Abstract
The ability to predict RNA secondary structure is fundamental for understanding and manipulating RNA function. The structural information obtained from selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) experiments greatly improves the accuracy of RNA secondary structure prediction. Recently, Das and colleagues [Kladwang et al., Biochemistry 50:8049 (2011)] proposed a “bootstrapping” approach to estimate the variance and helix-by-helix confidence levels of predicted secondary structures based on resampling (randomizing and summing) the measured SHAPE data. We show that the specific resampling approach described by Kladwang et al. introduces systematic errors and underestimates confidence in secondary structure prediction using SHAPE data. Instead, a leave-data-out jackknife approach better estimates the influence of a given experimental dataset on SHAPE-directed secondary structure modeling. Even when 35% of the data were left out in the jackknife approach, the confidence levels of SHAPE-directed secondary structure prediction were significantly higher than those calculated by Das and colleagues using bootstrapping. Helix confidence levels were thus significantly underestimated in the recent study, and resampling approach implemented by Kladwang et al. is not an appropriate metric for assigning confidences in SHAPE-directed secondary structure modeling.
Despite an explosion in discoveries of RNAs and their functional roles in biology, accurate knowledge of structures of these molecules is incomplete.1 Knowledge of information encoded in RNA structures, especially in RNA secondary structures (the pattern of base pairs) is necessary for understanding and manipulating RNA function. Computational RNA secondary structure prediction methods2,3 have been widely used to generate structural hypotheses in RNA research. Secondary structures predicted from sequence alone often have significant errors, however, including both falsely predicted and missing base pairs.1,4,5 Incorporation of experimental structural information derived from chemical probing experiments can significantly improve secondary structure predictions.6 For example, the comprehensive and quantitative information available from selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) probing experiments greatly improves the accuracy of RNA secondary structure prediction.5,7,8
With widespread adoption of SHAPE and other experimental approaches for directing RNA secondary structure prediction, rigorous a priori estimation of the confidence level of a given RNA secondary structure prediction would constitute a major and welcome advance. Recently, Das and colleagues proposed a “bootstrapping” (resampling) approach to estimate the variance and confidence level of predicted secondary structures based on resampling of measured SHAPE data.9 Based on their statistical study, Das and colleagues suggested that the confidence level of SHAPE-derived RNA secondary structure prediction is about 77%. Follow up analysis of the work of Das and colleagues revealed that important components of their experimental work were not consistent with recommended practices in using and evaluating SHAPE technologies.10 In this correspondence, we show that the specific resampling approach developed by Das and colleagues is unphysical, introduces systematic error into the resampled data, and results in a large underestimation of the confidence of SHAPE-directed secondary structure prediction. As detailed here, a leave-data-out jackknife approach more accurately estimates the influence of a given experimental dataset on SHAPE-directed secondary structure modeling.
MATERIALS AND METHODS
SHAPE-directed secondary structure prediction
We compared the resampling approach of Kladwang et al. and our jackknife approach on four RNA molecules: tRNAPhe (76 nucleotides), adenine riboswitch (71 nucleotides), cyclic-di-GMP riboswitch (97 nucleotides), and 5S rRNA (120 nucleotides). The SHAPE data for these RNAs are presented in the companion manuscript10 and were used to direct secondary structure prediction using RNAstructure as described.5
Kladwang resampling
For RNA molecules with nucleotide positions 1, 2, 3,…, N, bootstrap decoys were generated as described9. To summarize, N nucleotide positions were randomly picked with repetition. Thus, some nucleotide positions were picked multiple times whereas others were not picked at all. If a nucleotide position was picked three times, for example, the SHAPE reactivity of that position was multiplied by 3. In total, 400 such decoy SHAPE datasets were generated and the RNA secondary structure predicted for each decoy SHAPE dataset using RNAStructure.
