Abstract
Cerebral cavernous malformations (CCM) are neurovascular dysplasias affecting up to 0.5% of the population. Mutations in the CCM2 gene are associated with acquisition of CCM. We identify a previously uncharacterized domain at the C-terminus of CCM2 and determine its 1.9 Å resolution crystal structure. Because this domain is structurally homologous to the N-terminal domain of harmonin, we name it the CCM2 harmonin-homology domain or HHD. CCM2 HHD is observed in two conformations, and we employ analytical ultracentrifugation to test its oligomerization. Additionally, CCM2 HHD contains an unusually long 13-residue 310 helix. This study provides the first structural characterization of CCM2.
Keywords: Protein-protein interaction, Signal transduction, Cerebral cavernous malformation, X-ray crystallography, harmonin-homology domain
1. Introduction
Cerebral cavernous malformations (CCM) are neurovascular dysplasias that occur in up to 0.5% of the population[1]. Both the familial and sporadic forms of the disease most commonly result in the formation of hemorrhage-prone lesions in the brain[1] that are associated with stroke, seizure, or other neurological sequelae[1]. Approximately 20% of familial cases of CCM are associated with autosomal dominant inheritance of mutations in the gene CCM2 (malcavernin; osmosensing scaffold for MEKK3, OSM)[1]. The formation of lesions is thought to be associated with a ‘second-hit’ somatic mutation resulting in complete loss of functional CCM2[2,3]. CCM2 is also essential for embryonic angiogenesis, and its endothelial-specific loss is lethal mid-gestation[4,5]. The molecular and structural underpinnings of CCM2 function are therefore of significant cell biological and clinical interest.
CCM2 is a 444 amino acid protein predicted to contain a phosphotyrosine binding (PTB) domain at its N-terminus[6]. It is thought to interact directly with both KRIT1 (Krev interaction trapped-1; CCM1) and CCM3 (programmed cell death 10, PDCD10) to form the CCM complex[7]. The CCM2 PTB domain interacts with KRIT1 via conserved NPxY/F motifs in the KRIT1 N-terminus[8]. CCM2 also directly interacts with CCM3, but the interaction interface is not well characterized[7,9]. CCM2 is therefore the hub of the CCM complex, scaffolding to both KRIT1 and CCM3. CCM2 has also been implicated as an osmosensitive scaffold for p38 MAP kinase cascades[10,11] and in degradation of the GTPase RhoA[5,12].
The C-terminal portion of CCM2 has not been well described. This approximately 200 amino acid region cannot be homology modeled to known protein domains[13]. It is not yet known whether the CCM2 C-terminus contains a folded domain, nor how that domain may function in the cell. The C-terminus does, however, appear to be capable of inducing cell death through a pathway involving the neurotrophin receptor TrkA[13]. The molecular basis for this requirement is not understood.
In this study, we provide the first molecular level analysis and crystal structure of the C-terminus of CCM2. We find a helical domain that bears distinct structural similarity to the N-terminal domain of the Usher syndrome scaffolding protein harmonin[14,15] that we find can exist in two distinct conformational states.
2. Materials and Methods
2.1. Protein expression and purification
Four human CCM2 (UniProt ID:Q9BSQ5) constructs were subcloned into a modified pET32 vector with a TEV-cleavable N-terminal hexahistidine (6xHis) tag: CCM2-FL (residues 1–438), CCM2-CT (residues 231–438), CCM2-CT3 (residues 283–438) and CCM2-HHD (residues 283–379). These were expressed in E. coli Rosetta (DE3) cells (Novagen) and purified (Supplemental Materials).
2.2. Limited proteolysis
Limited proteolysis was performed on CCM2-FL and CCM2-CT as described in Supplemental Materials.
2.3. Crystallization and structure solution of CCM2 C-terminal domain
Optimized CCM2-HHD crystals were obtained with precipitant conditions of 2.072 M ammonium sulfate and 0.2M potassium formate (Supplemental Materials). Because CCM2-HHD has no discernable homology to other determined protein structures by sequence alignment, we determined the structure by the single-wavelength anomalous diffraction (SAD) method using an osmium derivative (Supplemental Materials). The structure is deposited in the PDB with accession code 4FQN.
2.4. Light Scattering and analytical ultracentrifugation
SEC-MALS was performed on purified CCM2-HHD or CCM2-HHD-E290R concentrated to 1 mg/ml. Data analysis was performed using ASTRA 6 software (Wyatt Technology). Sedimentation velocity experiments were performed to monitor possible mass action in the oligomerization behavior of CCM2. Both wild-type and CCM2-HHD-E290R were measured over 13–224µM for the wild-type and 11–239µM for CCM2-HHD-E290R (Supplemental Materials).
