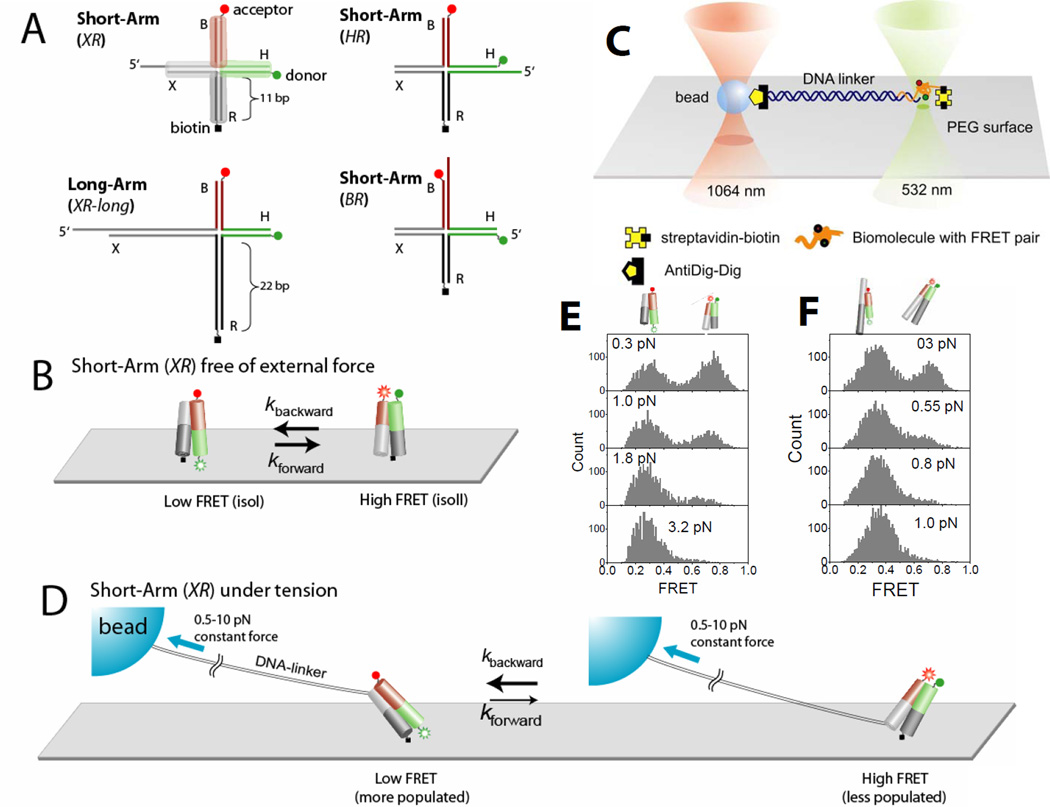
Holliday junction constructs and experimental scheme. (A) The HJ species studied. Junction XR comprises four arms of 11 bp, termed B (red), H (green), R (dark gray) and X (gray). Cy3 and Cy5 fluorophores are terminally attached to H and B arms respectively, and the molecule is tethered to the surface through biotin attached to the end of the R arm. Stretching force is applied through the λ-DNA linker hybridized to the X arm. In junction XR-long the lengths of arms R and X are increased to 21 bp. In junction HR the λ-DNA linker is hybridized to the H arm. In junctions HR and BR the λ-DNA linker is hybridized to the H and B arms respectively. (B) Junction XR is known to alternate between two different stacking conformers, isoI (Low FRET) and isoII (High FRET) with similar populations in both states. (C) A surface-immobilized biomolecule with FRET labeling is connected to a trapped bead via a long DNA linker. The linker DNA spatially separates the confocal beam (532 nm) from the trapping beam (1064 nm) such that enhanced photobleaching and an overwhelming background signal induced by the intense trapping laser are avoided. To apply force, the surface immobilized molecule was moved relative to the trapped bead. The confocal beam was programmed to follow the motion of the molecule using the mapping generated between sample scanning and beam scanning (Fig. S6). (D) Force is expected to bias the junction XR to isoI which possesses a larger separation between the two tether points than isoII. (E) FRET histograms of a single junction XR as a function of force. (F) FRET histograms of a single junction XR-long as a function of force.
