Abstract
Elk3/Net/Sap2 (here referred to as Elk3) is an Ets ternary complex transcriptional repressor known for its involvement in angiogenesis during embryonic development. Although Elk3 is expressed in various tissues, additional roles for the protein outside of vasculature development have yet to be reported. Here, we characterize the early spatiotemporal expression pattern of Elk3 in the avian embryo using whole mount in situ hybridization and quantitative RT-PCR and examine the effects of its loss of function on neural crest development. At early stages, Elk3 is expressed in the head folds, head mesenchyme, intersomitic vessels, and migratory cranial neural crest (NC) cells. Loss of the Elk3 protein results in the retention of Pax7+ precursors in the dorsal neural tube that fail to upregulate neural crest specifier genes, FoxD3, Sox10 and Snail2, resulting in embryos with severe migration defects. The results putatively place Elk3 downstream of neural plate border genes, but upstream of neural crest specifier genes in the neural crest gene regulatory network (NC-GRN), suggesting that it is critical for the progression from progenitor to definitive neural crest cell.
Keywords: Elk3, Neural crest specification, Progenitors, Pax7
Introduction
Neural crest (NC) cells are a transitory and vertebrate-specific cell type that are induced at the border of the neural plate and the non-neural ectoderm, and subsequently come to reside within the neural folds during neurulation. After neural tube closure, these cells delaminate from the neuroepithelium and migrate to distant locations in the embryo where they differentiate into multiple derivatives, including craniofacial cartilage and bone, neurons and glia of the peripheral nervous system, and melanocytes (Rogers et al., 2012). These cells are unique and interesting because they resemble stem cells in their ability to form multiple derivatives, as well as metastatic cancer cells in their ability to migrate and invade different tissues. Abnormal NC development can result in defective embryogenesis leading to birth defects.
A proposed neural crest gene regulatory network (NC-GRN) helps to explain some of the molecular interactions that are involved in neural crest cell specification, and their subsequent epithelial to mesenchymal transition (EMT) that leads to the formation of the migratory population (Sauka-Spengler and Bronner-Fraser, 2008). An important goal in the neural crest field is to identify additional factors that are involved in the NC-GRN, and to test their possible developmental roles. To aid in this goal, we previously conducted screens for genes that are upregulated during the process of neural crest formation (Adams et al., 2008; Gammill and Bronner-Fraser, 2002). Several transcription factors were identified that had not previously been known to function in the neural crest.
One of the genes upregulated as a consequence of neural crest induction is Elk3/Net/Sap2 (referred to hereafter as Elk3), a ternary complex factor that belongs to the Ets family of transcription factors. Elk3 is a transcriptional repressor that has been linked to cancer metastasis (Li et al., 2008; Sloan et al., 2009), wound healing, and vasculogenesis in cell lines and in mice (Ayadi et al., 2001b; Nozaki et al., 1996; Zheng et al., 2003). However, little is known about its role in normal embryonic development, although in vitro studies have suggested a role in cell migration (Buchwalter et al., 2005). Elk3 is expressed in the developing vasculature, the cranial mesenchyme and the limb buds of mouse embryos (Ayadi et al., 2001b). In addition, in vivo studies of the function of Elk3 during tumorigenesis in mice showed that individuals lacking Elk3 protein had smaller tumors due to their inability to become vascularized and oxygenated (Zheng et al., 2003).
Although well studied in vitro as a ternary complex factor, little is known about the function of Elk3 during early embryonic development. Here, we describe the spatiotemporal expression pattern and function of Elk3 during avian embryogenesis. In addition to its previously known distribution in the vasculature, we report that Elk3 is expressed in the neural folds and migratory cranial neural crest. The results show that loss of the Elk3 protein prevents proper neural crest specification, inhibits migration and results in the retention of precursor cells in the dorsal neural tube. Thus, in addition to its role in the vascular system, Elk3 also plays a critical role in neural crest development.
Materials and Methods
Embryos
Fertilized chicken eggs were obtained from local commercial sources (McIntyre Farms, San Diego, CA) and incubated at 37°C to the desired stages according to the criteria of Hamburger and Hamilton (HH). For whole mount in situ hybridization (WISH), embryos were fixed for one hour to overnight in 4% paraformaldehyde (PFA) at room temperature or 4°C, respectively, washed in PTW (PBS containing 0.1% Tween) and dehydrated in MeOH at room temperature before being stored at −20°C in 100% MeOH.
RNA Preparation and quantitative PCR
RNA was prepared from embryos (n = 10) using the RNAqueous-Micro isolation kit (Ambion) following the manufacturer's instructions. The obtained RNA was treated with DNaseI amplification grade (Invitrogen) and then reversed-transcribed to cDNA with SuperScript II (Invitrogen) using random hexamers. QPCR was performed using the 96-well plate ABI 7000 QPCR machine with Sybrgreen Itaq Supermix with ROX (Bio-Rad), 150–450 nM of each primer, and 200–500 ng of cDNA in a 25 µl reaction volume. During the exponential phase of the QPCR reaction, a threshold cycle (CT) and baseline was set according to the protocols of Applied Biosystems. The results for different samples were then analyzed using the Livak/ΔΔCt method. There were three replicates for every sample and GAPDH was used as a control.
Electroporation of Antisense Morpholinos and DNA Vectors
Two translation blocking antisense morpholinos to Elk3 (E3MO) were designed: E3MO1: 5’-TGTTCAGAGGGAGCTGGGTAAAACC-3’, E3MO2: 5’-TGATTGCACTCTCCATACCCAGAGC-3’ and one control morpholino (ContMO) was used: 5’-CCTCTTACCTCAGTTACAATTTATA-3’. Injections of the fluorescein-tagged morpholino (1–3 mM plus 0.5–1.5 mg/ml of PCI carrier plasmid DNA) (Voiculescu et al., 2008) were performed by air pressure using a glass micropipette targeted to the presumptive neural crest region at HH stages 4–5. Rescue experiments were performed in the same manner as E3MO electroporations except that the PCI-Elk3-RFP plasmid was used (.75 mg/ml) instead of the PCI carrier plasmid. Stage 4–5 electroporations were conducted on whole chick embryo explants placed ventral side up on filter paper rings. The Elk3 morpholinos and vectors were injected on the right side of the embryo, and the controls were injected on the left side when co-injected, and platinum electrodes were placed vertically across the chick embryos and electroporated with five pulses of 6.3 V in 50 ms at 100 ms intervals. The embryos were cultured in 1.0 ml of albumen in petri dishes until they reached the desired stages. The embryos were then removed and fixed for one hour to overnight in 4% PFA at room temperature or 4°C, respectively. The embryos were placed in PTW, viewed and photographed as whole mounts using a fluorescence stereomicroscope to show the electroporation efficiency.