3. Results
3.1. CCM2 contains a C-terminal domain
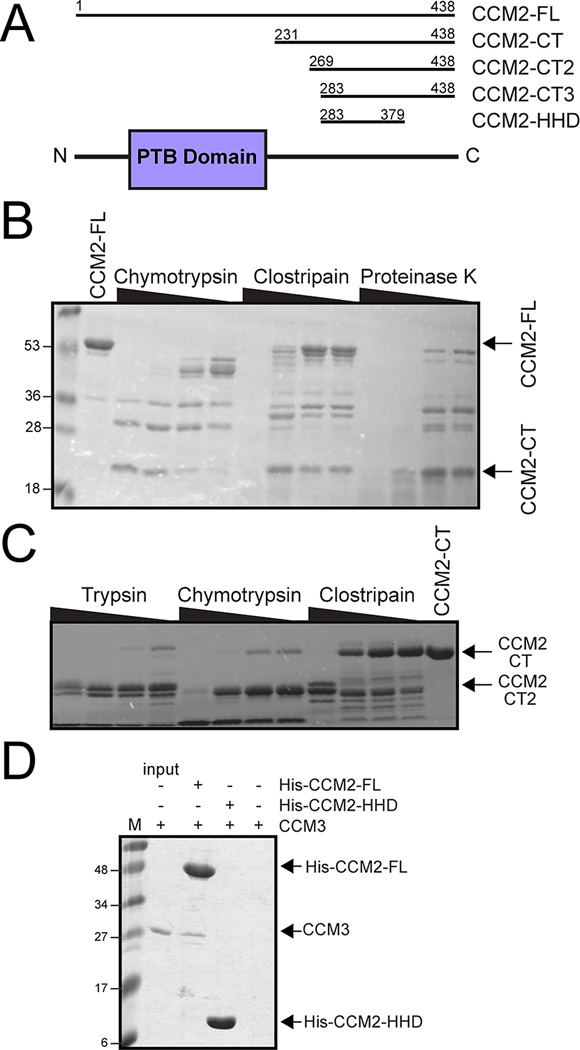
The CCM2 protein contains an N-terminal flexible loop of approximately 60 residues, a predicted PTB domain (encoded approximately by residues 60 to 220)[6], and a C-terminal region (residues ~220 to 444)(Fig 1A). No sequence homology to structural domains has previously been detected in the C-terminal region of CCM2, although it has been suggested to play a role in promoting apoptotic signaling by the TrkA receptor tyrosine kinase[13] and has predicted secondary structure. To identify whether a folded domain exists at the CCM2 C-terminus, we conducted limited proteolysis on full-length CCM2 (CCM2-FL) by incubating it with serial dilutions of three different proteases. A species of approximately 21 kDa molecular mass was present after treatment with all three proteases(Fig 1B) that N-terminal sequencing identified to begin at A232. This is slightly downstream from the predicted PTB domain, indicating the presence of a proteolytically stable C-terminal region of CCM2. Therefore, we produced a CCM2 construct encompassing residues R231-S438 (CCM2-CT) that we found by circular dichroism analysis to contain approximately 40% helical and 10% strand content, suggesting that the C-terminus of CCM2 can independently fold as a stable domain. Because this region of the protein was recalcitrant to crystallization, however, we conducted a second limited proteolysis analysis(Fig 1C). This yielded another region (CCM2-CT2) of approximately 17 kDa molecular mass that N-terminal sequencing determined to start at C269. Taking into account the limited proteolysis results and secondary structure predictions, we designed two further CCM2 constructs encompassing residues S283–S438 (CCM2-CT3) and residues S283-G379 (CCM2-HHD). This analysis provides direct experimental evidence in support of a folded domain at the C-terminus of CCM2(Fig 1A).
Figure 1. Domain organization of CCM2.
A) Domain schematic of CCM2. The predicted PTB domain is shown in purple and the C-terminal regions identified in this study are indicated. B) Limited proteolysis of full-length CCM2 (CCM2-FL) using a serial dilution of proteases reveals a proteolytically resistant portion of CCM2 (CCM2-CT). C) Limited proteolysis using CCM2-CT as input identifies a second proteolytically resistant region (CCM2-CT2). D) Pull-down assay. CCM2-FL or CCM2-HHD were bound to nickel-beads and incubated with CCM3. Right hand lane is CCM3 pull-down by empty beads.