Jackknife
The jackknife decoy was generated by picking (1 - f) × N nucleotide positions randomly where f is the fraction of data omitted. We performed jackknife analysis using f values of 0.1 and 0.35. Thus, for a 100-nucleotide RNA molecule, the jackknife decoy dataset will contain SHAPE values for 90 and 65 nucleotides, respectively, picked randomly out of the 100 values. Four hundred such decoy datasets were generated and the RNA secondary structure predicted for each decoy SHAPE dataset using RNAstructure. For both jackknife and bootstrapping approaches, we calculated the percentage of these 400 decoy RNA secondary structures that contained each base-pair observed in the secondary structure predicted by the original SHAPE data. The generation of a decoy dataset for each approach is illustrated in Figure 1. Control simulations performed with 100 decoy datasets converged with those using 400 decoys; we report the results of the 400 member decoy datasets here. Performing a jackknife analysis with 35% omitted data represents an extreme case, because it is uncommon to miss so many SHAPE measurements. Thus, the jackknifing results presented here represent an over-estimation of the variability of SHAPE-directed structure modeling.
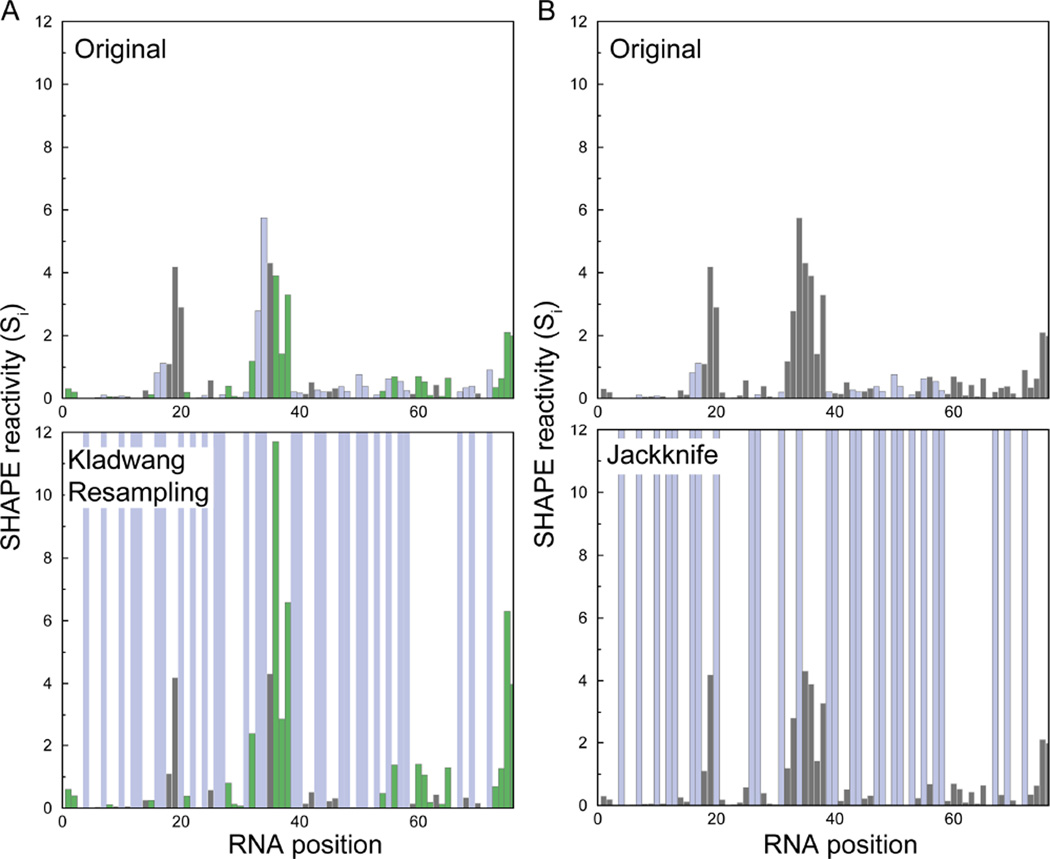
Figure 1. Generation of decoy SHAPE datasets via resampling-bootstrap and jackknife approaches.