To investigate the nature of such transient HJ structures and to understand how HJ conformational properties could depend on physiologically relevant forces, we built a hybrid instrument that combines smFRET with optical trapping via a long linker (bacteriophage λ DNA)(17). The trapping and fluorescence excitation beams in our confocal microscope are spatially separated (minimum 13 µm, Fig. 1C) such that fluorescence and force processes can operate without mutual interference. The long linker acts as a loose spring that dampens the random forces generated by Brownian motion of the trapped bead and reduces force variations due to the nanometer-scale conformational change of the HJ. The effective stiffness of the λ DNA at 2 pN of force is about 0.002 pN/nm such that 5 nm movement of the HJ causes negligible force fluctuations (~0.01 pN) at the trapped bead. Therefore, the measurements can be performed under effectively constant force without the need for active force clamping. The relaxation time scales of the λ DNA under tension are faster than the time scale of conformational fluctuations we investigate here(19). The trapping beam (1,064 nm) was fixed along the optical axis of the microscope, and force was applied by moving the surface-tethered HJ using a piezoelectric sample scanner. The confocal excitation beam (532 nm) was programmed to follow the HJ using a piezo-controlled mirror to maintain uniform excitation and detection efficiencies regardless of the specimen location (and therefore force) (17)
To determine comprehensively the force response of the HJ, we used the following four constructs (Fig. 1A). The four helices comprising the HJ are named B (red), H (green), R (dark gray), and X (gray). Helix R was labeled at its 5’ terminus with biotin for surface immobilization, and helices X, H, or B were extended by a 12 nt ssDNA 5’-overhang to permit annealign to a cohesive end of λ-DNA (named junctions XR, HR and BR respectively). The other end of the λ-DNA was attached to a bead via digoxigenin/anti-digoxigenin coupling in order to pull on the DNA using optical tweezers in three different directions, between X and R arms for junction XR etc. Junctions XR and XR-long differ in the length of the X and R arms (11 bp vs. 21 bp). Cy3 (FRET donor), was attached