3.2. The CCM2 C-terminal domain does not interact with CCM3
CCM2 has been shown to interact with another cerebral cavernous malformation-associated protein, CCM3[7,9], through a small region C-terminal to the predicted PTB domain potentially corresponding to residues 220–245[16]. Indeed, purified constructs including this region can bind CCM3[9]. To confirm that the newly identified C-terminal domain of CCM2 does not bind CCM3, we conducted pull-down experiments using immobilized CCM2-FL or CCM2-HHD with purified CCM3 as input. We observed that while CCM2-FL pulls down CCM3, CCM2-HHD does not (Fig 1D).
3.3. Overall structure of the CCM2 C-terminal domain
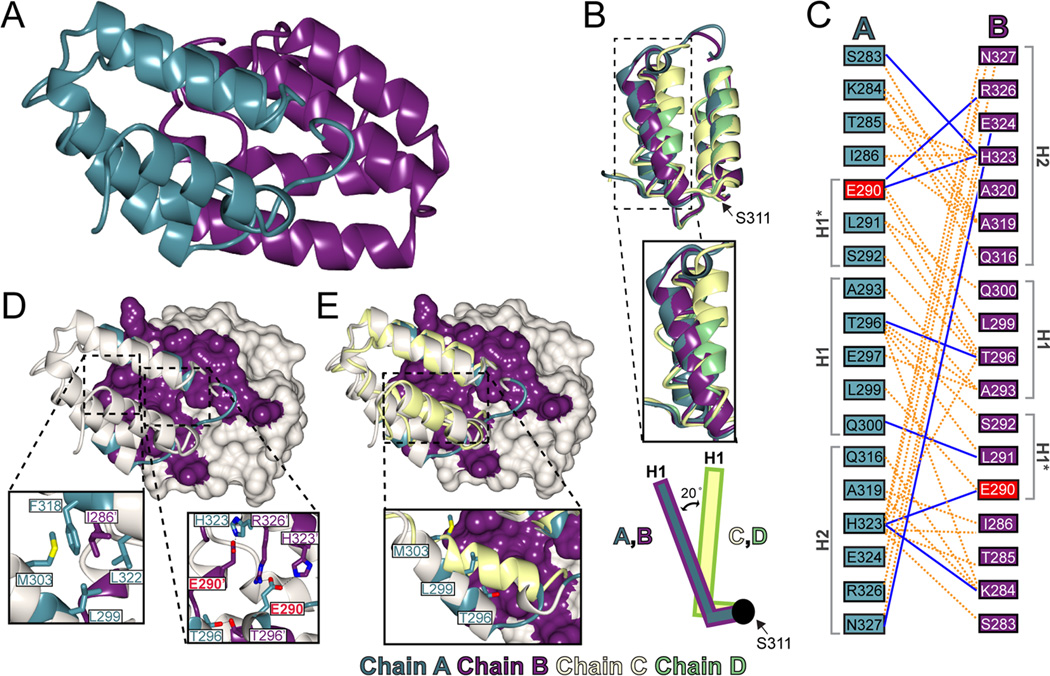
The proteolytically resistant C-terminal domain of CCM2 has low primary sequence similarity to previously determined structures, suggesting that it may comprise a novel fold. We therefore determined its structure by X-ray crystallography by using the SAD method (Fig S1), and refined against a 1.9Å native dataset (Table S1). Overall, we found good electron density throughout the structure (Fig S1), with four copies of CCM2 per asymmetric unit. The CCM2 C-terminus folds as a globular domain comprised of a five-helical bundle that we denote as helices H1 to H5 with a 4-residue helix (H1*) present at the N-terminus of chains A and B. Although helices H1, H2, H3, H5, and H1* are α-helices, helix H4 is a long 13-residue 310 helix (Fig 2A and B). A 310 helix of this length is uncommon, as the majority of 310 helices observed in protein structures are typically only 3 or 4 residues[17]. Occasionally, however, helices of comparable length are found, as discussed in a 2006 survey of 310 helices in the Protein Data Bank[17] that cited a 15 residue helix in PDB 1PZ4[18]. The low frequency of longer 310 helices is likely due to decreased structural stability, resulting in more stringent packing requirements[19]. The 310 helix of CCM2 packs against helices H1 and H5 and is anchored by two salt bridges at its N- and C-termini made by residues R346 and R354 binding helix H1 residue E314 and helix H5 residue E366, respectively. The conserved P355-F356 pair at the C-terminus of H4 is also potentially important for stabilizing this helix (Fig 2C). The overall structure of CCM2-HHD encompasses residues S283-K375.