Whole Mount In Situ Hybridization (WISH)
Chick WISH was performed as previously described (Acloque et al., 2008) with the following modifications: embryos were not treated with proteinase K, triethanolamine, or acetic anhydride, on day one, and the day two washes were shortened, no CHAPS was used and MABT was used instead of KTBT. The linearized plasmid DNA templates that were used for digoxigenin- and fluorescein-labeled RNA probes were Elk3, FoxD3, Sox10, Snail2 and Sox2. The embryos were imaged in whole mount and after were transverse sectioned at 14–25 µm in a Microm HM 550 cryostat.
Immunohistochemistry
Immunohistochemistry for Pax7 (Developmental Studies Hybridoma Bank (DSHB), Pax7), HNK1 (DSHB, 3H5), Snail2 (DSHB, 62.1E6), Sox2 (R&D Systems, AF-2018), Pax6 (Covance, PRB-2184), Caspase (R&D Systems, AF835) and PH3 (Upstate/Millipore, 06-570) was performed as follows. Embryos were fixed in 4% paraformaldehyde in phosphate buffer for twenty minutes at room temperature. All washes were performed in TBST + Ca2++ with .5% triton x-100. Blocking was performed with 10% donkey serum in the same buffer. The primary antibodies (1:10 for all hybridoma antibodies and 1:1000 for all others) were incubated in the TBST buffer from overnight to two days at 4° and secondary antibodies (1:500 to 1:1000) were applied in the same buffer for either three hours at room temperature or overnight at 4°.
Cell Death and Proliferation
Embryos were immunostained with the proliferation marker phosphorylated histone H3 (rabbit anti-PH3 IgG, 1:1000) or the apoptosis marker, caspase (rabbit anti-caspase IgG, 1:1000), and then detected using the secondary Alexa Fluor goat anti-rabbit 350 (1:1000).
Results
Sequence of chick Elk3 protein
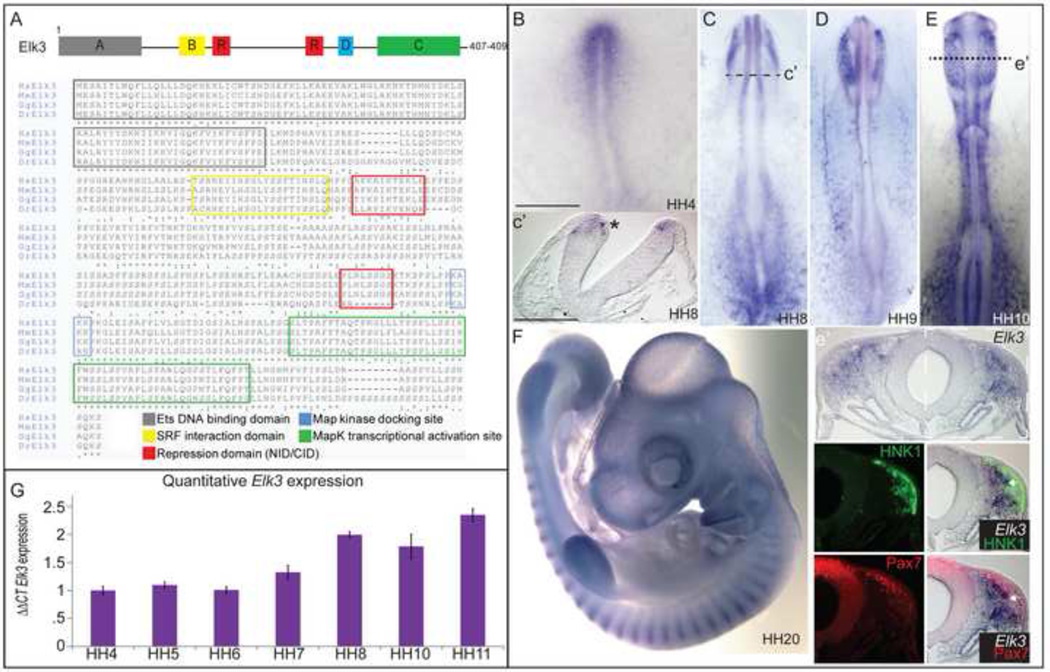
The chick Elk3 protein is highly conserved with human, mouse and zebrafish (Fig. 1A), exhibiting an overall amino acid sequence similarity of 86%-88% with mouse and human, respectively, 60% similarity with zebrafish and approximately 98% conservation in the functional domains between organisms. The chick Elk3 protein contains an Ets binding domain (amino acid (a.a.) residues 1–89), a serum response factor interaction domain (a.a. 141–162), two repressor domains (a.a. 166–176, 283–293), a mitogen activated protein (MAP) kinase docking site (a.a. 299–302) and a MAP kinase transcriptional activation site (a.a. 335–388).
Figure 1. Expression of Elk3 during avian development.
(A) Schematic diagram of the Elk3 protein with the six functional domains (identified by color) and the amino acid sequence. (B–F) Whole mount in situ hybridization (WISH) of Elk3 at stages HH4 (B), HH8-(C), HH9 (D), HH11 (D), and HH20 (F). Elk3 is expressed in the developing head mesenchyme, neural folds and migratory neural crest cells, intersomitic vessels and the developing limb buds. (E’) Section of HH11 embryo from (E). WISH of Elk3 expression (top panel), immunohistochemistry of HNK1 (middle panel), and overlay (bottom panel) showing that Elk3 expression overlaps with migratory cells. (G) Quantitative PCR showing the expression of the Elk3 transcript compared with HH4 embryos and normalized with GAPDH expression. The Livak/ΔΔCt method was used for the quantitative analysis. Scale bars: 200 µm (B–F), 100 µm (C’, E’).