to the 5’-terminus of helix H, and Cy5 (acceptor) to the 5’-end of helix B. For junctions XR and XR-long, the stretching force should favor isoI (low FRET), in which there is a larger separation between the two tether points, over isoII (high FRET) (Fig. 1D). Indeed, single molecule FRET histograms as a function of force show that the low FRET state is significantly favored at forces exceeding 0.5 and 1.0 pN for junctions XR-long and XR respectively (Fig. 1E and 1F). Likewise, isoII (high FRET) would be favored at high forces for junction HR. In contrast, the two tether points would have similar distances for isoI and isoII in the case of junction BRand force-induced bias should be minimal.
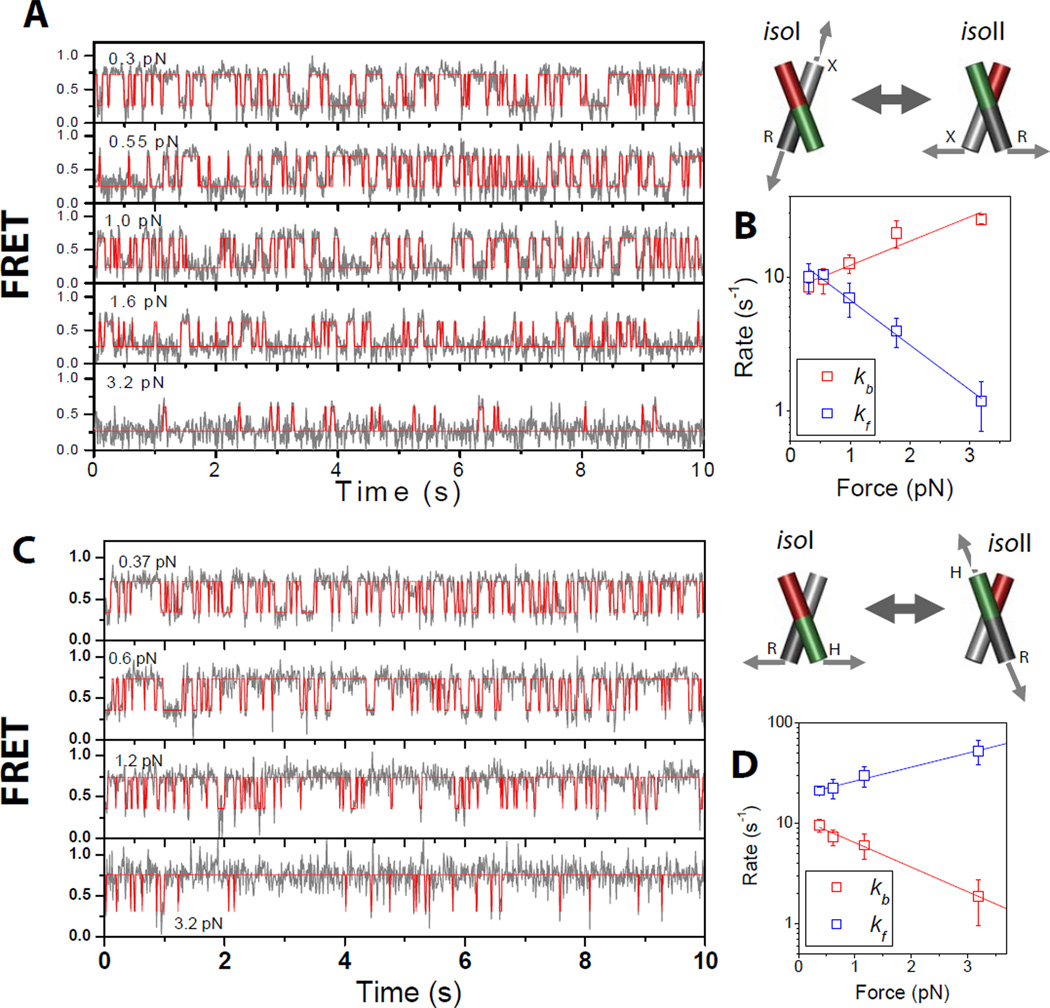
Fig. 2A shows smFRET time traces at five different forces (gray lines, 10 s duration each with 10 ms integration time) obtained from a single molecule of junction XR. Enhanced photostability by means of the use of Trolox (20) allowed us to obtain one to five cycles of force data from a single molecule before photobleaching, corresponding to observation over 50 to 250 s. Idealized FRET trajectories generated by hidden Markov modeling (red lines) (21) are also shown. At the lowest force (0.3 pN), the junction switches between the high and low FRET states with similar populations. As the force exceeds 1 pN, the dynamics become clearly biased to the low FRET state. Fig. 2B shows the transition rates determined from hidden Markov modeling as a function of force. The transition rate kf for the forward reaction from the low FRET state (isoI) to the high FRET state (isoII) decreases with increasing force (blue), while the transition rate for the backward reaction kb (isoII to isoI) increases with force (red) as expected. Both changes were linear in the log-linear scale but interestingly, kf had twice the slope of kb. If the reaction is viewed as possessing a single transition state, the slope reflects the distance to the transition state (1). Therefore, the transition state lies closer to isoII than to isoI when force is applied via the XR vector. Averaged over five molecules, the distance from isoII to the transition state (Δxb‡) is 1.5 ± 0.3 nm and the distance from isoI to the transition state (Δxf‡) is 2.9 ± 0.6 nm (Table 1).
Figure 2.