Figure 2. Overall structure of CCM2 harmonin-homology domain (HHD).
A) Cartoon diagram of the CCM2 HHD (Chain A) with helices labeled. Structural images generated using CCP4MG [20]. B) Topology diagram of the CCM2 HHD generated using TopDraw. C) Cartoon diagram of CCM2 with the 310 helix (H4) highlighted in teal. Insets show regions that stabilize the 310 helix: a salt bridge between R354 and E346 (upper left), a salt bridge between R346 and E314 (lower right), and conserved residues P355 and F356 (upper right). D) Superposition of the N-domain of harmonin (PDB ID: 3K1R)[14] in orange and the CCM2 HHD in teal. E) Structure-based alignment of CCM2 HHD to the harmonin N-domain generated using Aline. The secondary structure of CCM2 is depicted above its sequence and the secondary structure of harmonin below its sequence. α-helices are shown as cylinders and 310 helices as spirals. Residues highlighted in yellow are conserved between CCM2 and harmonin. Red triangles denote harmonin residues that bind cadherin 23.
3.4. The CCM2 C-terminal domain is homologous to the N-terminal domain of harmonin
To investigate whether the C-terminal domain of CCM2 folds in a similar fashion to previously determined structures, we submitted each of the four chains to the Dali server. We found that there is distinct structural similarity to the Usher syndrome scaffolding protein harmonin (PDB IDs:3K1R, 2KBQ, 2KBR)[14,15], a protein involved in mechanotransduction in hair cell bundles (Fig 2D). Although sequence identity between the CCM2 C-terminal domain and the harmonin N-terminal domain is only 14% over 80 residues (Fig 2E), the Dali server yielded Z-scores between CCM2 and harmonin that ranged from 8.6 to 11.9 and root-mean-square deviations (RMSD) from 1.7Å to 2.6Å over 71 to 80 Cα atoms. Based on this structural similarity, we therefore denote this domain in CCM2 the harmonin-homology domain, or HHD.
3.5. The CCM2 HHD crystal structure indicates two conformational states
The CCM2 HHD crystallizes with four copies per asymmetric unit. Analysis of the crystal packing reveals two distinct conformations. Two of the copies (chains A and B) have an extensive interaction interface between them (Fig 3A), but this interface is not observed for the other two copies (chains C and D) either in the asymmetric unit or with symmetry-related molecules. Chains A and B are experimentally identical (RMSD is 0.55Å over 93 Cα[20]) and chains C and D are structurally very similar (RMSD is 0.75Å over 76 Cα). The two classes (A/B and C/D), however, are conformationally more divergent from one another (e.g. the RMSD between chain A and chain D is 1.39Å over 76 Cα). This divergence is centered on the N-terminal portion of the CCM2 HHD up to residue S311 (RMSD for residues 311–375 between chains A and D is 0.87Å over 65 Cα)(Fig 3B). Although there is strong electron density for residues S283–S311 in both chains A and B, this N-terminal region is partly disordered in chains C and D, for which S292 and L299 are the first visible residues, respectively. Furthermore, in the visible N-terminal portions of chains C and D, helix H1 is conformationally hinged closer to the hydrophobic core of CCM2 HHD by approximately 20° (by HELIXANG) and pivots around residue S311(Fig 3B).
Figure 3. The CCM2 harmonin-homology domain putative dimerization interface.
A) Crystallographically observed interaction interface between CCM2 HHD chains A and B. Chain A is colored in teal and chain B in purple. B) Superposition of the four molecules in the ASU. Chain A is colored in teal, B in purple, C in yellow, and D in green. The hinge residue S311 is indicated by an arrow. Inset shows the two conformations of helix H1. Schematic illustrates the conformational change in helix H1. C) Map of residues involved in the CCM2 HHD interface. Residue E290 is mutated in this study and indicated in red. D) Chain B shown in surface view and A as a ribbon diagram. Residues involved in mediating the interface are depicted in purple or teal for chain B or A respectively. Insets are close-ups of key residues involved in the hydrophobic pocket (left) and charge cluster (right). E) Superposition of chain C (yellow) on chain A, with chain B shown as a surface. The two CCM2 HHD conformations are incompatible with each other.