(A) tRNAPhe SHAPE reactivities determined using a standard10 approach (top) and representative decoy dataset generated by specific resampling algorithm of Kladwang et al.9 (bottom). The SHAPE reactivities not present in the decoy datasets (top) are shown as blue bars in the original dataset and are shaded blue in the decoy dataset (bottom). Kladwang resampling also results in modified SHAPE reactivities (green bars, notice the increase in SHAPE reactivities at these positions in the decoy dataset). (B) SHAPE reactivities (top) and representative decoy dataset as generated by jackknifing (bottom). The jackknife approach results in SHAPE reactivities that are not picked in the decoy dataset (blue bars).
RESULTS AND DISCUSSION
SHAPE reactivity at a position (Si) in an RNA reports the conformational flexibility of a given nucleotide and is inversely related to the propensity of the position to form a base pair or tertiary contact.7,11 The measured SHAPE reactivity can be used as an experimentally-based correction to bias an RNA secondary structure prediction.5 To estimate confidences for individual helices in a given secondary structure prediction, Das and colleagues proposed a bootstrapping approach to resample the measured SHAPE data.9 This approach was motivated by the demonstrated utility of bootstrapping analyses in evaluation of phylogenetic trees.12 Bootstrapping entails reshuffling a given set of data with repetition; critically, for bootstrapping to be statistically justified the calculated quantity usually needs to be independent of the order of the shuffled data, an assumption that is generally valid in the construction of phylogenetic trees.
In the approach taken by Das and colleagues, SHAPE reactivities were generated for an RNA molecule by “resampling with repetition” the original SHAPE reactivities. Given SHAPE reactivities Si for nucleotides i = 1, 2, 3,…, N, resampling with repetition entails picking N indices randomly, whereby an index value can be selected multiple times for a given position, such that some nucleotides are not picked at all, whereas some are picked multiple times. This specific implementation is not appropriate for treatment of SHAPE data as SHAPE reactivities are not independent: both the actual reactivity and the position associated with a given reactivity are required for secondary structure modeling. As SHAPE data are thus inherently unsuitable for “resampling with replacement”, Das and colleagues retained the positional information of SHAPE reactivity after resampling (hence, there was no shuffling per se).9 In addition, when a position was sampled multiple times during bootstrapping, the SHAPE reactivity of the position was increased by the amount equal to the original reactivity of the position each time (Das, personal communication). Hence, the relative error introduced at each position in this resampling approach is n × Si, where n is the number of repetitions, (0, 1, 2,…; Fig. 1A, B). This treatment of repetition is intrinsically different from the repetition introduced by a standard bootstrapping approach as implemented for multiple sequence alignments, which increases the weight of the repeated column in the alignment and thus enhances both sequence similarity and differences. In contrast, in the approach used by Das and colleagues, repetition of high SHAPE values increases the probability of breaking a base pair, whereas repetition of low SHAPE data does not increase the probability of a forming a base pair.
This problem is most significant in regions of low, but non-zero, SHAPE reactivities that are often base paired and was compounded by SHAPE signal processing errors in the prior work.10 Several occurrences of multiplying the SHAPE reactivity value under this approach would effectively destabilize a helix in which the original SHAPE data would otherwise clearly score as unreactive, and likely paired, overall. Thus, the bootstrapping approach as implemented by Das and colleagues9 results in introduction of systematic error because the errors are integer multiples of the original data itself. In addition, the decoy datasets generated were not the same size because, after each round of shuffling to retain positional information, the number of data points differs from the number in the original dataset. Since the errors added to the dataset are integer multiples of the data itself, the perturbation is high, resulting in many datasets that are highly dissimilar to the experimental data. Overall, this approach results in an underestimation of the confidence of secondary structure prediction from the SHAPE data.