Expression of Elk3 in the chick embryo
Using in situ hybridization to visualize the spatiotemporal expression pattern of transcripts, the Elk3 expression pattern appears very dynamic throughout the early stages of chick development. Elk3 is expressed in the anterior primitive streak by Hamburger-Hamilton (HH) stage 4 (Fig. 1B) and in the developing head folds and head mesenchyme at HH8 (Fig. 1C, 1C’). Elk3 is expressed at low levels in the early neural folds (Fig. 1C’), and transcript levels appear to decrease around the time of neural tube closure. At HH9, Elk3 remains in the developing head mesenchyme (Fig. 1D), which will eventually contribute to portions of the craniofacial bone, vasculature and muscle populations. Elk3 transcripts then appear to be elevated in a subpopulation of migrating cranial NC cells at HH10- 11, as well as in the R4 migratory stream and the cranial mesenchyme (Fig. 1E, 1E’). Elk3 transcripts partially overlap with the migratory neural crest cell marker, HNK1 (Fig. 1E’, green), and with Pax7 in the migratory crest (Fig. 1E’, red), but are below the level of detection by in situ hybridization in the dorsal neural tube (Fig. 1E’). By HH stage 20, Elk3 is expressed in the developing limb buds, the branchial arches, and the intersomitic blood vessels (Fig. 1F) similar to the patterns described in mouse embryos (Ayadi et al., 2001a).
To determine the quantitative expression levels of Elk3, we performed quantitative PCR (QPCR) on 10 whole embryos from stages HH4- HH11. When normalized to GAPDH expression and compared to the Elk3 expression levels at HH4 using the Livak/ΔΔCt method, Elk3 appears to be expressed at low levels through stages HH4–6, and the expression increases at HH7 just prior to the onset of neural crest specifiers like FoxD3 and Snail2. The expression peaks between stages HH8–11, as neural crest cells commence migration, and levels remain high as development proceeds (Fig. 1G).
Elk3 is required for cranial NC specification
Because Elk3 is expressed in the developing head folds beginning at HH7, we blocked the expression of the Elk3 protein during gastrulation at HH4 by electroporating one of two translation blocking morpholinos, referred to throughout as “E3MO”, prior to the onset of Elk3 expression in the neural folds. To examine the effects of Elk3 knockdown on cranial NC specification, we performed whole mount in situ hybridization (WISH) for the neural crest specifier genes, FoxD3 and Sox10 (Figure 2A–2E). Loss of the Elk3 protein greatly reduced the number of FoxD3-expressing cells on the injected side of the embryo at HH8 (Fig. 2A, arrow, N= 11/14), consistent with a requirement for Elk3 in cranial NC specification. In addition, the expression of Sox10 was greatly reduced on the Elk3-morphant side of the embryo at the same stage (Fig. 2C, arrow, N= 9/10). In transverse section, only cells lacking E3MO are seen to express Sox10 (Fig. 2E). Overexpression of the pCI-RFP-Elk3 DNA construct in concert with E3MO at HH4 was sufficient to rescue the expression of both FoxD3 and Sox10 at HH8 (Fig. 2B, 2D, N= 6/8 and 10/14, respectively), demonstrating the specificity of the effect. In addition, we examined Snail2 protein expression which, similar to FoxD3 and Sox10, was reduced at HH9 after Elk3 knockdown (Fig. 2F, 2F’, arrow, N= 15/16).
Figure 2. Elk3 is required for cranial NC specification.
(A–E) Whole mount in situ hybridization of FoxD3 (A–B) and Sox10 (C–E) in embryos electroporated at HH4 with either the E3MO alone (A, C, 2.5-3mM, arrows show loss of expression) or the E3MO with Elk3-RFP DNA (B, D, 3mM and 3µg/µl). (E) Section of embryo fixed at HH9 showing WISH for Sox10 expression (top panel) which is downregulated in the cells with E3MO (middle panel) and the overlay (bottom panel). (F, G) Immunohistochemistry of embryos electroporated at HH4 with ContMO on left and E3MO on right showing Snail2 (downregulation on right side compared with control side) and Pax7 protein expression. Dashed lines are sections of these embryos shown in F’ and G’. (H, I) Sections of HH9 embryo electroporated with RFP on the left side and Elk3 over-expression construct on the right side, whole mount immunohistochemistry for Snail2 (green, left panel) and Pax7 (green, right panel) was performed. Sections of these embryos are shown in H’ and I’ and overlay is below showing RFP from DNA constructs in red. 200 µm (A-D, F, G, H, I), 100 µm (E, F’-I’).
The effects of Elk3-knockdown on the neural crest specifier genes, FoxD3, Sox10, and Snail2 prompted us to examine the requirement of Elk3 for the expression of the neural plate border gene, Pax7, which is required for neural crest formation during gastrulation and lies upstream of neural crest specifier genes in the neural crest gene regulatory network (Basch et al., 2006; Betancur et al., 2010). During normal development, Pax7 is expressed in the newly induced crest population residing in the neural folds and is also expressed by migratory neural crest cells. Therefore, Pax7 represents a good marker for assessing whether Elk3 is required for maintenance of the neural crest precursor pool. Surprisingly, we noted that the loss of Elk3 resulted in a maintenance or increase of Pax7 protein in dorsal neural tube cells (Fig. 2G, 2G’, asterisk, N= 13/14) concomitant with the loss of FoxD3 and Sox10. Sections of an HH8–9 embryo immunostained for Pax7 expression show that Pax7 levels are unaffected in the presumptive midbrain at this stage (Fig. 2G’, asterisk), and may be increased in more posterior and “younger” dorsal neural tube cells (data not shown). This phenotype is consistent with the possibility that Elk3 may be required for the transition of NC precursors to NC cells, and that the loss of Elk3 maintains newly induced precursors in a “stem-like” state prior to the onset of definitive neural crest markers.