3.6. Structural analysis of the interface between chains A and B
The crystallographically observed interface between chains A and B encompasses 18 residues from each chain, and comprises 9 hydrogen bonds and 109 non-bonded contacts (as defined by PDBSum) (Fig 3C). The interface between molecules A and B buries a total surface area of 1911Å2 (956.9Å2 from chain A and 954.1Å2 from chain B) as defined by the PISA server and has a shape complementarity of 0.64[21] (Fig 3D). The PISA server also shows this interface to have a Complex Formation Significance Score (CSS), a measure of interface relevance, of 1.000. The interaction is almost two-fold symmetric and encompasses the N-terminus through residue Q300 and residues Q316-N327. This includes helix H1*, the N-terminal portion of helix H1, and the C-terminal portion of helix H2. The interaction surface is predominantly comprised of three regions: two hydrophobic pockets and a charge cluster. Because of the almost two-fold symmetry, the charge cluster is found at the center of the interface and the hydrophobic pockets on the edge (Fig 3E) (Supplemental Materials).
3.7. Determination of the oligomeric state of the CCM2 HHD by size exclusion chromatography with light scattering and analytical ultracentrifugation
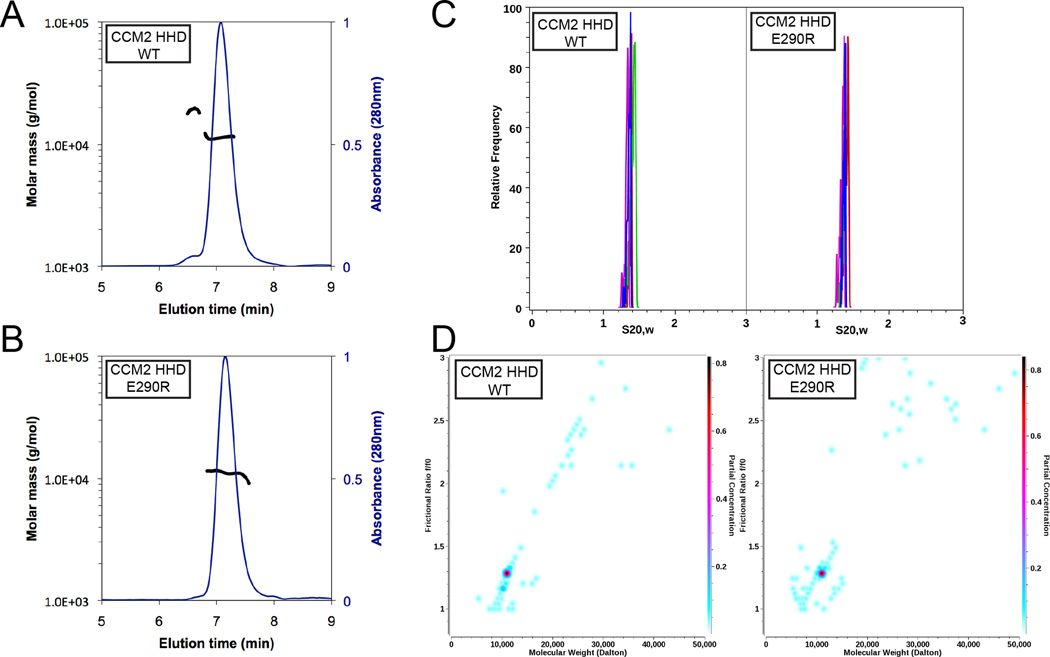
Because of the presence of both monomeric and dimeric forms of the CCM2 HHD in the crystal structure and the fact that the dimeric interface is evolutionarily conserved (Fig S2), we tested the oligomeric state of the CCM2 HHD by size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS). We found that both dimeric and monomeric forms of CCM2-HHD are present in solution as determined by SEC-MALS, with the monomeric species making up the majority of the protein (Fig 4A). We also confirmed that the dimeric peak was CCM2-HHD by N-terminal sequencing and that dimerization was not due to disulfide formation by analysis of non-reducing SDS-PAGE (data not shown). We next generated a point mutation at the dimer interface, E290R, and found by SEC-MALS that the small dimeric peak was no longer observed (Fig 4B).
Figure 4. Size exclusion chromatography with multi-angle light scattering (SEC-MALS) and analytical ultracentrifugation (AUC) analysis of CCM2 HHD.
