Abstract
Ubiquitin-dependent proteolysis plays a pivotal role in stress responses. To investigate the mechanisms of these cellular processes, we have been studying Schizosaccharomyces pombe mutants that have altered sensitivities to various stress conditions. Here, we showed that Lub1, a homologue of Ufd3p/Zzz4p/Doa1p in budding yeast, is involved in the regulation of ubiquitin contents. Disruption of the lub1+ gene resulted in monoubiquitin as well as multiubiquitin depletion without change in mRNA level and in hypersensitivity to various stress conditions. Consistently, overexpression of genes encoding ubiquitin suppressed the defects associated with lub1 mutation, indicating that the phenotypes of the lub1 mutants under stress conditions were due to cellular ubiquitin shortage at the posttranscriptional level. In addition, the lub1-deleted cells showed aberrant functions in ubiquitin/proteasome-dependent proteolysis, with accelerated degradation of ubiquitin. Also Cdc48, a stress-induced chaperon-like essential ATPase, was found to interact with Lub1, and this association might contribute to the stabilization of Lub1. Our results indicated that Lub1 is responsible for ubiquitin homeostasis at the protein level through a negative regulation of ubiquitin degradation.
Ubiquitin is a highly conserved 76-amino-acid protein found in all eukaryotes and is widely distributed throughout the cell. A major function of ubiquitin is to label proteins destined for selective degradation, which plays crucial roles in the regulation of protein levels responsible for various cellular processes (13, 14, 25, 38).
In fission yeast, several genes encoding ubiquitin were identified (24). The ubi1+ (SPAC11G7.04) and ubi3+ (SPAC6G10.11c) genes, encoding an N-terminal ubiquitin fused to the ribosomal proteins L40 and S37, respectively, belong to the class of ubiquitin genes whose translation product is a ubiquitin-ribosome fusion protein. The ubi4+ (GenBank accession no. AF095794) gene,encoding a multiubiquitin linear repeat of eight ubiquitins linked in tandem, belongs to another class of gene whose translation products are multiubiquitin fusion proteins. The products of ubiquitin genes are posttranslationally processed to ubiquitin monomers (24).
Most functions of ubiquitin can be understood in the context of this small protein acting as a tag. Ubiquitin is covalently attached to substrate proteins through an isopeptide bond between the C-terminal glycine of ubiquitin and an ɛ-amino group of a lysine residue on a substrate protein, which can be followed by further addition of ubiquitins within the linked ubiquitin itself to generate a multiubiquitin chain (25, 38). The resulting configuration of a multiubiquitin chain depends on which lysine residue (K48 or K63) within ubiquitin is used for isopeptide bond formation. The modification of proteins by ubiquitin has long been associated with the tagging of cytosolic and nuclear proteins for degradation by proteasomes. More recently, additional functions of ubiquitin have been described (25, 38). The ubiquitination of plasma membrane proteins for endocytosis is generally followed by lysosome/vacuole-mediated degradation. Whereas the K48-linked multiubiquitin chains are usually associated with proteasomal degradation, the K63 linkages are reportedly involved in various processes, including endocytosis. Thus, the linkage also provides an additional layer of specificity (25, 38).
In budding yeast several lines of evidence have demonstrated that ubiquitin, especially Ubi4p, is an essential component in the stress response. The transcription of UBI4 is strongly induced by a variety of environmental stresses, including heat shock, nutrient depletion, and exposure to DNA-damaging agents (31, 34, 37). In addition, UBI4 mutants are hypersensitive to several kinds of stress conditions (4, 8), and the degradation of abnormal proteins generated by stress conditions was targeted to the 26S proteasome through the K48-linked multiubiquitin modification (20). Also, based on altered degradation of ubiquitinated proteins in budding yeast, UFD3/ZZZ4/DOA1 was identified (15). This gene is required for ubiquitin-dependent proteolysis (9, 15) and plays a role in protein acetylation and ubiquitination (29), as well as in the determination of volatile anesthetic sensitivity (39). Although Ufd3p was shown to interact with Cdc48p, a chaperon-like ATPase (9), the significance of their association in relation to the function of Ufd3p has not been well elucidated. Cdc48 and its complex partners (Cdc48p, Npl4p, and Ufd1p), which constitute a ubiquitin-specific chaperon in the budding yeast, are critical components of the stress response, and it has been postulated that they effect the removal of ubiquitinated proteins from the endoplasmic reticulum and/or multiprotein complexes (12).
Decreased expression of ubiquitin could confer defects in ubiquitin/proteasome- and/or vacuole-dependent proteolysis (9, 30). In addition, the accumulation of large amounts of damaged or denatured proteins can lead to aberrant cell function or eventual cell death in yeast and mammalian cells (18, 26, 28), and this has been implicated in the pathogenesis of several diseases, including neurodegenerative disorder, hypertension, and uterine cervical carcinoma among others (5). Thus, an appropriate ubiquitin supply is essential for cellular homeostasis and cellular response to stress. However, the mechanisms underlying the regulation of cellular ubiquitin contents, especially at the protein level, need to be unraveled. Elucidation of these mechanisms may lead to a better understanding of the regulatory proteins in the control of cellular processes and may lead to the development of new strategies or drugs for the management of the diseases.
In an attempt to identify genes involved in the stress response, we screened for fission yeast mutants that showed increased sensitivity to UV and high temperature and isolated a mutant cell, identified as lub1-1 (low ubiquitin content). Here, we report that Lub1, together with Cdc48, participates in the maintenance of cellular ubiquitin contents. Disruption of the lub1+ gene resulted in an accelerated degradation of ubiquitin and caused defects in ubiquitin/proteasome-dependent proteolysis, rendering cells sensitive to several stress conditions. Furthermore, we show that the WD domain of Lub1 is essential for the stability of Lub1, probably through its interaction with Cdc48.