Since the loss of Elk3 led to a decrease in NC cell specifier genes and a maintenance or upregulation of Pax7, we examined whether the addition of exogenous Elk3 had an effect on neural crest specification. To this end, we electroporated an overexpression construct (pCI-Elk3-RFP, here referred to as “Elk3”) at HH4. The results suggest that Elk3 overexpression reduces the numbers of migratory Snail2+ neural crest cells while causing a minor upregulation of Snail in the premigratory neural crest (Fig. 2H, 2H’, N= 6/8). Pax7 appeared to be slightly decreased in a subset of the embryos (Fig. 2I, 2I’, N= 4/7) on the Elk3-injected side. The results suggest that over-expression of Elk3 can effect neural crest development, though not exactly opposite to that of its knock-down. These data are consistent with the possibility that maintenance of appropriate levels of this transcription factor in the neural crest is critical.
To determine when Elk3 activity is required during neural crest specification, we electroporated the E3MO from stages HH4- HH8. Elk3 knockdown between stages HH4- HH6 yielded similar results to those described in previous experiments; Sox10 expression and neural crest migration were reduced at both stages (Fig. 3A–3B, 3E–3F, N= 16/20). When the E3MO was electroporated at HH5, Pax7 and HNK1 expression were lost in migratory neural crest cells similar to effects of electroporations at HH4 (Fig. 3E, 3E’ compared to Fig. 2G, 2G’, N= 4/5). Electroporations performed at HH6 also affected Pax7 and HNK1 expression in migratory cells, but to a lesser extent (Fig. 3F, F’, N= 5/6). Thereafter, the effects were less penetrant with embryos appearing morphologically normal at stages 7/8 (Fig. 3C– 3D, 3G, N= 8/8), but somewhat delayed neural crest migration that was only detectable after sectioning (Fig. 3G’, arrow).
Figure 3. Loss of Elk3 affects cranial NC development in a stage-dependent fashion.
(A–D) Whole mount in situ hybridization of Sox10 (A–B) in embryos electroporated at stages HH5- HH8 with the E3MO on the right side (asterisks) and ContMO on the left side. E3MO affects neural crest development at all stages, but the severity declines with time and is not obvious by whole mount when electroporated at stage 8 (D). (E–G) Whole mount immunohistochemistry with antibodies to Pax7 (red) and HNK1 (blue) in embryos electroporated at stages HH5- HH8 with the E3MO on the right side and ContMO on the left side (Fitc, green). Similar effects are seen when E3MO is electroporated at stages HH5–6 as at earlier stages (see Fig. 2, 5). At HH7–8, the effects are less dramatic, but a delay in neural crest migration is still evident. (E’-G’) Cryosections of embryos from E–G showing overlays of Pax7/Fitc (top panel) and HNK1/Fitc (bottom panel). Scale bars: 200 µm (A–G), 100 µm (E’–G’).
To determine if the reduction of FoxD3 and Sox10 expression was due to a decrease in proliferation or an increase in cell death, we performed immunohistochemistry using a proliferation marker, phosphorylated histone H3 (PH3), and a marker of apoptosis, caspase, after knockdown of Elk3 using the E3MO. The results show that loss of the Elk3 protein had little effect on proliferation as evidenced by similar levels of PH3 on the injected and uninjected internal control side in both the whole mount and section (Fig. S1A, S1C–D, N= 9/11). This suggests that the loss of specifier genes is not due to a lack of proliferation. However, knocking down Elk3 at HH4 did cause a slight increase in cell death mainly localized to the ventral neural tissue (Fig. S1B, S1E, N= 12/15). Curiously, the ventral neural tube lacks Elk3 expression suggesting that this affect may be non-cell autonomous. In addition, we examined the effects of Elk3-RFP overexpression to determine if it is sufficient to induce cell proliferation or if excess Elk3 affects cell death. Overexpression of Elk3 had little effect on PH3 (Fig. S1F, S1H, N= 6/6) or caspase (Fig. S1G, S1I, N=4/4) expression when compared to the control (RFP, left side).