A) SEC-MALS analysis of the 11.1 kDa CCM2-HHD. Two peaks are observed: a monomeric peak with a molecular weight of 11.1 kDa ±2% and a dimeric peak with a molecular weight of 19 kDa ±2%. Molar mass is plotted on the primary y-axis, and normalized A280 on the secondary y-axis. B) SEC-MALS analysis of CCM2–HHD-E290R. One peak is observed, with molecular weight of 11.1 kDa ±1.4%. C) van Holde – Weischet sedimentation coefficient distributions for CCM2-HHD (left), measured at 0.59 OD230 (red), 1.14 OD230 (green), 0.4 OD280 (magenta), and 1.0 OD280 (blue), and CCM2–HHD-E290R (right), measured at 0.49 OD230 (red), 1.1 OD230 (green), 0.37 OD280 (magenta), and 1.07 OD280 (blue). The narrow sedimentation distribution of both CCM2–HHD and CCM2–HHD-E290R forms indicates homogeneity, and suggests that all concentrations reflect the same oligomeric state. D) 2-dimensional spectrum analysis fit combined with a 50 iteration Monte Carlo analysis. CCM2-HHD (left) and CCM2–HHD-E290R (right). The plots show a major species with a molecular weight consistent with the monomer (10.9 kDa, 95% confidence intervals: 9.3 kDa, 12.6 kDa). The y-axis denotes the anisotropy of the molecule, which suggests a mostly globular structure with a value of 1.35 (95% confidence interval: 1.25, 1.44). The color scale on the right y-axis denotes relative concentration.
Since our light scattering data neither completely rule out the existence of a CCM2-HHD dimer nor fully validate it, we next sought to study the dimerization equilibrium of the putative dimer by analytical ultracentrifugation (AUC). Sedimentation velocity experiments (SV) were carried out to determine the oligmeric state of CCM2-HHD and CCM2-HHD-E290R. We examined both wild-type and mutant over a range of loading concentrations to determine any possible mass action effects (Table S2). These experiments showed homogeneous compositions for both wild-type and mutant (Fig 4C), resulting in molecular weights consistent only with a monomer of CCM2-HHD (Fig 4D). Sedimentation profiles from multiple loading concentrations did not produce any change in the sedimentation behavior, suggesting the absence of any mass action. Both wild-type and mutant displayed essentially identical sedimentation coefficients at all investigated concentrations. While the sedimentation resulted in observations of monomeric protein, these experiments were carried out at concentrations substantially lower than the crystallization experiments. These results do not preclude a monomer-dimer affinity in the concentration range present during crystallizations. However, AUC experiments conducted at such high concentrations may be problematic.
Together, the SEC-MALS and AUC results suggest that the homodimer observed in the crystal structure may represent a weak interface primarily observable under crystallization conditions. These studies do not, however, preclude the potential that other regions of CCM2 or CCM2 binding partners could facilitate formation of this dimerization interface.
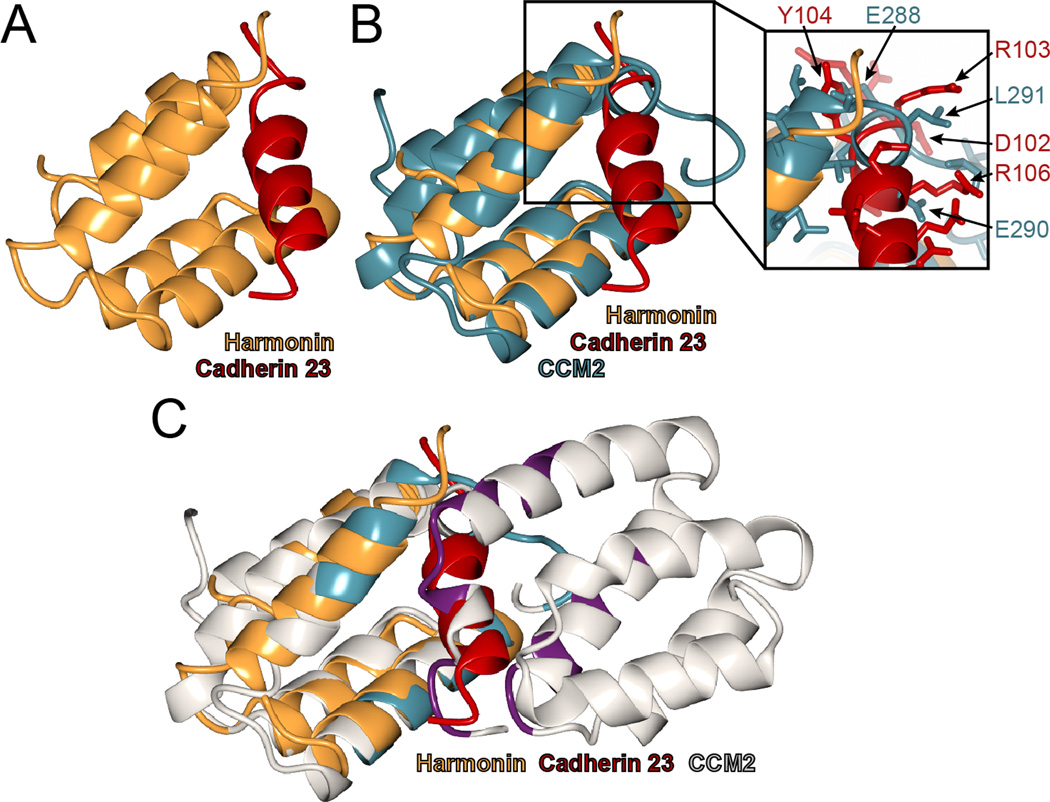
3.8. The crystallographic CCM2 HHD dimerization interface resembles the harmonin cadherin 23 binding site