An alternate statistical method for estimation of the influence of a specific experimental dataset on secondary structure modeling is the jackknife approach. Resampling by the jackknife approach has also been used to estimate the confidence of phylogenetic trees.13 With jackknifing, the fluctuation is introduced simply through omission of a fraction of the data, and the number of data points in each decoy dataset is identical. The key variable in jackknife resampling is the fraction of excluded data.
To quantify the extent of underestimation of the confidence of SHAPE prediction by the method used by Das and colleagues, we used the jackknife method to evaluate recovery of SHAPE-predicted base pairs for four RNA molecules – tRNAPhe, the adenine and cyclic di-GMP riboswitches, and 5S rRNA by generating decoys with up to 35% of the SHAPE data randomly removed. SHAPE data were measured using the documented approach for performing the SHAPE experiment,8,14 which is substantially different from that introduced by Das and colleagues.9,10 Experimentally, it is uncommon to have 35% of SHAPE missing and few experimentalists would try to predict a structure with such a high level of missing information, we therefore also performed a jackknife analysis with 10% of the data omitted, the latter corresponding to a more realistic practical occurrence.
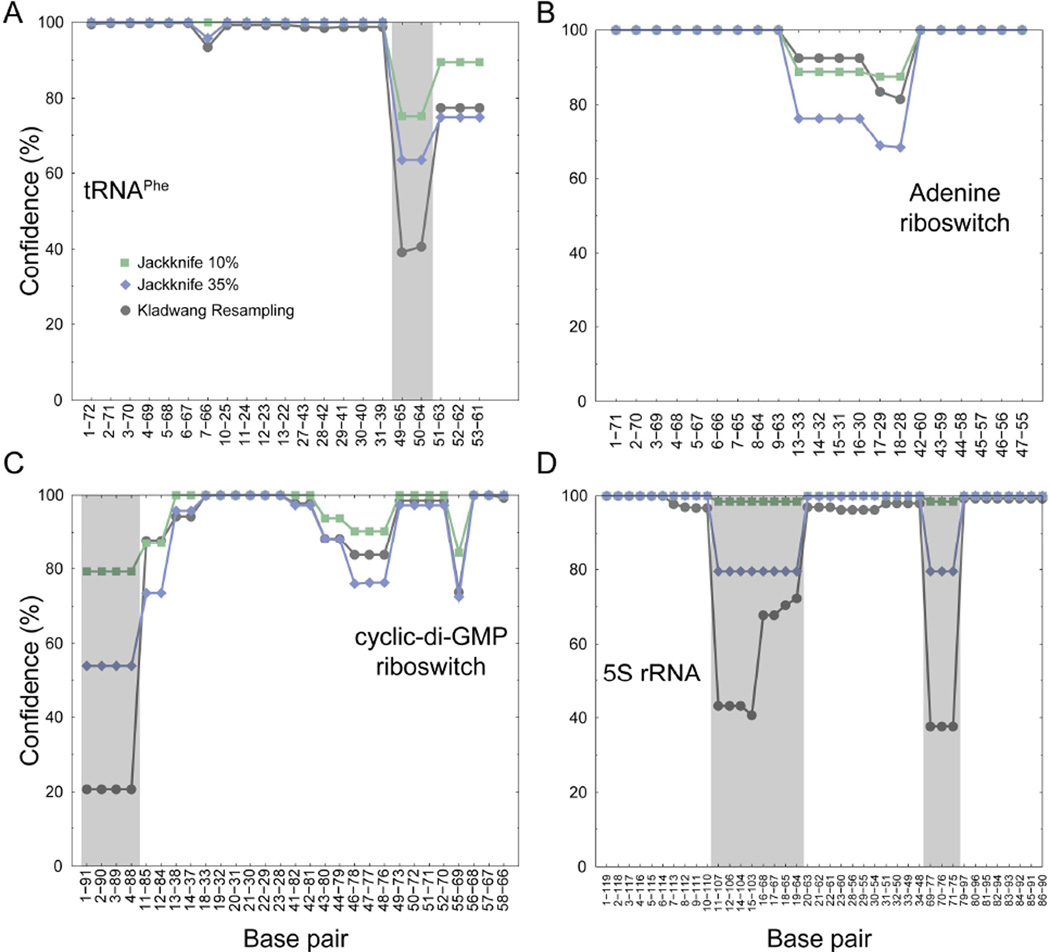
In general, both the Kladwang et al. resampling approach and jackknifing identified the same helices as being less well-defined by the SHAPE data (Fig. 2). However, the Kladwang et al. approach grossly underestimated the confidence of the helices in three of the SHAPE-directed structure models, those of tRNAPhe, the cyclic-di-GMP riboswitch, and 5S rRNA. Estimated confidences based on bootstrapping were less than confidence estimates obtained using decoy datasets that were missing (an extreme) 35% of the experimental data (Fig. 2). In contrast, using the jackknife approach with a more physically and experimentally realistic 10% omitted data, the majority of the base pairs have 100% prediction confidences, and the lowest is ~80%, supporting the general robustness of the SHAPE-directed secondary structure models.