Elk3 is not required for neural development
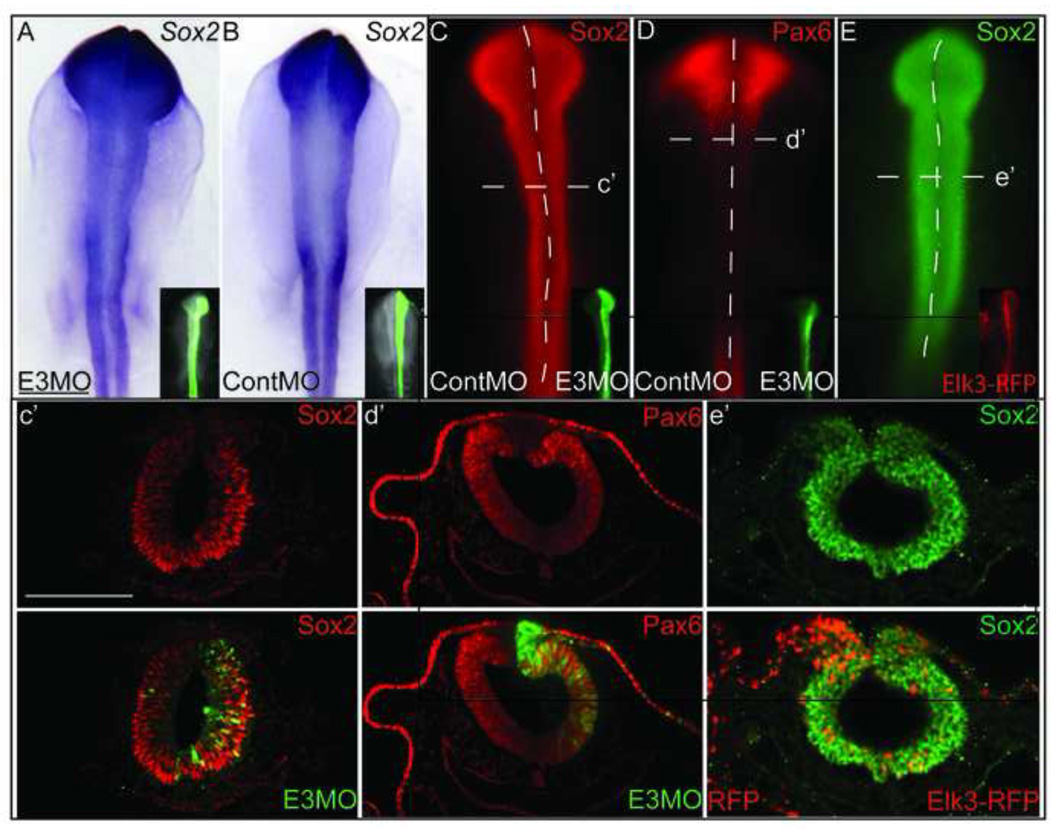
The neural plate border region can give rise to either neural crest or dorsal neural tube cells, which become part of the CNS (Krispin et al., 2010). Therefore, to verify that the loss of the neural crest specifier expression that was observed after Elk3 knockdown was not due to the inhibition of neural development, we electroporated the E3MO into HH4 embryos and performed WISH for the neural gene, Sox2, and performed immunohistochemistry for Sox2 and Pax6 proteins, both of which mark neural cells. The loss of Elk3 did not affect the expression of Sox2 (Fig. 4A, N= 5/6) when compared to the ContMO (Fig. 4B). In addition, immunohistochemistry for Sox2 (neural progenitors) and Pax6 (neural and ectodermal tissues) proteins, showed that there is no decrease in the expression of either Sox2 (Fig. 4C, 4C’, N= 12/13) or Pax6 (Fig. 4D, 4D’, N= 10/10) in the neural tube, and the ectoderm marked by Pax6 is unaffected in the Elk3-morphant cells (Fig. 4D’). Additional experiments were performed to determine if Elk3 overexpression affected neural development. Embryos injected with RFP (left side) and Elk3 (right side) had little change in Sox2 protein expression (Fig. 4E, 4E’, N= 4/4). These data support a role for Elk3 specifically in the development of cranial NC cells independent of non-NC ectodermal derivatives.
Figure 4. Loss of Elk3 does not affect neural or ectodermal development.
(A, B) Whole mount in situ hybridization of Sox2 and (C, D) immunohistochemistry showing no change in expression for Sox2 and Pax6 in embryos electroporated with the E3MO alone (A, B, right side) or with the ContMO and E3MO (C, D, ContMO is on left side and E3MO on right) at HH4. (E) Whole mount immunohistochemistry for Sox2 using an embryo electroporated with Elk3 at HH4 showing no effect. (C’-E’) Sections showing that loss and gain of Elk3 does not lead to a loss of neural tissue marked by Sox2 (C’, E’) or dorsal neural tube and ectoderm marked by Pax6 (D’). Scale bars: 200 µm (A–E), 100 µm (C’-E’).
Elk3 knockdown reduces cranial NC migratory streams and anterior cranial structures
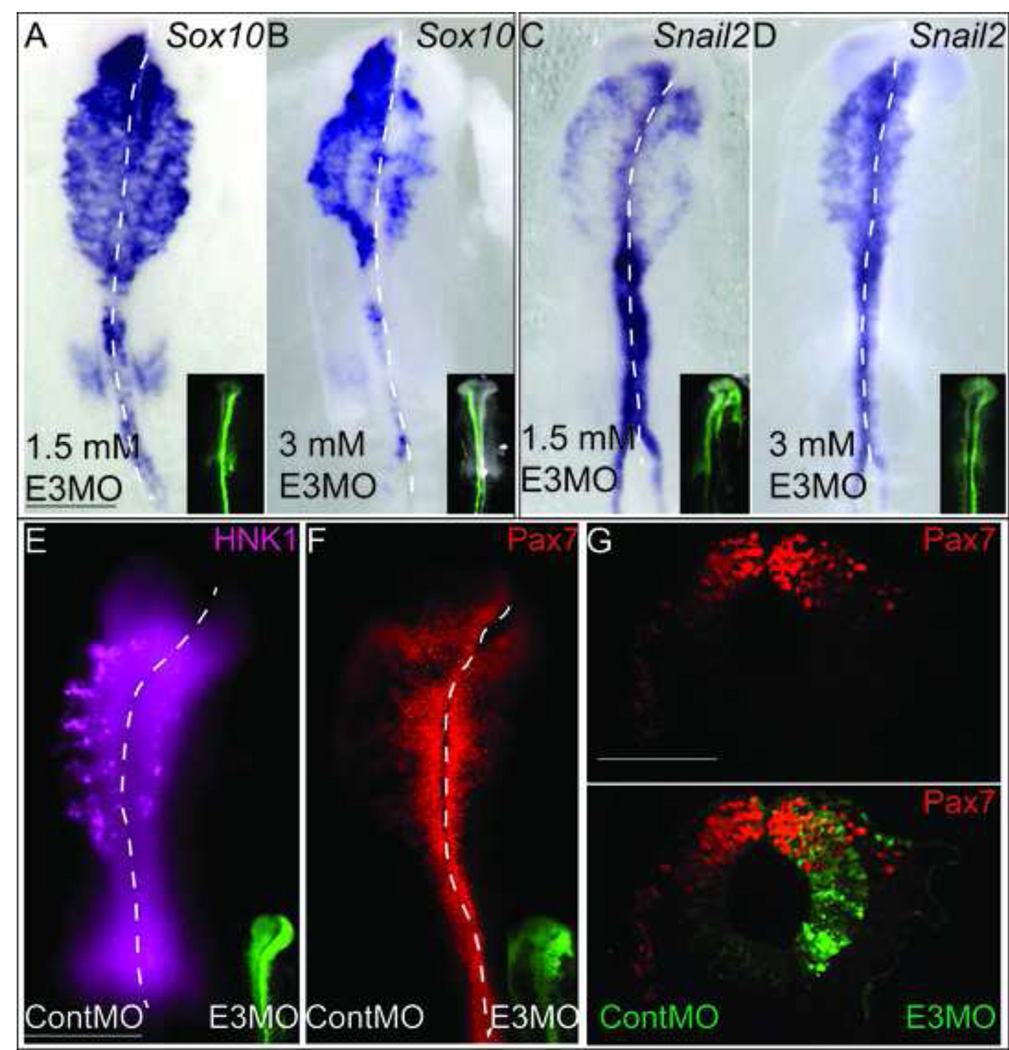
Although Elk3 plays a clear role in cranial NC cell specification, the Elk3 transcript is also expressed in migratory cranial NC cells. To examine if Elk3 is required for EMT or migration, we electroporated E3MO into HH4 embryos and performed WISH for Sox10 (Fig. 5A–5B) and Snail2 (Fig. 5C–5D) expression at migratory stages. The results show that loss of Elk3 has a dose dependent effect on both Sox10 (N= 17/22) and Snail2 (N= 9/12) expression in the migratory crest. At 1.5 mM, the E3MO reduces the migratory field marked by both Sox10 and Snail2 in stage HH9-HH10 embryos (Fig. 5A, 5C). At 3mM, the cranial NCs expressing Sox10 and Snail2 appeared to be even more reduced (Fig. 5B, 5D). We performed immunohistochemistry with the HNK1 antibody, which labels migrating neural crest cells, and observed a profound decrease in the HNK1 positive cells as well as the distance traveled by these cells on the E3MO-injected side compared to the control MO injected side (Fig. 5E, compare right side with left side, N= 11/11). Despite the reduction in Sox10, Snail2 and HNK1-positive migratory cells, prominent Pax7-expressing cells remained in the dorsal neural tube at HH9 (Fig. 5F, N= 9/9). In fact, the only Pax7-positive migratory cells are E3MO negative and hence are able to migrate laterally at HH10 (Fig. 5G) whereas the Pax7+/E3MO+ cells remain in the dorsal neural tube. These data support a role for Elk3 in the transition from Pax7-positive progenitor cells at the onset of NC cell specification to FoxD3/Snail2/Sox10-positive cells that form bona fide migratory neural crest cells.
Figure 5. Knockdown of Elk3 severely effects cranial NC migration.