The CCM2 HHD is structurally similar to the N-terminal domain of the Usher syndrome protein harmonin (Fig 2D). Harmonin binds directly to the cell adhesion protein cadherin 23 via two domains: its N-terminal domain and a PDZ domain. The structural basis for the harmonin N-terminal domain binding to cadherin 23 has been determined by NMR spectroscopy[15] (Fig 5A), so we compared this mode of binding to the crystallographically-observed CCM2 HHD dimer. We found that the CCM2 HHD dimer interface occupies a similar location to that observed for the harmonin interaction with cadherin 23 (Fig 5B and C). Although we did not observe direct interaction between CCM2-HHD and cadherin 23 (data not shown) we suggest that the equivalent surface in CCM2 HHD could potentially mediate other protein-protein interactions.
Figure 5. CCM2 HHD crystallographic dimerization interface is analogous to harmonin-cadherin 23 interface.
A) Structure of harmonin N-terminal domain (orange) bound to a cadherin 23 peptide in red (PDB ID: 2KBR)[15]. B) Superposition of CCM2-HHD (teal) and harmonin (orange). Cadherin 23 is shown (red). Exploded view shows specific residues that clash between CCM2-HHD and Cadherin 23 in the superposition. C) Superposition of the CCM2-HHD dimer onto harmonin. The cadherin binding site is equivalent to the dimer interface between CCM2 HHD chains A (left, interface residues teal) and chain B (right, interface residues purple).
4. Discussion
Understanding the function and role of proteins associated with cerebral cavernous malformations has recently become an area of intense interest, as KRIT1, CCM2, and CCM3 have been found to be involved in diverse signaling pathways. These include regulation of integrin activation, degradation of Rho-family GTPases and receptor tyrosine kinases, stabilization of VEGFR2, interactions with MAP and sterile-20 kinase, and TrkA signaling. The discovery of new regions within these proteins that can provide a functional basis for understanding their signaling pathways could therefore allow for significant further studies. Previously, we discovered that CCM3 contains an N-terminal dimerization domain of a novel fold, and a C-terminal focal adhesion targeting (FAT)-homology domain[9], allowing us to discover interesting new functions and roles for CCM3[22,23]. Likewise, the current study provides the first molecular level description of CCM2: the discovery of a previously uncharacterized folded domain at its C-terminus that we term the CCM2 harmonin homology domain (HHD). Interestingly we observe two conformations for the CCM2 HHD in our structure.
The similarities between the CCM2 HHD dimerization interface and the harmonin-cadherin 23 interface are interesting. The non-overlapping interactions of the harmonin N-terminal and PDZ domains with cadherin 23 are both of moderate affinity (KD of approximately 10–20 µM). Together, however, these facilitate a tight harmonin-cadherin 23 complex[15]. Similarly, CCM2 could also potentially utilize bidentate moderate affinity interactions to specific partners, perhaps via both the PTB and HHD domains. Therefore, the discovery of the HHD provides an exciting new level of understanding to these important proteins and will likely spur new functional analyses of CCM2, its binding partners, and their function.
Supplementary Material
Highlights.
Limited proteolysis reveals a C-terminal domain in CCM2.
The 1.9 Å crystal structure of the CCM2 C-terminal domain is determined.
CCM2 C-terminal domain is structurally homologous to harmonin.
We term this domain the CCM2 harmonin-homology domain, HHD.
This domain may represent a regulated site of protein-protein interaction for CCM2.