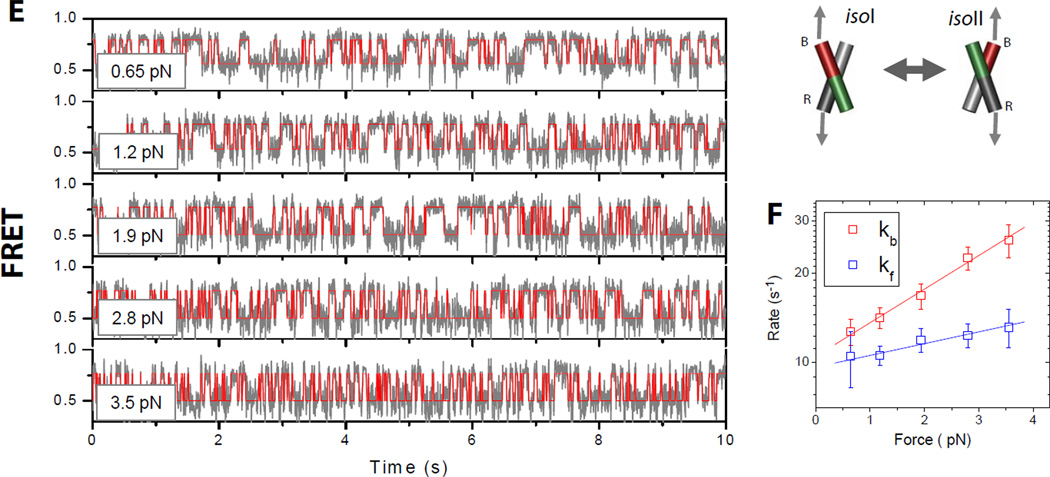
Conformer exchange dynamics of the HJ as a function of applied force. (A) FRET time traces (gray lines) of a single junction XR molecule at different forces. FRET efficiency is approximated by the acceptor intensity divided by the sum of the donor and acceptor intensities. Red lines are the most likely FRET trajectories generated via hidden Markov modeling. The imposed force (indicated on the top left of each plot) increases top to bottom. (B) Log-linear plot of rate constants of conformer exchange as a function of force. Rates of transitions from states isoII to isoI (kb red) and isoI to isoII (kfblue) are differentiated by color. Error bars represent standard deviations obtained from repeated measurements of the same molecule. From linear fitting, we found that the transition state is closer to isoII (1.8 nm) than to isoI (3.3 nm). (C) Same as (A) but for a single junction HR molecule. (D) Same as (B) for a single junction HR molecule. (E) Same as (A) and (C) but for a single junction BR molecule. (F) Same as (B) and (D) but for a single junction BR molecule.
Table 1.
Distance to the transition state from isoI (Δxf‡) and from isoII (Δxb‡) measured from the force-dependent transition rates between isoI and isoII for four different junctions. Errors represent standard deviation from five different molecules each. Also shown is Δxeq, the distance between isoI and isoII determined from force-dependent changes in the equilibrium constant. For junction BR, Δxeq deviates significantly from (Δxb‡+Δxf‡) showing that dBR is not a valid reaction coordinate connecting isoI and isoII. In contrast, Δxeq=Δxb‡+Δxf‡ within error for junctions XR and HR showing that dXR and dHR are reaction coordinates valid from isoI to isoII.
| XR | XR-long | HR | BR | |
|---|---|---|---|---|
| Δxb‡ (nm) | 1.5 (±0.3) | 2.6 (±0.6) | 2.4 (±0.5) | 1.1 (±0.2) |
| Δxf‡ (nm) | 2.9 (±0.6) | 7.7 (±1.5) | 1.3 (±0.3) | 0.37 (±0.2) |
| Δxeq (nm) | 4.4 (±0.8) | 9.9 (±2.6) | 3.1 (±0.8) | 0.7 (±0.2) |
| Δxb‡+Δxf‡ (nm) | 4.4 (±0.8) | 10.3 (±2.0) | 3.6 (±0.5) | 1.5 (±0.3) |
We next studied junction HR where the λ DNA tether has been transferred from the X to the H arm. In this construct, the force is expected to bias the HJ to the high FRET isoII state, and indeed this was the result (Fig. 2C). kb decreased and kf increased with stronger forces, but with two-fold higher slope for kb than for kf (Fig. 2D). Averaged over five molecules, Δxb‡=2.4 ± 0.5 nm and Δxf‡=1.3 ± 0.3 nm. In both junctions, (Δxb‡+Δxf‡) is equal to the distance between isoI and isoII, Δxeqcalculated from equilibrium population vs. force data (Table I). Therefore, the distances between the ends of the pulled arms, dXR for junction XR and dHR for junction HRare suitable reaction coordinates spanning the complete trajectory from isoI to isoII (Fig. 3A).
Figure 3.