MATERIALS AND METHOD
Cell culture, treatments, and nomenclature
Fission yeast strains used in this study are listed in Table 1. The complete medium (yeast extract-peptone-dextrose [YPD]) and essential minimal medium (EMM) have been described previously (32). Standard methods for Schizosaccharomyces pombe genetics, as described by Moreno et al. (22), were followed. Gene disruptions are denoted by lowercase letters representing the disrupted gene followed by two colons and the wild-type gene marker used for disruption (e.g., lub1::ura4+) and are also abbreviated by the gene preceded by Δ (e.g., Δlub1). Proteins are denoted by roman letters with only the first letter capitalized (e.g., Lub1).
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| HM123 | h− leu1-32 | Our stock |
| HM528 | h+ his2 | Our stock |
| KP456 | h− leu1-32 ura4-D18 | Our stock |
| KP433 | h− leu1-32 lub1-1 | This study |
| KP1648 | h− leu1-32 ura4-D18 lub1-1 | This study |
| KP717 | h− leu1-32 ura4-D18 lub1::ura4+ | This study |
| KP970 | h− leu1-32 ura4-D18 mts2-1 | 11 |
| KP778 | h+ leu1-32 ubi4-H45 | 24 |
| 5A/1D | h−/h+ leu1-32/leu1-32 ura4- D18/ura4-D18 his2/+ ade6-M210/ade6-M216 | Our stock |
Isolation of lub1-1 mutants
The lub1-1 mutants were isolated in a screen of cells that had been mutagenized with nitrosoguanidine (22). The wild-type HM123 cells were mutagenized with 300 μM nitrosoguanidine (Sigma) for 1 h (approximately 10% survival). Mutants, in appropriate dilutions, were seeded onto YPD plates and incubated at 27°C. After 4 days, the mutants were replica plated onto YPD plates and were incubated at 36°C or UV irradiated (100 J/m2, 254 nm; UV linker FS-800; Funa) and incubated at 27°C. Mutants that showed both temperature and UV sensitivities were selected. The original mutants isolated were backcrossed three times to wild-type strain HM123 or HM528.
Cloning and disruption of the lub1+ gene
To identify the mutated gene, lub1-1 mutants were transformed with an S. pombe genomic DNA library constructed in pDB248 (22). The Leu+ transformants were replica plated onto YPD plates and incubated at 36°C, and plasmid DNA was recovered from 12 transformants that showed plasmid-dependent rescue. All 12 plasmids had identical or overlapping inserts, as judged from the restriction digests, and all complemented both UV and temperature sensitivities of the lub1-1 mutants. A one-step gene disruption by homologous recombination was performed (27). Construction of the lub1::ura4+ disruption was performed as follows. The PstI-SacI fragments (3.3 kb) containing the full-length lub1+ gene generated by PCR from genome DNA of wild-type cells were subcloned into the PstI-SacI site of pGEM-5Zf (+) (Promega). Then, HindIII fragments containing the ura4+ gene (1.8 kb) were inserted into the HindIII sites of the previous constructs. The PstI-SacI fragments containing the disrupted lub1+ gene (2.9 kb) were transformed into diploid cells (5A/1D). Stable integrants were subsequently cloned on plates coated with medium lacking uracil, and gene disruption by the lub1+ derivative containing the ura4+ insertion was confirmed by genomic Southern blotting (data not shown).
Plasmids
The DNA fragments containing the coding sequences of lub1+, cdc48+, and the ubiquitin gene were amplified by PCR with genomic DNA as a template. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene) for the construction of K48R and K63R mutant ubiquitins. PCR fragments were inserted into the pREP1 expression vector (21), containing a thiamine-repressible nmt1 promoter, producing a protein fusion with glutathione S-transferase (GST) or FLAG, so that the protein was tagged with epitope sequences at the N terminus. Similarly, the coding sequences for the N-terminal half containing the WD domain [Lub1 (1-279)], C-terminal region [Lub1 (230-713)], and other truncated mutant proteins were amplified by PCR with the above construct containing the full-length lub1+ gene as a template and were subcloned into the pREP1-GST vectors. The constructs were confirmed by sequencing. GST-Lub1 was characterized to be fully functional as demonstrated by the suppression of Δlub1 phenotypes (data not shown). The expression vector for the polypeptide including the destruction box of Cdc13 (TA13), B-type cyclin in fission yeast, was a generous gift from Y. Hiraoka (7).
Western blot analysis
Cells were cultured for 18 h to logarithmic phase. In measuring protein stability, cells were placed in fresh medium supplemented with 5 μM thiamine (for FLAG-ubiquitin) or 100 μg of cycloheximide/ml (for Cdc13-green fluorescent protein [GFP] and endogenous ubiquitin). Total-cell lysates were prepared as follows. Approximately 2 × 107 cells were resuspended in 500 μl of NaOH lysis buffer (1.85 M NaOH, 7.5% 2-mercaptoethanol). After incubation on ice for 10 min, proteins were precipitated by the addition of 500 μl of 50% trichloroacetic acid. The pellets were washed and solubilized in sodium dodecyl sulfate (SDS) sample buffer (10% glycerol, 5% 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8). Equal amounts of proteins (30 μg/lane) were subjected to SDS-polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). Membranes were probed with mouse monoclonal antibodies against FLAG (Sigma) and multiubiquitin (MBL, Nagoya, Japan) or rabbit polyclonal antibodies against GST, GFP, and ubiquitin (MBL). The specific signals were detected with an ECL system (Amersham Pharmacia Biotech, Tokyo, Japan). Positions of molecular mass markers (in kilodaltons) were indicated.