(A–D) Whole mount in situ hybridization of Sox10 (A, B) and Snail2 (C, D) in embryos injected with the E3MO at HH4 showing a dose-dependent response (1.5mM and 3mM as indicated on figure). (E, F) Immunohistochemistry for HNK1 and Pax7 in HH9 embryos electroporated with ContMO on the left side and E3MO on the right side at HH4 (FITC in inset). (G) Section of an HH10 embryo immunostained for Pax7 (top panel) and overlayed with E3MO/FITC (bottom panel). Scale bars: 200 µm (A–F), 100 µm (G).
To determine if the effects of Elk3 knockdown recovered over time, E3MO-electroporated embryos were allowed to survive until 2–3 days post-electroporation (HH16–17). In all cases (n=7), the embryos exhibited severe anterioposterior head axis shortening (Fig. S2, asterisks) and craniofacial defects characterized by the loss of the most anterior structures, like the olfactory placode, a posterior to anterior shift of the eyes, and more dispersed and anteriorly-localized Sox10+ neural crest cells. In addition, the embryos exhibited abnormal branchial arch development, lack of trigeminal ganglion development and a failure to close the frontonasal process (Fig. S2, arrow). All embryos also had posterior blood vessel edema and heart developmental defects when compared to uninjected controls (data not shown). Thus, they failed to recover from the early specification and migratory effects after loss of Elk3.
Discussion
Elk3 is known to function as a transcriptional repressor that is required for wound healing, tumor growth and angiogenesis in mouse models, and also functions as an anti-oncogenic factor in some cancers (Li et al., 2008; Nakade et al., 2004; Zheng et al., 2003). Here, we show that during development, Elk3 is expressed in the cranial mesenchyme and the neural folds at the onset of neural crest specification as well as in migratory crest cells at later stages. Based on its conserved expression pattern in chick (Fig. 1B), mouse (Ayadi et al., 2001a), and frog (Bowes et al., 2010), together with high conservation in functional domains (98%) across species (Fig. 1A), we speculate that Elk3 may play a conserved role during vertebrate neural crest development.
Loss of function experiments demonstrate that Elk3 is critical for normal neural crest development. The results show that Elk3 is required for the expression of the neural crest specifier genes FoxD3, Sox10 and Snail2 (Fig. 2), and that its knockdown leads to a loss of migratory neural crest cells (Fig. 5) and later to abnormal craniofacial development (Fig. S2). Interestingly, Elk3-morphant cells in the dorsal neural tube maintain expression of Pax7 protein (Fig. 2G–2J, Fig. 5F, 5G). These cells are unable to upregulate FoxD3, Sox10 and Snail2 (Fig. 2A–2F), or to migrate out of the neural tube (Fig. 3, 5), whereas neural development proceeds normally (Fig. 4).
These results raise the intriguing possibility that cells receiving the E3MO may be arrested in a progenitor or stem-cell like stage prior to the expression of bona fide neural crest markers, like Sox10, Snail2 and FoxD3, suggesting that Elk3 may be a prerequisite for their subsequent expression. After Elk3 knock-down, the cells are maintained in a Pax7+ but bona fide neural crest marker negative state, which we speculate represents a “progenitor” state. Although there is an apparent reduction in Elk3 transcript levels around the time of neural tube closure, we cannot rule out the possibility that even low levels of transcript, perhaps below the level easily detectable by in situ hybridization, still produce enough protein to have a critical effect on downstream genes. In the context of the cranial neural crest gene regulatory network, this places Elk3 in a critical position between neural plate border and neural crest specifier genes, functioning downstream of the former, but upstream of the latter. Taken together, the results reveal a novel developmental role for Elk3 in neural crest cell specification, outside of its known roles in angiogenesis and cancer progression.