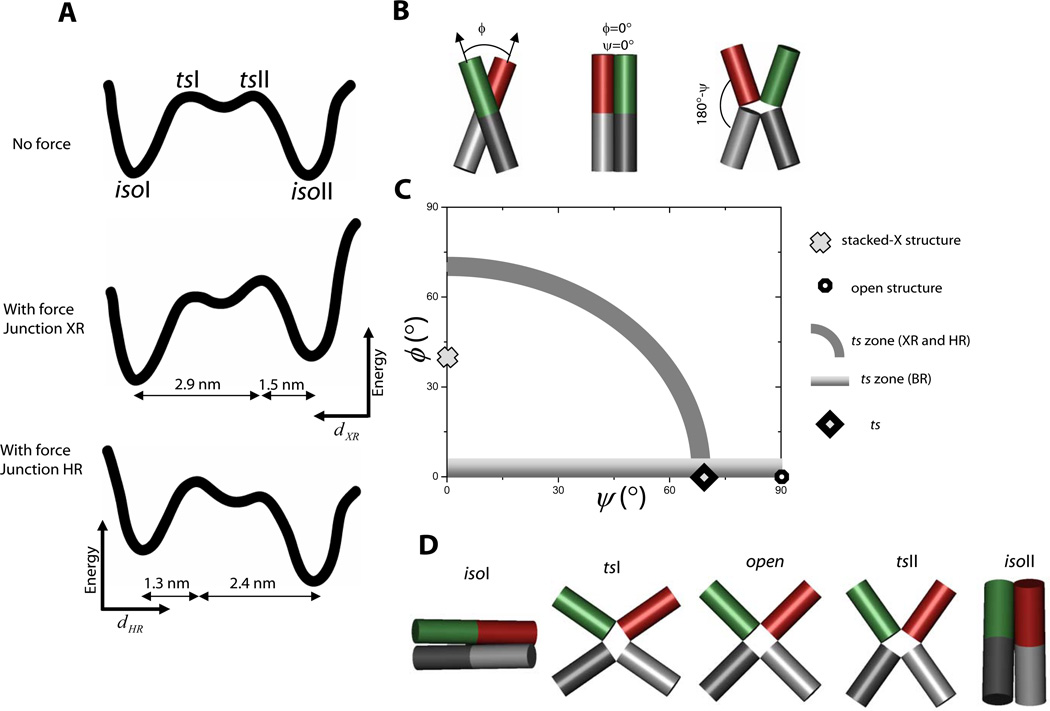
Mapping the reaction landscape and determining the transition state structure. (A) A proposed reaction landscape with two distinct transition states with nearly identical energies (top). In junction XRthe applied force would tilt the energy landscape toward isoI so that the transition state, tsII, nearer to isoII would become the state of highest energy along the entire coordinate (middle). The reaction coordinate here is the distance between the ends of X and R arms, dXRwhich increases to the left as shown. Similarly, in junction XRthe transition state, tsI, nearer to isoI would become the single transition state upon application of force. The reaction coordinate here is the distance between the ends of H and R arms, dHRwhich increases to the right. (B) Two angular coordinates ϕ and ψ define the global conformation of the HJ. (C) Two-dimensional conformational space of HJ conformations. The stacked-X structure and open structure are marked. The gray arc represents a zone that satisfies experimental constraints derived from XR and HR data, and the gradient zone is derived from BR data. The consensus location of the transition state is marked with a diamond. (D) Global structures of isoI, isoII and two transition states, tsI and tsII, plus an open structure.
In one pulling direction represented by dXRthe transition state lies closer to isoII (Fig. 3A, middle panel) while for the other pulling direction along dHRthe transition state more closely resembles isoI (Fig. 3A, bottom panel). These two transition states can not represent a single structure because then both dXR and dHR must be relatively small, and by symmetry so must be dXB and dHB. Such a structure would have all four helices in the same hemisphere relative to the junction core which is highly unlikely considering the symmetry of the HJ. Instead, we favor a model where there are at least two different transition states, tsI and tsII, equal in energy but corresponding to different values of dXR (or dHR), such that force would elevate one of them into the single highest energy barrier via the tilting of the energy landscape (Fig. 3A).