Northern blot analysis
Total RNA was isolated as described previously (17). The total-RNA samples (20 μg/lane) were separated by 1% formaldehyde-agarose gel electrophoresis and blotted onto a nylon membrane (Amersham Pharmacia Biotech). Membranes were subsequently hybridized with digoxigenin (DIG)-labeled full-length ubi4+ cRNA as a probe for ubi4+ mRNA. The specific signals were detected with a DIG system (Roche Applied Science, Tokyo, Japan), according to the instruction manual from the manufacturer. Equality of the loading mRNA was confirmed by hybridization with leu1+ cRNA.
Immunoprecipitation
Cells were suspended in immunoprecipitation buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM EGTA, 0.2 mM sodium orthovanadate, 0.2 mM phenylmethylsulfonyl fluoride) containing glass beads. Cells were broken mechanically by a bead beater (BioSpec Products Inc.) for 1 min, after which tubes were placed on ice for 1 min. This process was repeated three times. Insoluble materials including glass beads were removed by centrifugation. Equal amounts of proteins were immunoprecipitated with an agarose-immobilized anti-FLAG antibody (Sigma). The immunoprecipitates were washed three times with 1 ml of the immunoprecipitation buffer and then were resuspended in SDS sample buffer. After boiling for 5 min, the complexes were evaluated by Western blot analysis.
Measurement of proteasome activity
Preparation of cellular extracts and measurement of proteasome activities were carried out as described previously with some modifications (23). Cell extracts were prepared from immunoprecipitation buffer containing glass beads as described above. For the activity assay, cellular extracts (see Fig. 5B, lane 1, 10 μg; lane 2, 20 μg; lanes 3 to 6, 40 μg) and 20 μM fluorogenic proteasome substrate Suc-LLVY-MCA (Peptide Institute Inc., Osaka, Japan), which is cleaved by purified proteasome (36), were incubated at 37°C for 1 h. Then the reaction was stopped with the addition of 2% SDS, and the proteasome activity was determined with a spectrofluorometer (Hitachi, Tokyo, Japan; excitation at 380 nm and emission at 460 nm). For inhibition studies, the extract was first incubated with 50 μM MG132 (Peptide Institute Inc.) for 30 min at 37°C and then the substrate was added to the assay mixture.
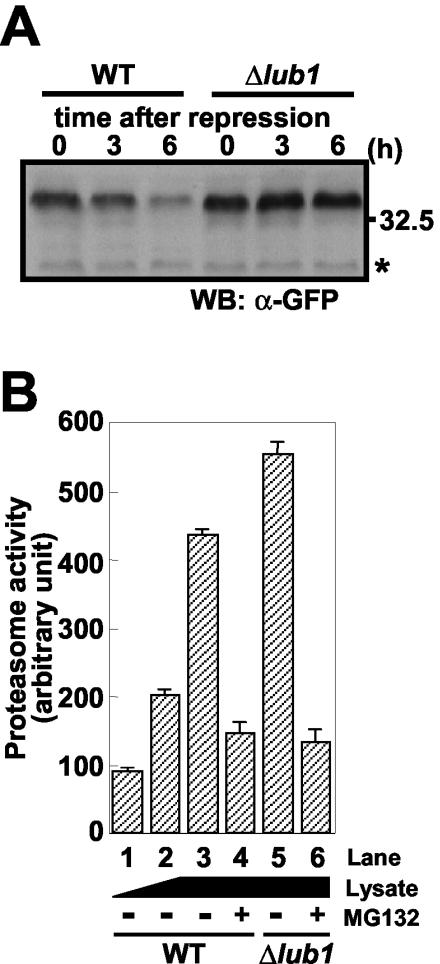
FIG. 5.
Defects in ubiquitin/proteasome proteolysis. (A) Wild-type (WT) and Δlub1 cells harboring the expression vector for Cdc13-GFP were cultured, 100 μg of cycloheximide/ml was added to the medium, and then the cells were cultured for the indicated times. The total lysates were analyzed by immunoblotting with an anti-GFP antibody. The equality of the loading protein was confirmed by the nonspecific band (asterisk). WB, Western blotting. (B) Δlub1 cells showed intact proteasome activities. The extracts from the wild-type and Δlub1 cells were prepared. Equal amounts of proteins were first incubated with the inhibitor MG132 at 37°C for 30 min, and the substrate for the proteasome (Suc-LLVY-MCA) was added to the assay mixtures. After a 1-h incubation at 37°C, the proteolytic activities were determined, as described in Materials and Methods. Proteasome activity dependent on the amounts of cellular protein (lane 1 to 3) and the effects of lub1 deletion (lanes 3 and 5) and the proteasome inhibitor (lanes 4 and 6) on proteasome activity are indicated. The data represent mean values, and bars indicate the standard deviations of triplicate determinations.
RESULT
Isolation and disruption of lub1+ gene
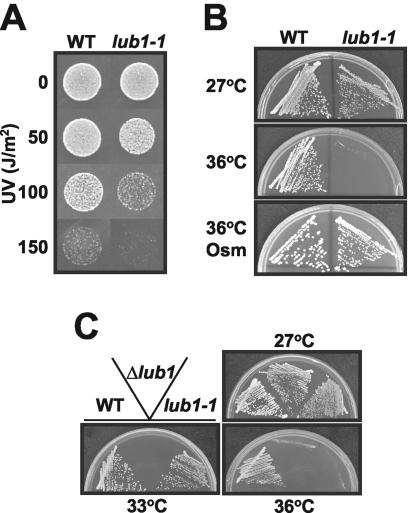
In an attempt to identify genes involved in the stress responses, we obtained a fission yeast mutant cell, identified as lub1-1, that showed increased sensitivity to both UV and high temperature. Growth rates of lub1-1 cells were almost the same as those of wild-type cells at the permissive temperature (27°C), whereas lub1-1 cells showed very high sensitivity to UV irradiation (Fig. 1A) and high temperature (Fig. 1B, middle), as well as to several stress-inducing agents, such as Ca2+, methyl methanesulfonate, sodium arsenite, hydrogen peroxide, caffeine, and ethanol (data not shown). The temperature-sensitive phenotype of lub1-1 cells was suppressed by hyperosmolarity (Fig. 1B, bottom), suggesting that lub1-1 cells had defects in cell wall integrity (32). Indeed, lub1-1 cells showed hypersensitivity to β-glucanase treatments compared with wild-type cells (data not shown).