The neural plate border gene, Pax7, is required during gastrulation for NC cell fate specification in chicken (Basch et al., 2006) and later in development, Pax7-positive cells contribute to NC cell derivatives (Murdoch et al., 2012). However, Pax7-null mice exhibit very mild NC-related defects, but die soon after birth (Mansouri et al., 1996). Mansouri et al. (1996) suggest that in the mouse, Pax7 and its paralog, Pax3, may compensate for the loss of one another. In contrast, at early stages of chick development, Pax7 appears to be the dominant paralog since it is expressed earlier than Pax3, and knockdown of Pax3 during gastrulation has no effect on neural crest development (Basch et al., 2006).
The position of the E3MO containing cells in the dorsal neural tube and their maintenance of Pax7 expression suggests that they may represent either neural or neural crest progenitor cells. Given that they have low levels of Sox2 expression (Fig. 4A–4E), a known marker for neural progenitor cells (Graham et al., 2003), we speculate that they represent neural crest progenitors prior to the expression of neural crest specifier genes like FoxD3, Sox10 and Snail2. A similar situation has been observed for Pax7 expressing cells in the olfactory epithelium. Pax7 is expressed in olfactory precursor cells, which are multipotent and can give rise to multiple olfactory cell types (Murdoch et al., 2010). Our results suggest that in the neural crest, Elk3 may be required for the Pax7-positive dorsal neural tube cells to progress along the NC program; such that they subsequently upregulate NC cell specifiers enabling them to emigrate from the neural tube. Interestingly, we noted some cell death in the neural tube, which does not express detectable Elk3 by in situ hybridization. This neural cell death could be a non-cell autonomous effect due to a role of Elk3 in the mesenchyme adjacent to the neural tube (Fig. 1E’). Elk3 is also known to be a downstream effector of FGF signaling, which is involved in neural development.
Our data show that loss of Elk3 at HH4 leads to a severe reduction in migrating neural crest cells (Fig. 3, 5). One possible explanation for the abrogation of NC cell migration observed in Elk3-morphant embryos is that it may be a secondary consequence of the block to NC cell specification resulting in a failure to exit the neural tube and initiate migration. Consistent with this possibility, we observed numerous Pax7-positive cells retained in the dorsal neural tube of morphant, but not control embryos, at stages of active cranial NC migration (Fig. 5F, 5G). These Pax7+/Elk3-morphant cells apparently failed to migrate as the numbers of Pax7+ cells were much reduced in the periphery. Thus, we speculate that the lack of migration may be due to the maintenance of progenitor status (Pax7-positive precursor cells) that are unable to progress to definitive neural crest cells due to their failure to upregulate transcription factors like FoxD3 and Sox10, which are required for NC cell specification and migration (Drerup et al., 2009; Stewart et al., 2006).
An alternative possibility is that Elk3 may be directly required for the emigration of Pax7+ neural crest precursors cells. Elk3 is expressed in migratory NC cells (Fig. 1E, 1E’) consistent with a possible role in migration. However, Elk3 expression levels are reduced in the dorsal neural tube just prior to EMT. It is possible that Elk3 may regulate cell adhesion during NC cell migration in a similar manner to its function in mouse embryonic fibroblast migration (Buchwalter et al., 2005), where it acts as a transcriptional repressor. Thus, it would be interesting to test whether the loss of Elk3 in the premigratory NC cell population leads to a dysregulation of adhesion molecules, which are tightly regulated during the process of EMT (Kerosuo and Bronner-Fraser, 2012). The finding that Elk3 knockdown has a less severe, but still apparent, effect on NC cell migration when injected just prior to EMT (Fig. 3) seems consistent with a primary role in specification with an additional, but less penetrant role in migration.
Conclusions
In summary, our study reveals a novel function for the transcriptional regulator, Elk3, in neural crest development. The results place Elk3 downstream of neural plate border genes and upstream of neural crest specifiers in the NC gene regulatory network (Betancur et al., 2010) and establish Elk3 as a protein required for the progression of the NC program from progenitor to definitive neural crest cell. This research has allowed us to elaborate the NC-GRN and establish a new tier of regulation that may be critical for maintenance of neural crest “stemness.”
Supplementary Material
Figure S1. Elk3 knockdown does not affect proliferation, but leads to an increase cell death in the neural tube. (A–E) ContMO was injected on the left side and E3MO was injected on the right side of HH4 embryos (A, B) or Elk3 MO or ContMO were injected alone in the right side of HH4 embryos (C–E). Immunohistochemistry was performed for phosphorylated histone H3 (PH3) protein expression (A, C–D,F) and caspase (B, E, G–H). PH3 expression remained unchanged in the E3MO injected embryos while caspase levels increased slightly in the NT cells. (F–H) Immunohistochemistry for PH3 and caspase after Elk3 overexpression. RFP was injected into the left side of the embryos and Elk3-RFP in the right side. PH3 and caspase are shown in green and the RFP/Elk3 is shown in red (inset). Scale bars: 200 µm (A, B, F, G), 100 µm (C–E, H, I).
Figure. S2. Neural crest cells exhibit severe craniofacial defects after Elk3 knockdown at HH4. (A, B) Bright field images of embryos 48 hours after E3MO electroporation reveal severe malformations of the head, including failure to close the frontonasal process, branchial arch defects, missing olfactory placode and anterior shift of the eyes. (C, D) Whole mount in situ hybridization for Sox10 at HH16 in either an uninjected embryo or one injected with the E3MO at HH4 (1.5 mM) showing abnormal cranial neural crest development, with cells that are more dispersed and displaced anteriorly. The trigeminal ganglion fails to condense. Eye is circled with dotted line. Asterisks indicate anterior-posterior cranial axis shortening and arrows indicate the failure of the frontonasal portion of the face to close and branchial arch defects (N= 7/7). Scale bars: 200 µm (A-D).
Elk3 is expressed in the neural foldsprior to neural crest EMT, and in the migratory crest during early avian development.
Elk3 is required for neural crest specification marked by FoxD3 and Sox10.
Loss of Elk3 leads to the maintenance of Pax7 positive, but FoxD3/Sox10 negative cells in the dorsal neural tube.