The data presented so far show that the distance change upon stacking conformer transitions is about 4 nm. Since thermal energy is about 4 pN-nm, a force on the order of 1 pN would consequently change the equilibrium between the two states by 2–3 fold. Such small scale conformational fluctuations at these low forces are probably impossible to detect in a purely mechanical measurement, especially at our time resolution (10 ms).
What determines the force sensitivity of the junction? Is it an intrinsic property of the junction core or is it dependent on the length of helical arms on which the force is applied? Since the four arms of the HJ meet at its center, we may recast the experimental configuration as a torque being applied around the central pivot point. The torque is proportional to the product of the magnitude of force and the distance between the point of application of the force and the pivot point (i.e., the length of the arm). Therefore, it could be expected that increasing arm length would result in a greater torque for the same force. We tested such a lever arm effect using junction XR-long, where the X and R arms are lengthened by about a factor of two (from 11 bp to 21 bp) compared to junction XR. FRET histograms as a function of applied force (Fig. 1E and Fig. 1F) show that increasing the lever arm length has magnified the force effect such that much lower force is needed for junction XR-long to achieve the same conformational bias. Fig. S4 compares the transition rates vs. force between five molecules each of junctions XR and XR-long each and shows that junction XR-long exhibits much greater changes in rates for the same magnitude of force (also compare Δxf‡ and Δxb‡ in Table I). Since the persistence length of double stranded DNA is about 50 nm (~ 150 bp) (22) the lever arm effect can probably be extended by another factor of five for arms of ≥ 100 bp. That is, forces as low as 0.1 pN would be enough to influence the junction conformations, illustrating the exquisite force sensitivity of the HJ.
Since the effect of force depends on the arm lengths, the most natural reaction coordinates are angular. The angles that define the global shape of the junction are ϕ, the interhelical angle between two stacked pairs of helices, and ψ, the angle that measures the degree of unstacking of stacked helices (23) (Fig. 3B). For example, for a stacked-X structure ϕ=40° and ψ=0° (15), while for an open structure, ϕ is 0° and ψ is 90° (Fig. 3C). These two angles are well-defined within the angular space in which identities of stacking pairs are maintained. Our aim here is to deduce the structure of the transition state by determining the ϕ and ψ values of the transition state using a geometrical analysis. The analysis below estimates the angles (ϕIIψII) of the transition state tsII in the isoII half of the conformational reaction coordinate, but the same conclusions hold for tsI.
tsII lies a third of the way from isoII to isoI along the dXR coordinate (Table 1, Fig. 3A). We can show that this condition is satisfied for a collection of (ϕIIψII) values, starting from (70°, 0°) at one extreme and arriving at (0°, 70°) at the other (Fig. 3C, gray zone) (18). In order to obtain an additional constraint, we performed an equivalent force analysis on junction BR (Fig. 2E, 2F, Table 1). Junction BR exhibited much reduced (by 5–6 fold) force dependence of the equilibrium populations compared to junctions XR and HR (compare Δxeq values in Table 1). The residual force dependence of the equilibrium populations may be attributed to the finite diameter of the DNA duplex (18). In contrast to junctions XR and HRapplication of force on junction BR accelerated both forward and backward transitions (Fig. 2F). Therefore, the distance between the ends of B and R arms, dBR, must be larger in the transition state than in the stacked-X structures. This condition is satified only if ϕII in the transition state is smaller than the 40° of the stacked-X structure. Furthermore, the distance to the transition state is 0.37 nm at minimum which constrains ϕII to be essentially zero (18). In combination, our best estimate is (ϕIIψII)ts= (0°, 70°) for tsII (Fig. 3C). This transition state is similar to the open state, but with arms deviating by about 20° from the ideal open state while displaying signatures on which pairs of helices are nearly stacked over each other (Fig. 3D). The structure bears a strong resemblance to the HJ structure bound to the Cre recombinase (24). Following the same argument, we can deduce that the transition state in the isoI-like conformational space, tsI, also has (ϕIψI)ts= (0°, 70°).
By probing the HJ dynamics in response to pulling forces in three different directions, we mapped the location of the transition states in the two-dimensional reaction landscape and deduced the global structure of the transient species populated during the HJ conformational changes. Our simplest model envisions a shallow minimum between the two transition states, depicted as the open structure (Fig. 3A and 3D), but it is also possible that a continuum of conformations exist, spanning from tsI and tsII with nearly identical free energies, instead of having a single well-defined minimum.