FIG. 1.
The lub1-1 mutants and Δlub1 cells showed a hypersensitivity to stress conditions. (A) The wild-type and lub1-1 cells were cultured at 27°C to exponential phase and seeded with appropriate dilutions onto YPD plates. Cells were exposed to UV irradiation at the indicated doses and incubated at 27°C. (B) The wild-type and lub1-1 cells were streaked onto YPD plates and incubated at 27 (top) or 36°C (middle), respectively. Cells were streaked onto YPD plates containing 1.2 M sorbitol (Osm) and incubated at 36°C (bottom). (C) Wild-type, lub1-1, and Δlub1 cells were streaked onto YPD plates and incubated at 27, 33, and 36°C, respectively.
The lub1+ gene was identified by complementation of the temperature-sensitive growth defects of lub1-1 cells. Nucleotide sequencing of the cloned DNA fragments revealed that the lub1+ gene is SPBC887.04c (National Center for Biotechnology Information accession number, NP_596478). It encodes a novel highly conserved 713-amino-acid protein similar to the product of budding yeast UFD3/DOA1/ZZZ4 (9, 29, 39) and to human PLAP (6). To investigate the gene functions, the lub1+ gene was knocked out in a diploid by homologous recombination with the ura4+ marker gene, as described in Materials and Methods. The tetrad analysis of the heterozygous diploid showed four viable spores, indicating that the lub1+ gene is nonessential for cell growth (data not shown). Notably, the Δlub1 cells exhibited pleiotropic defects more severe than that of the lub1-1 cells (Fig. 1C). Microscopic observations revealed that the majority of the Δlub1 cells were enormously enlarged and round even at the permissive temperature. In addition, the experiments using GFP-tagged Lub1 demonstrated that Lub1 was localized to the whole cytoplasm and nucleus regardless of their culture conditions (data not shown).
The C terminus of Lub1 is responsible for its functions
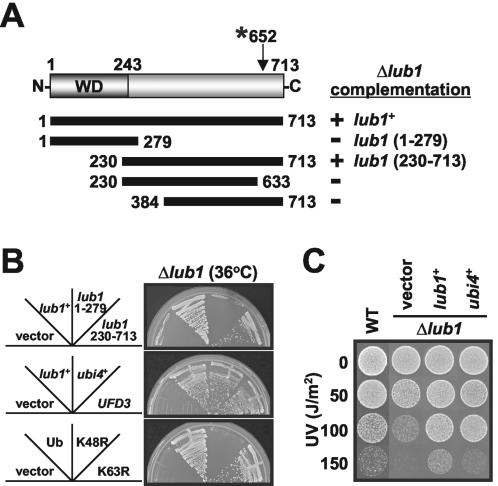
To determine the region responsible for Lub1 function, we examined the Δlub1 cells expressing a series of truncated mutant proteins. As shown in Fig. 2B, top, defects caused by the loss of the lub1+ gene were suppressed by the expression of Lub1 (full length) or Lub1 (230-713), whereas Lub1 (1-279) had no effects. Note that the effects of Lub1 (230-713) on the restoration of the Δlub1 phenotype were not comparable to those of Lub1 (full length). Further analysis revealed that the functional domain of Lub1 is located at the region comprising amino acids 230 to 713. Specifically, the regions comprising amino acids 230 to 384 and 633 to 713 were indispensable for Lub1 function (Fig. 2A). In addition, the same results were observed with GST-tagged Lub1s (data not shown). These results indicate that the C terminus of Lub1, rather than the N-terminal WD domain, is important for its functions. Consistent with these results, the mutation site of the lub1-1 allele is at codon 652 (proline), and the mutation results in a stop codon (Fig. 2A). Since the amino acid sequence of Lub1 is similar to that of Ufd3p/Zzz4p/Doa1p in budding yeast, we examined whether Ufd3p could substitute for Lub1 functions. As shown in Fig. 2B, middle, expression of Ufd3p completely suppressed the Δlub1 phenotypes, as observed for Lub1 expression, indicating that Ufd3p is a functional homologue of Lub1.
FIG. 2.
The C-terminal region is responsible for Lub1 functions. (A) Schematic representation of Lub1 truncated mutants. The ability of each Lub1 deletion mutant to suppress the temperature sensitivity of Δlub1 cells is summarized. +, positive; −, negative; WD, WD repeat domain; *, mutation site. (B and C) Identification of the dosage-dependent suppressor of the Δlub1 phenotype. Δlub1 cells harboring the expression vector for the indicated proteins were streaked onto EMM plates and incubated at 36°C (B) or UV irradiated at the doses indicated and incubated at 27°C (C) as described for Fig. 1. Ub, FLAG-ubiquitin; K48R and K63R, FLAG-ubiquitin (K48R and K63R) mutants; WT, wild type.
Overexpression of ubiquitin rendered Δlub1 cells resistant to stress conditions
To further investigate Lub1 function, we next identified dosage-dependent suppressors of the Δlub1 phenotype. The ubi4+ gene (GenBank accession no. AF095794 [24]),encoding a multiubiquitin fusion protein, was identified as a multicopy suppressor for the temperature sensitivity phenotype of Δlub1 cells (Fig. 2B, middle), and UV sensitivity was also recovered by ubi4+ overexpression (Fig. 2C). The Δlub1 cells carrying the ubi1+ and ubi3+ genes in multicopy plasmids were also able to reverse the pleiotropic phenotypes and morphological defects (data not shown).