Acknowledgements
We thank V.Schirf, W.Min, B. Turk, H.J.Lou, K. Draheim, A.Stiegler, W.Liu, N.Alicea-Velázquez, B.H.Ha, J.Chacon, D.Calderwood, J.Ferrullo, and beamlines X25, X29, and NECAT. Grants from NSF(OSF), AHA(XL), NIH CA054174,RR022200(BD), and NSF TG-MCB070038(BD). TJB funded by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Structured summary of protein interactions:
CCM2 binds to CCM3 by pull down (View interaction)
CCM2 and CCM2 bind by x-ray crystallography (View interaction)
CCM2 and CCM2 bind by molecular sieving (View interaction)
References
- 1.Cavalcanti DD, Kalani MY, Martirosyan NL, Eales J, Spetzler RF, Preul MC. Cerebral cavernous malformations: from genes to proteins to disease. J Neurosurg. 2012;116:122–132. doi: 10.3171/2011.8.JNS101241. [DOI] [PubMed] [Google Scholar]
- 2.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18:911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulday G, Blecon A, Petit N, Chareyre F, Garcia LA, Niwa-Kawakita M, Giovannini M, Tournier-Lasserve E. Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech. 2009;2:168–177. doi: 10.1242/dmm.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehead KJ, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15:177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liquori CL, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl S, et al. Novel CCM1, CCM2, and CCM3 mutations in patients with cerebral cavernous malformations: in-frame deletion in CCM2 prevents formation of a CCM1/CCM2/CCM3 protein complex. Hum Mutat. 2008;29:709–717. doi: 10.1002/humu.20712. [DOI] [PubMed] [Google Scholar]
- 8.Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14:2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Zhang R, Zhang H, He Y, Ji W, Min W, Boggon TJ. Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J Biol Chem. 2010;285:24099–24107. doi: 10.1074/jbc.M110.128470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Izumi Y, Burg MB, Ferraris JD. Rac1/osmosensing scaffold for MEKK3 contributes via phospholipase C-gamma1 to activation of the osmoprotective transcription factor NFAT5. Proc Natl Acad Sci USA. 2011;108:12155–12160. doi: 10.1073/pnas.1108107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlik MT, et al. Rac-MEKK3-MKK3 scaffolding for p38 MAPK activation during hyperosmotic shock. Nat Cell Biol. 2003;5:1104–1110. doi: 10.1038/ncb1071. [DOI] [PubMed] [Google Scholar]
- 12.Crose LE, Hilder TL, Sciaky N, Johnson GL. Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J Biol Chem. 2009;284:13301–13305. doi: 10.1074/jbc.C900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harel L, et al. CCM2 mediates death signaling by the TrkA receptor tyrosine kinase. Neuron. 2009;63:585–591. doi: 10.1016/j.neuron.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, Pan L, Chen X, Wu L, Zhang M. The structure of the harmonin/sans complex reveals an unexpected interaction mode of the two Usher syndrome proteins. Proc Natl Acad Sci U S A. 2010;107:4040–4045. doi: 10.1073/pnas.0911385107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan L, Yan J, Wu L, Zhang M. Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23. Proc Natl Acad Sci U S A. 2009;106:5575–5580. doi: 10.1073/pnas.0901819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kean MJ, et al. Structure-Function Analysis of Core STRIPAK Proteins: A Signaling Complex Implicated In Golgi Polarization. J Biol Chem. 2011;286:25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enkhbayar P, Hikichi K, Osaki M, Kretsinger RH, Matsushima N. 3(10)-helices in proteins are parahelices. Proteins. 2006;64:691–699. doi: 10.1002/prot.21026. [DOI] [PubMed] [Google Scholar]
- 18.Dyer DH, Lovell S, Thoden JB, Holden HM, Rayment I, Lan Q. The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-A resolution. J Biol Chem. 2003;278:39085–39091. doi: 10.1074/jbc.M306214200. [DOI] [PubMed] [Google Scholar]
- 19.Vieira-Pires RS, Morais-Cabral JH. 3(10) helices in channels and other membrane proteins. J Gen Physiol. 2010;136:585–592. doi: 10.1085/jgp.201010508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Ji W, Zhang R, Folta-Stogniew E, Min W, Boggon TJ. Molecular recognition of LD motifs by the FAT-homology domain of cerebral cavernous malformation 3 (CCM3) J Biol Chem. 2011;286:26138–26147. doi: 10.1074/jbc.M110.211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H, Min W. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000722. ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.