The development reported here expands on the current arsenal of hybrid single molecule techniques combining force and other observables (8, 25–27). Unlike DNA or RNA hairpins, where forces on the order of 15 pN are necessary to induce mechanical unzipping (10, 11), the conformations of HJs could be biased at 0.5 pN or lower. The lever arm effect makes it unlikely that a purely mechanical tool could have probed the force effect on HJ conformations because if the arms are lengthened to magnify the distance change, the force effect will occur at even lower forces. FRET can also report on vectors other than the end-to-end distances which we exploited here by pulling on XR, HR or BR arms while simultaneously measuring the same HB vector via FRET, which led to the two dimensional mapping of reaction landscapes. Our method is readily applicable to other nucleic acids systems and their interaction with proteins and enzymes, and with the advent of new orthogonal labeling techniques, should be extendable to proteins and protein complexes. The next technical challenge would be to obtain time evolution of the end-to-end distance by force, for example due to the action of DNA processing enzymes (28), and correlate it with the enzyme conformational changes simultaneously measured via fluorescence.
Supplementary Material
Acknowledgments
We thank Wei Cheng at UC Berkeley for providing the protocol for the preparation of anti-Dig coated bead, Michelle Wang at Cornell University for giving generous advices about building optical tweezers, Yann Chemla at University of Illinois for helpful discussion, and Chirlmin Joo for generous help in preparation of illustrations. Funding was provided by the National Sciences Foundation CAREER Award (PHY 0134916) and the National Institutes of Health (GM065367).T.H. is an investigator with the Howard Hughes Medical Institute.
References and notes
- 1.Bustamante C, Chemla YR, Forde NR, Izhaky D. Annu Rev Biochem. 2004;73:705. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 2.Stryer L, Haugland RP. Proc. Natl. Acad. Sci., USA. 1967;58:719. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ha T, et al. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6264. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha T. Methods. 2001;25:78. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 5.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Optics Letters. 1986;11:288. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 6.Kapanidis AN, et al. Science. 2006 Nov 17;314:1144. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat Struct Mol Biol. 2004 Oct;11:1008. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 8.Lang MJ, Fordyce PM, Block SM. Journal of Biology. 2003;2:6. doi: 10.1186/1475-4924-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarsa PB, et al. Angew Chem Int Ed Engl. 2007;46:1999. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Science. 2001;292:733. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 11.Woodside MT, et al. Science. 2006 Nov 10;314:1001. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holliday R. Genet. Res. 1964;5:282. [Google Scholar]
- 13.Duckett DR, et al. Cell. 1988;55:79. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 14.Lilley DMJ. Quarterly Reviews of Biophysics. 2000;33:109. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 15.Eichman BF, Vargason JM, Mooers BHM, Ho PS. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3971. doi: 10.1073/pnas.97.8.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grainger RJ, Murchie AIH, Lilley DMJ. Biochemistry. 1998;37:23. doi: 10.1021/bi9721492. [DOI] [PubMed] [Google Scholar]
- 17.Materials and methods are available as supporting material on Science Online.
- 18.McKinney SA, Declais AC, Lilley DMJ, Ha T. Nature Structural Biology. 2003;10:93. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 19.Meiners JC, Quake SR. Physical Review Letters. 2000;84:5014. doi: 10.1103/PhysRevLett.84.5014. [DOI] [PubMed] [Google Scholar]
- 20.Rasnik I, McKinney SA, Ha T. Nat Methods. 2006 Nov;3:891. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 21.McKinney SA, Joo C, Ha T. Biophys J. 2006 Sep;91:1941. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265:1599. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Ha T, Schulten K. Nucleic Acids Res. 2004;32:6683. doi: 10.1093/nar/gkh1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Duyne GD. Annu Rev Biophys Biomol Struct. 2001;30:87. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 25.Ishijima A, et al. Cell. 1998;92:161. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 26.Shroff H, et al. Nano Lett. 2005 Jul;5:1509. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- 27.Gore J, et al. Nature. 2006 Jan 5;439:100. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JB, et al. Nature. 2006 Feb 2;439:621. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.