Recent studies demonstrated that K48-linked chains are usually associated with proteasomal degradation, whereas K63 linkage is involved in a variety of cellular events including endocytosis and DNA repair (38). To determine which configuration of the ubiquitin linkages is responsible for the complementation of Δlub1 phenotypes, we evaluated the effects of K48R and K63R mutant ubiquitins. These mutant proteins have the invariant lysine in position 48 or 63 mutated to arginine, respectively, and expression of these mutant ubiquitins has a chain-terminating effect, resulting in the premature termination of ubiquitin chains. As shown in Fig. 2B, bottom, K48R ubiquitin, unlike K63R ubiquitin, failed to suppress the temperature sensitivity of Δlub1 cells. These results suggested that the temperature sensitivity phenotype of Δlub1 cells is due to the lack of ubiquitins that are needed to tag proteins for proteasomal degradation.
The Δlub1 cells showed defects in ubiquitin/proteasome-dependent proteolysis associated with acceleration of ubiquitin degradation
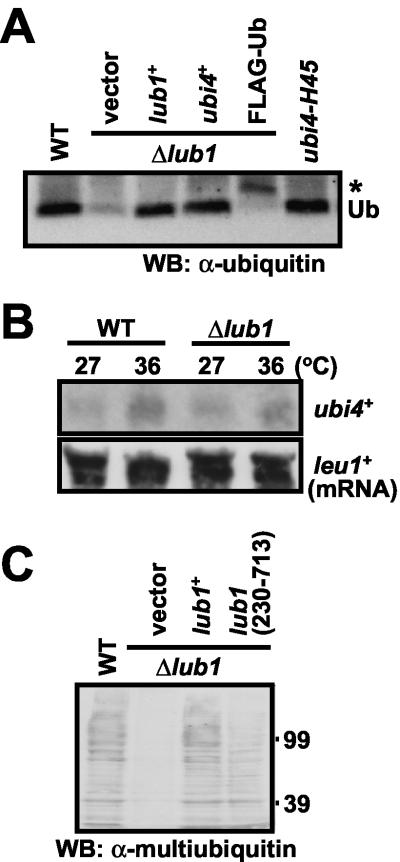
Since the insufficient expression of ubiquitins was expected in Δlub1 cells, we next examined the cellular ubiquitin contents in Δlub1 cells. As shown by immunoblot analysis using an antiubiquitin antibody (Fig. 3A), the expression level of monoubiquitin was dramatically decreased in Δlub1 cells compared with those of wild-type cells and ubi4-H45 mutants, which are defective in Ubi4 expression (24). Consistently, the expression level of multiubiquitin in Δlub1 cells was also decreased (Fig. 3C). On the other hand, no difference in the expression and induction of ubi4+ mRNA between wild-type and Δlub1 cells was observed (Fig. 3B).
FIG. 3.
Decreased expression of cellular ubiquitin contents in Δlub1 cells. (A) Intracellular expression of monoubiquitin. Wild-type (WT), ubi4-H45, and Δlub1 cells harboring the expression vector alone or the vector for Lub1, Ubi4, or FLAG-ubiquitin were cultured for 18 h. The whole-cell lysates were analyzed by immunoblotting with an antiubiquitin antibody to determine the monoubiquitin expression level. Positions of monoubiquitin (Ub) and FLAG-tagged ubiquitin (asterisk) are indicated. WB, Western blotting. (B) Northern blot analysis of ubi4+ mRNA. The wild-type and Δlub1 cells were cultured at 27 or 36°C, respectively. The total RNA was isolated and analyzed by Northern blotting with DIG-labeled ubi4+ cRNA. Equality of the loading mRNA was confirmed with leu1+ cRNA. (C) Immunoblot analysis of multiubiquitin. The wild-type and Δlub1 cells harboring the expression vector for Lub1 (full length) or Lub1 (230-713) were cultured for 18 h. The determination of multiubiquitin expression level is as described for panel A.
To clarify the relationship between Lub1 and ubiquitin expression, we examined whether the expression of Lub1 would reverse the intracellular ubiquitin levels. As shown in Fig. 3A, depletion of monoubiquitin in Δlub1 cells was blocked completely by the overexpression of the lub1+ gene. Moreover, decreased levels of multiubiquitin were also reversed by the expression of Lub1 (full length) or truncated mutant Lub1 (230-713) (Fig. 3C). However, Lub1 (230-713) expression did not completely restore the levels of multiubiquitin, and this is consistent with the results observed in Fig. 2B, top. These findings suggested that the decrease in cellular levels of ubiquitin in Δlub1 cells occurred at the posttranscriptional level, presumably by the promotion of ubiquitin degradation.
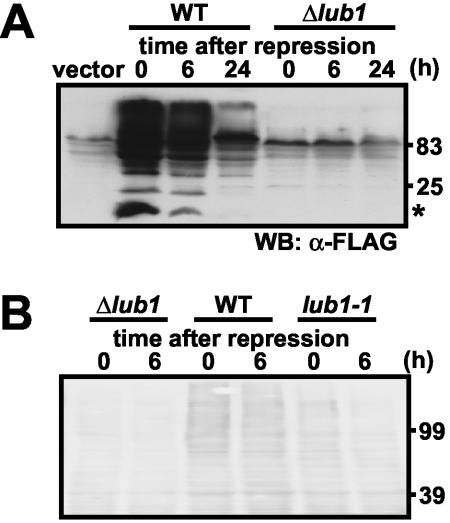
To elucidate this further, we examined the turnover of exogenously expressed ubiquitin. Time course immunoblot analysis after the repression of the nmt1 promoter by thiamine revealed that the levels of mono- and multiubiquitin in Δlub1 cells were dramatically decreased even in the time zero point after repression (Fig. 4A). Consistently, expression levels of endogenous ubiquitin in Δlub1 cells were also very low (Fig. 4B and as already demonstrated in Fig. 3). These results strongly suggested that we could not detect ubiquitin expression in Δlub1 cells because degradation of ubiquitin appeared to occur very rapidly. To clarify this point, we next examined the ubiquitin turnover in lub1-1 cells. It is important that the lub1-1 cells exhibited a milder growth defect than Δlub1 cells (Fig. 1C). Then, we found that ubiquitin levels in lub1-1 cells were marginal compared to those in wild-type cells under the same culture conditions (Fig. 4B). In addition, ubiquitin turnover in lub1-1 cells was much faster than that in wild-type cells (Fig. 4B). These findings collectively suggested that increased ubiquitin turnover was an explanation for the low ubiquitin contents in lub1-deleted cells.