Figure 2. Confidence estimation for SHAPE-directed secondary structure modeling.
Confidence estimates for SHAPE-predicted base pairs, calculated using the specific resampling algorithm of Kladwang et al.9 (gray circles) and using jackknife procedures that omit either 10% (green squares) or 35% (blue diamonds) of the data for (A) tRNAPhe, (B) adenine riboswitch, (C) cyclic-di-GMP riboswitch, (D) 5S rRNA. Gray shading emphasizes regions in which the resampling-bootstrap approach underestimated confidences as compared to removing (an extreme) 35% of the experimental SHAPE data.
A second problem with the specific resampling approach created by Das and coworkers lies in their helix-by-helix interpretation. The bootstrap resampling of the SHAPE data introduces noise that perturbs the relative free energy of each RNA structure. Longer helices with lower free energies are less sensitive to this perturbation and are more likely to have high confidence in prediction, especially given that Das and colleagues define the bootstrap value of a helix as the maximum of the bootstrap values of base pairs across that helix.9 By this definition, longer helices have more base pairs and are more likely to have at least one base pair with a high “bootstrap value”. In contrast, shorter and less stable helices are more sensitive to perturbations and thus more prone to break under perturbation. Therefore, the estimated bootstrap value of a helix primarily reflects the stability of the helix rather than the underlying SHAPE data.
In summary, the specific bootstrapping procedure proposed by Das and colleagues to resample the SHAPE data is unphysical, introduces systematic error into the resampled data, and results in an underestimation of the confidence of SHAPE-directed secondary structure modeling. The calculated confidence level obtained via this approach is not an appropriate metric for estimation of the accuracy of experimentally-directed RNA secondary structure prediction. Instead, this work supports use of the jackknife approach to generate resampled SHAPE data and to estimate the sensitivity of predicted secondary structures to the underlying SHAPE dataset. The more general issue of a priori identification of individual highly probable helices within a given experimentally-directed RNA structure model remains a major research challenge.
Acknowledgments
Funding: This work was supported by grants from the NIH (GM080742 to N.V.D. and AI068462 to K.M.W.).
REFERENCES
- 1.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Mathews DH. Revolutions in RNA secondary structure prediction. J Mol Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- 3.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 4.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by Selective 2'-Hydroxyl Acylation and Primer Extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 9.Kladwang W, VanLang CC, Cordero P, Das R. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard CW, Hajdin CE, Karabiber F, Mathews DH, Favorov OV, Dokholyan NV, Weeks KM. Principles for understanding the accuracy of SHAPE-directed RNA structure modeling. Biochemistry. 2013;52 doi: 10.1021/bi300755u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J Am Chem Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efron B, Halloran E, Holmes S. Bootstrap confidence levels for phylogenetic trees. Proc Natl Acad Sci U S A. 1996;93:7085–7090. doi: 10.1073/pnas.93.14.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soltis PS, Soltis DE. Applying the bootstrap in phylogeny reconstruction. Statistical Science. 2003;18:256–267. [Google Scholar]
- 14.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]