Elk3 is required in the NC-GRN downstream of NC border genes and upstream of NC specifiers
Acknowledgements
We would like to thank Laura Gammill and Meghan Adams for previous identification of Elk3 and the Bronner lab for helpful discussions. This work was supported by NIH grant HD037105 to MEB, NIH minority supplement P01 HD037105 to CDR, and an Amgen summer fellowship and SURF fellowship to JLP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acloque H, Wilkinson DG, Nieto MA. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods in cell biology. 2008;87:169–185. doi: 10.1016/S0091-679X(08)00209-4. [DOI] [PubMed] [Google Scholar]
- Adams MS, Gammill LS, Bronner-Fraser M. Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn. 2008;237:1021–1033. doi: 10.1002/dvdy.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, Suelves M, Dolle P, Wasylyk B. Net, an Ets ternary complex transcription factor, is expressed in sites of vasculogenesis, angiogenesis, and chondrogenesis during mouse development. Mechanisms of development. 2001a;102:205–208. doi: 10.1016/s0925-4773(01)00289-1. [DOI] [PubMed] [Google Scholar]
- Ayadi A, Zheng H, Sobieszczuk P, Buchwalter G, Moerman P, Alitalo K, Wasylyk B. Net-targeted mutant mice develop a vascular phenotype and up-regulate eg-1. The EMBO journal. 2001b;20:5139–5152. doi: 10.1093/emboj/20.18.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annual review of cell and developmental biology. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. The ternary complex factor Net regulates cell migration through inhibition of PAI-1 expression. Molecular and cellular biology. 2005;25:10853–10862. doi: 10.1128/MCB.25.24.10853-10862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136:2623–2632. doi: 10.1242/dev.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Seminars in cell & developmental biology. 2012;23:320–332. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krispin S, Nitzan E, Kalcheim C. The dorsal neural tube: a dynamic setting for cell fate decisions. Developmental neurobiology. 2010;70:796–812. doi: 10.1002/dneu.20826. [DOI] [PubMed] [Google Scholar]
- Li B, Ni P, Zhu Q, Cao H, Xu H, Zhang S, Au C, Zhang Y. Growth inhibitory effect of the ternary complex factor Net on human pancreatic carcinoma cell lines. The Tohoku journal of experimental medicine. 2008;216:139–147. doi: 10.1620/tjem.216.139. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- Murdoch B, DelConte C, Garcia-Castro MI. Embryonic Pax7-expressing progenitors contribute multiple cell types to the postnatal olfactory epithelium. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:9523–9532. doi: 10.1523/JNEUROSCI.0867-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, Delconte C, Garcia-Castro MI. Pax7 lineage contributions to the Mammalian neural crest. PLoS One. 2012;7:e41089. doi: 10.1371/journal.pone.0041089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakade K, Zheng H, Ganguli G, Buchwalter G, Gross C, Wasylyk B. The tumor suppressor p53 inhibits Net, an effector of Ras/extracellular signal-regulated kinase signaling. Molecular and cellular biology. 2004;24:1132–1142. doi: 10.1128/MCB.24.3.1132-1142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M, Onishi Y, Kanno N, Ono Y, Fujimura Y. Molecular cloning of Elk-3, a new member of the Ets family expressed during mouse embryogenesis and analysis of its transcriptional repression activity. DNA and cell biology. 1996;15:855–862. doi: 10.1089/dna.1996.15.855. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Jayasena CS, Nie S, Bronner ME. Neural crest specification: tissues, signals, and transcription factors. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1:52–68. doi: 10.1002/wdev.8. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sloan KA, Marquez HA, Li J, Cao Y, Hinds A, O'Hara CJ, Kathuria S, Ramirez MI, Williams MC, Kathuria H. Increased PEA3/E1AF and decreased Net/Elk-3, both ETS proteins, characterize human NSCLC progression and regulate caveolin-1 transcription in Calu-1 and NCI-H23 NSCLC cell lines. Carcinogenesis. 2009;30:1433–1442. doi: 10.1093/carcin/bgp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Voiculescu O, Papanayotou C, Stern CD. Spatially and temporally controlled electroporation of early chick embryos. Nature protocols. 2008;3:419–426. doi: 10.1038/nprot.2008.10. [DOI] [PubMed] [Google Scholar]
- Zheng H, Wasylyk C, Ayadi A, Abecassis J, Schalken JA, Rogatsch H, Wernert N, Maira SM, Multon MC, Wasylyk B. The transcription factor Net regulates the angiogenic switch. Genes Dev. 2003;17:2283–2297. doi: 10.1101/gad.272503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Elk3 knockdown does not affect proliferation, but leads to an increase cell death in the neural tube. (A–E) ContMO was injected on the left side and E3MO was injected on the right side of HH4 embryos (A, B) or Elk3 MO or ContMO were injected alone in the right side of HH4 embryos (C–E). Immunohistochemistry was performed for phosphorylated histone H3 (PH3) protein expression (A, C–D,F) and caspase (B, E, G–H). PH3 expression remained unchanged in the E3MO injected embryos while caspase levels increased slightly in the NT cells. (F–H) Immunohistochemistry for PH3 and caspase after Elk3 overexpression. RFP was injected into the left side of the embryos and Elk3-RFP in the right side. PH3 and caspase are shown in green and the RFP/Elk3 is shown in red (inset). Scale bars: 200 µm (A, B, F, G), 100 µm (C–E, H, I).
Figure. S2. Neural crest cells exhibit severe craniofacial defects after Elk3 knockdown at HH4. (A, B) Bright field images of embryos 48 hours after E3MO electroporation reveal severe malformations of the head, including failure to close the frontonasal process, branchial arch defects, missing olfactory placode and anterior shift of the eyes. (C, D) Whole mount in situ hybridization for Sox10 at HH16 in either an uninjected embryo or one injected with the E3MO at HH4 (1.5 mM) showing abnormal cranial neural crest development, with cells that are more dispersed and displaced anteriorly. The trigeminal ganglion fails to condense. Eye is circled with dotted line. Asterisks indicate anterior-posterior cranial axis shortening and arrows indicate the failure of the frontonasal portion of the face to close and branchial arch defects (N= 7/7). Scale bars: 200 µm (A-D).