FIG. 4.
Accelerated degradation of ubiquitin by lub1 deletion. (A) Ubiquitin failed to accumulate in Δlub1 cells. Wild-type (WT) and Δlub1 cells harboring the expression vector for FLAG-ubiquitin were cultured and then were treated with 5 μM thiamine for the indicated times. The whole-cell lysates were prepared and subjected to immunoblot analysis. Positions of monoubiquitin (asterisk) are indicated. WB, Western blotting. (B) Accelerated degradation of endogenous ubiquitin. Wild-type and Δlub1 and lub1-1 mutant cells were cultured and then were treated with 100 μg of cycloheximide/ml for the indicated times. The total lysates were analyzed by immunoblotting with an antimultiubiquitin antibody.
Notably, the degradation of Cdc13-GFP, containing a B-type cyclin destruction box (7), which is a well known substrate in ubiquitin/proteasome-dependent proteolysis, was impaired in Δlub1 cells (Fig. 5A). This was similarly observed in mts2-1 cells (11) (data not shown), and the overexpression of Lub1 suppressed the temperature sensitivity of mts2-1 cells (data not shown). These results demonstrated that Δlub1 cells had defects in ubiquitin/proteasome-mediated proteolysis.
The Δlub1 cells showed intact proteasome activity
The ubiquitin/proteasome pathway is composed of the ubiquitin-conjugating system and the 26S proteasome as the proteolytic core. To clarify which process is impaired, we measured the proteasome activities in Δlub1 cells. Proteasome activity in the extract was assessed by determining the cleavage of Suc-LLVY-MCA fluorogenic peptides. The increase in cleavage activity depended on the amounts of protein extracts (Fig. 5B, lanes 1 to 3). To further examine the proteasome activity, we studied the effects of proteasome inhibitors on the peptide cleavage activities. As shown in Fig. 5B, lanes 4 and 6, cleavage activities were inhibited by MG132, a selective proteasome inhibitor (19). However, a strong calpain inhibitor (ZLLal [35]) with a structure related to that of MG132 (ZLLLal), showed little effect at the same concentration as MG132 (data not shown). The cleavage activities in Δlub1 cells were slightly enhanced compared with that in wild-type cells (Fig. 5B, lane 5), indicating that the proteasome itself is, at least, intact. These results suggested that defects in ubiquitin/proteasome-mediated proteolysis associated with lub1 deletion are caused by accelerated ubiquitin degradation and not by the proteasome itself.
The WD domain of Lub1 is required for its stability
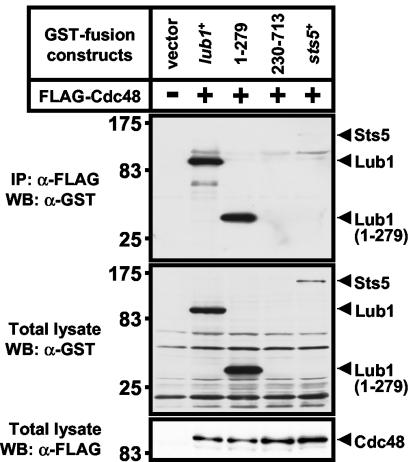
A previous study demonstrated that Ufd3p showed direct binding with Cdc48p in budding yeast (9). To elucidate the Lub1-Cdc48 interaction, we performed an immunoblot analysis with immunoprecipitation. Lub1 (full length), Lub1 (1-279), and Lub1 (230-713) were individually coexpressed with FLAG-Cdc48 in KP456 cells. As shown in Fig. 6, we found that Lub1 (full length) and Lub1 (1-279) were associated with Cdc48, whereas the binding between Cdc48 and Sts5 (33) could not be detected (Fig. 6, top). In contrast to Lub1 (full length) and Lub1 (1-279) expression levels in total lysates, expression of Lub1 (230-713), lacking the WD domain, was hardly detected (Fig. 6, middle). These results demonstrated that the WD domain of Lub1 is important for the interaction with Cdc48 and suggested that Cdc48 plays a crucial role in the stability of Lub1.
FIG. 6.
Interaction between Lub1 and Cdc48 in vivo. The GST-tagged Lub1 (full length) and truncated mutant proteins Lub1 (1-279) and Lub1 (230-713) or Sts5 (for negative control) were coexpressed with FLAG-Cdc48 in KP456 cells, respectively. The cell lysates were immunoprecipitated (IP) by an agarose-immobilized anti-FLAG antibody, and the immunoprecipitates were subjected to immunoblot analysis using an anti-GST antibody (top). Total lysates were also subjected to immunoblot analysis using an anti-GST antibody (middle) and an anti-FLAG antibody (bottom), respectively. WB, Western blotting.
DISCUSSION
The ability of cells to adapt to environmental stresses such as fluctuations in nutrients, temperature, and exposure to UV irradiation or to other toxic components is important for maintaining cellular homeostasis in yeast and mammalian cells. Ubiquitin/proteasome-mediated protein degradation is one of the major components of the stress response, which includes degradation of abnormal or misfolded proteins and the regulation of stress response pathways (13, 14, 25, 38). Specifically, the induction of ubi4+ mRNA by stress conditions is thought likely to be the primary mechanism for increasing the pool of ubiquitin for the degradation of stress-damaged proteins (8).
In a screen for S. pombe mutants that showed increased sensitivity to both UV and high temperature, we isolated the lub1+ gene, which encodes a novel and highly conserved protein containing the WD repeat motif. The Δlub1 cells showed a decreased expression of endogenous monomeric ubiquitins as well as multiubiquitins (Fig. 3), rendering the cells sensitive to various stress conditions (Fig. 1; data not shown). Notably, the decreased level of ubiquitin was not accompanied by a corresponding change in ubi4+ mRNA level (Fig. 3B), indicating that Lub1 is involved in the posttranscriptional regulation of cellular ubiquitin contents and stress responses. Supporting this was the finding that genes encoding ubiquitin complemented the phenotypes caused by lub1 deletion (Fig. 2B). In addition, Δlub1 cells showed synthetic lethality with ubi4+ gene deletion (data not shown). These results clearly indicate that Lub1 is involved in ubiquitin homeostasis, and we propose that Lub1 might be a negative regulator of ubiquitin degradation. Ubiquitin homeostasis, except for transcriptional regulation, has not been well documented. Here, it is notable that Lub1 is involved in the posttranscriptional regulation of ubiquitin homeostasis.
The two main proteolytic systems are the vacuole/lysosome and the proteasome. Although the type of proteolytic pathway involved in ubiquitin degradation is not known, Lub1 could be involved in vacuole-dependent ubiquitin degradation, including autophagy, because loss of ubiquitin/proteasome proteolysis leads to the activation of vacuole-dependent proteolysis (10). It is noteworthy that vacuole-dependent protein degradation is not necessarily accompanied by nonselective degradation of the substrate proteins. Indeed, aminopeptidase I is efficiently captured by the autophagosome and is selectively degraded under stress conditions by an unknown mechanism (16). In addition, protein degradation mediated by the vacuole is thought to be a vital process for the degradation of long-lived proteins, such as ubiquitin (20). At present, we cannot rule out the involvement of the proteasome in the degradation of ubiquitin because the proteasome itself is intact even in lub1-deleted cells (Fig. 5B). Alternatively, it is possible that the translation efficiency in Δlub1 cells is decreased. However, this seems less likely to be the case because other endogenous (data not shown) as well as exogenous proteins, such as GST-Sts5 and GFP-Cdc13, showed intact expression in Δlub1 cells. Still another possibility is the loss of deubiquitinating enzyme (DUB) activity (30), but this is less probable because the overexpression of the gene encoding Ubi4 was able to release ubiquitin and thus increase the level of ubiquitin, indicating that DUB, which is involved in the cleavage of the ubiquitin precursor, was at least not impaired (data not shown). In addition, decreased DUB activity was not detected in Δlub1 cells by experiments using ubiquitin-β-galactosidase substrate (2, 30) (data not shown).
Notably, the K48R mutant ubiquitin could not restore the temperature sensitivity of Δlub1 cells (Fig. 2B). The role of the Lys48-linked multiubiquitin chains, unlike the Lys63 linkage, is to serve as 26S proteasome recognition motifs for degradation. These results suggest that Δlub1 cells are short of ubiquitin, which tags proteins for proteasome-mediated protein degradation. Consistently, degradation of Cdc13, a B-type cyclin in fission yeast, was impaired in Δlub1 cells (Fig. 5A), suggesting that ubiquitin shortage caused a dysfunction in ubiquitin/proteasome-dependent proteolysis. This hypothesis is supported by the results indicating that the proteasome activities in Δlub1 cells were not impaired but rather were high compared with those of wild-type cells (Fig. 5B). The increased activities of the proteasome may be partially explained by the compensation of the ubiquitin shortage. In contrast, it has been reported that the linkage through Lys63 plays a critical role in stress resistance to UBI4 mutants in budding yeast (1) and that it is implicated in the degradation of membrane-anchored proteins in the lysosome/vacuole (25, 38). Although the diverse roles of these ubiquitin linkages in the stress response remain to be established, our results show that Lub1 participates in the ubiquitin homeostasis and stress response associated with Lys48 proteasome-dependent proteolysis.
To investigate the precise targets for Lub1 and to address how ubiquitin contents are regulated, we conducted further experiments. Our findings indicate that Lub1 forms a complex with Cdc48 through the WD domain of Lub1 and that the WD domain is required for Lub1 stability (Fig. 6). It has also been reported that the transcription of lub1+ and cdc48+ genes is enhanced under stress conditions (3). Taking these results together, we hypothesized that Cdc48 is implicated in the stabilization of Lub1, thereby contributing to the rapid and significant induction of cellular ubiquitin under stress conditions at the protein level. This notion is supported by the results that the coexpression of Lub1 (230-713) with Cdc48 resulted in binding that was hardly detected (Fig. 6). Consistently, the attenuated restoration of ubiquitin contents (Fig. 3C), as well as the temperature-sensitive phenotype of Δlub1 cells (Fig. 2B), could be explained by the expression of a low level of Lub1 (230-713) compared with that of Lub1 (full length). Although a physical interaction between Lub1 with Cdc48 was shown, it is still unclear how Cdc48 function is related to the defect in ubiquitin levels in Δlub1 cells. Further studies are needed to understand the mechanisms of regulation of ubiquitin content by Lub1, which functions cooperatively with Cdc48.
As mentioned previously, the lub1+ gene encodes a highly conserved WD repeat protein with high sequence similarity to the budding yeast Ufd3p/Doa1p/Zzz4p (9, 29, 39) and human PLAP (6). Studies have shown that the UFD3 mutants have a low content of cellular ubiquitin and are defective in protein degradation (9, 15). Similar to UFD3 mutants, the Δlub1 cells have low levels of ubiquitin and Lub1 is found to be a functional homologue of Ufd3p (Fig. 2B). Unlike UFD3 mutants in which the UFD3-2 allele encodes a protein with a mutation in the fifth WD repeat (9), the product of the lub1-1 allele has a mutation localized in the region outside the WD domain at the C terminus, resulting in a truncated mutant (Fig. 2A). Consistently, truncation experiments revealed that the C-terminal region is necessary for Lub1 function whereas the WD domain at the N terminus is not essential (Fig. 2A). As the WD domain is important for the association with Cdc48, it is possible that a mutation in the WD domain in UFD3-2 cells could cause the defect in their association, rendering low cellular levels of Ufd3p.
In summary, we have demonstrated that Lub1 is responsible for the maintenance of cellular ubiquitin contents, presumably by a negative regulation of vacuole-dependent ubiquitin degradation. Moreover, Lub1 and Cdc48 cooperatively play key roles in the stress response to maintain the supply of ubiquitin at the posttranscriptional level. These findings may provide new insights in the understanding of the regulatory mechanism of cellular ubiquitin homeostasis.
Acknowledgments
We thank Susie O. Sio for critical reading of the manuscript.
This work was supported by 21st Century COE Program and research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCE
- 1.Arnason, T., and M. J. Ellison. 1994. Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14:7876-7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, R. T., J. W. Tobias, and A. Varshavsky. 1992. Ubiquitin-specific proteases of Saccharomyces cerevisiae. J. Biol. Chem. 267:23364-23375. [PubMed] [Google Scholar]
- 3.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, L., R. Watt, and P. W. Piper. 1994. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol. Gen. Genet. 243:358-362. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 6.Clark, M. A., L. E. Ozgur, T. M. Conway, J. Dispoto, S. T. Crooke, and J. S. Bomalaski. 1991. Cloning of a phospholipase A2-activating protein. Proc. Natl. Acad. Sci. USA 88:5418-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, D. Q., Y. Tomita, A. Yamamoto, Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5:169-190. [DOI] [PubMed] [Google Scholar]
- 8.Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035-1046. [DOI] [PubMed] [Google Scholar]
- 9.Ghislain, M., R. J. Dohmen, F. Levy, and A. Varshavsky. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884-4899. [PMC free article] [PubMed] [Google Scholar]
- 10.Glas, R., M. Bogyo, J. S. McMaster, M. Gaczynska, and H. L. Ploegh. 1998. A proteolytic system that compensates for loss of proteasome function. Nature 392:618-622. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, C., G. McGurk, P. Dillon, C. Rosen, and N. D. Hastie. 1993. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature 366:355-357. [DOI] [PubMed] [Google Scholar]
- 12.Hampton, R. Y. 2002. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14:476-482. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, E. S., P. C. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky, D. J., and Y. Ohsumi. 1999. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 17.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 18.Kostova, Z., and D. H. Wolf. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280-27284. [DOI] [PubMed] [Google Scholar]
- 21.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 22.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 23.Ogiso, Y., A. Tomida, and T. Tsuruo. 2002. Nuclear localization of proteasomes participates in stress-inducible resistance of solid tumor cells to topoisomerase II-directed drugs. Cancer Res. 62:5008-5012. [PubMed] [Google Scholar]
- 24.Okazaki, K., H. Okayama, and O. Niwa. 2000. The polyubiquitin gene is essential for meiosis in fission yeast. Exp. Cell Res. 254:143-152. [DOI] [PubMed] [Google Scholar]
- 25.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 26.Plemper, R. K., and D. H. Wolf. 1999. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24:266-270. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz, A. L., and A. Ciechanover. 1999. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 50:57-74. [DOI] [PubMed] [Google Scholar]
- 29.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, K., K. Matsumoto, and A. Toh-e. 1988. Dual regulation of the expression of the polyubiquitin gene by cyclic AMP and heat shock in yeast. EMBO J. 7:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toda, T., S. Dhut, G. Superti-Furga, Y. Gotoh, E. Nishida, R. Sugiura, and T. Kuno. 1996. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16:6752-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toda, T., H. Niwa, T. Nemoto, S. Dhut, M. Eddison, T. Matsusaka, M. Yanagida, and D. Hirata. 1996. The fission yeast sts5+ gene is required for maintenance of growth polarity and functionally interacts with protein kinase C and an osmosensing MAP-kinase pathway. J. Cell Sci. 109:2331-2342. [DOI] [PubMed] [Google Scholar]
- 34.Treger, J. M., K. A. Heichman, and K. McEntee. 1988. Expression of the yeast UBI4 gene increases in response to DNA-damaging agents and in meiosis. Mol. Cell. Biol. 8:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsubuki, S., Y. Saito, M. Tomioka, H. Ito, and S. Kawashima. 1996. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J. Biochem. (Tokyo) 119:572-576. [DOI] [PubMed] [Google Scholar]
- 36.Tsukahara, T., S. Ishiura, and H. Sugita. 1988. An ATP-dependent protease and ingensin, the multicatalytic proteinase, in K562 cells. Eur. J. Biochem. 177:261-266. [DOI] [PubMed] [Google Scholar]
- 37.Watt, R., and P. W. Piper. 1997. UBI4, the polyubiquitin gene of Saccharomyces cerevisiae, is a heat shock gene that is also subject to catabolite derepression control. Mol. Gen. Genet. 253:439-447. [DOI] [PubMed] [Google Scholar]
- 38.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 39.Wolfe, D., T. Reiner, J. L. Keeley, M. Pizzini, and R. L. Keil. 1999. Ubiquitin metabolism affects cellular response to volatile anesthetics in yeast. Mol. Cell. Biol. 19:8254-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]